Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech
Abstract
1. Introduction
2. Results and Discussion
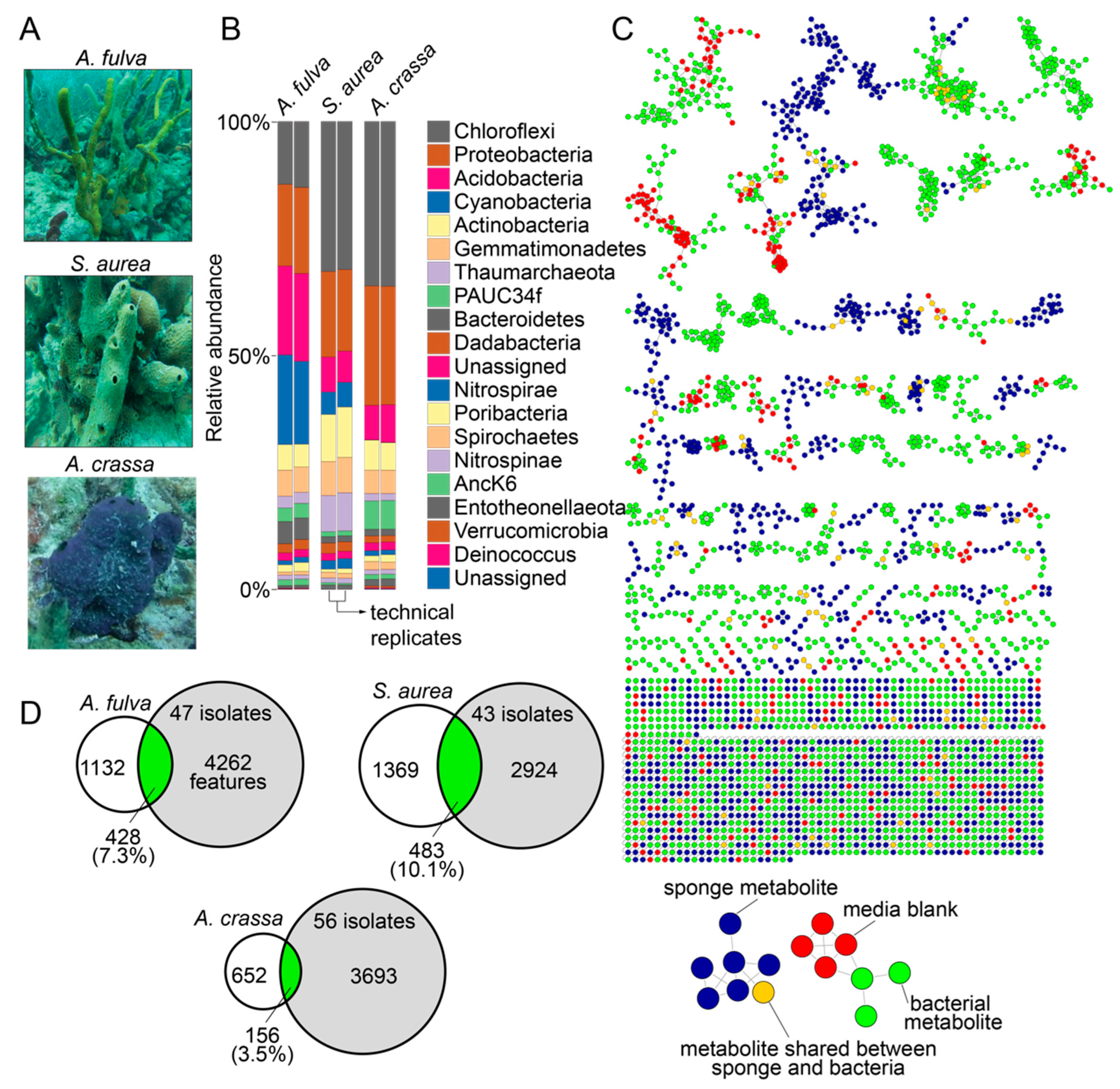
2.1. Marine Sponge Specimens
2.2. Metabolomes of Sponges and Bacterial Isolates
2.3. Phylogeny of Bacterial Isolates
2.4. Dependence of Isolation Media and Source Sponge on Bacterial Metabolomes
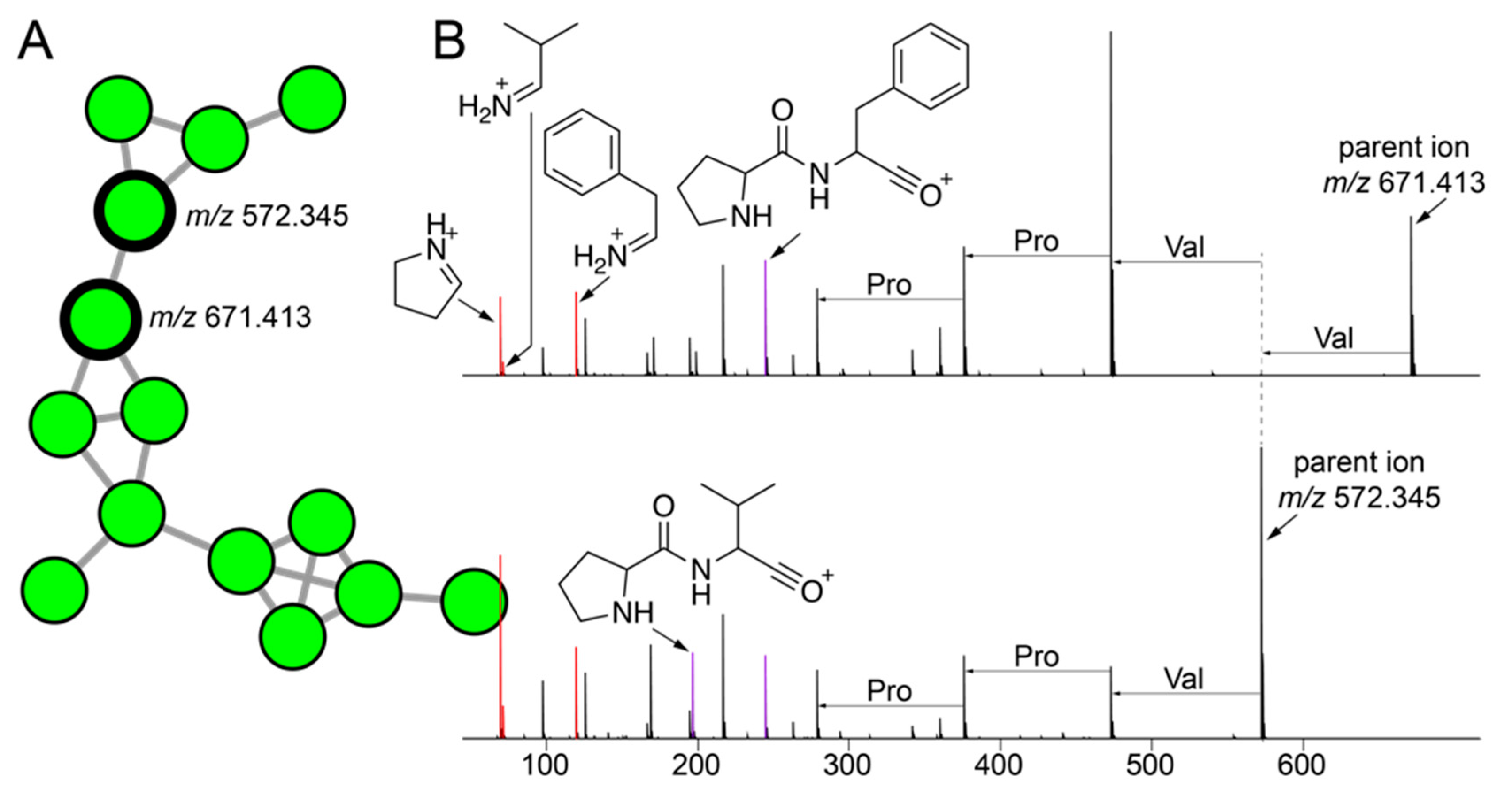
2.5. Cryptic Metabolites Are Shared between Sponges and Commensal Bacteria
2.6. Commensal Bacteria Are Sources of Novel Natural Products
2.7. Pedagogic Deliverables
3. Materials and Methods
3.1. Collection of Sponge Specimens
3.2. Bacterial Culturing
3.3. Preparation of Extracts for LC/MS
3.4. LC/MS Data Collection, Preprocessing, and Statistical Analysis
3.5. Genomic DNA Isolation
S rRNA Gene Amplicon Sequencing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Goeij, J.M.; van Oevelen, D.; Vermeij, M.J.A.; Osinga, R.; Middelburg, J.J.; de Goeij, A.F.P.M.; Admiraal, W. Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science 2013, 342, 108. [Google Scholar] [CrossRef]
- McMurray, S.E.; Stubler, A.D.; Erwin, P.M.; Finelli, C.M.; Pawlik, J.R. A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Prog. Ser. 2018, 588, 1–14. [Google Scholar] [CrossRef]
- Bart, M.C.; Hudspith, M.; Rapp, H.T.; Verdonschot, P.F.M.; de Goeij, J.M. A deep-sea sponge loop? Sponges transfer dissolved and particulate organic carbon and nitrogen to associated fauna. Front. Mar. Sci. 2021, 8, 604879. [Google Scholar] [CrossRef]
- Loh, T.-L.; Pawlik, J.R. Chemical defenses and resource trade-offs structure sponge communities on Caribbean coral reefs. Proc. Natl. Acad. Sci. USA 2014, 111, 4151. [Google Scholar] [CrossRef] [PubMed]
- Helber, S.B.; Hoeijmakers, D.J.J.; Muhando, C.A.; Rohde, S.; Schupp, P.J. Sponge chemical defenses are a possible mechanism for increasing sponge abundance on reefs in Zanzibar. PLoS ONE 2018, 13, e0197617. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, M.P.; Sneed, J.M.; Ritson-Williams, R.; Young, R. Marine chemical ecology in benthic environments. Nat. Prod. Rep. 2019, 36, 410–429. [Google Scholar] [CrossRef] [PubMed]
- Abdelaleem, E.R.; Samy, M.N.; Desoukey, S.Y.; Liu, M.; Quinn, R.J.; Abdelmohsen, U.R. Marine natural products from sponges (Porifera) of the order Dictyoceratida (2013 to 2019); a promising source for drug discovery. RSC Adv. 2020, 10, 34959–34976. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Jiménez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, V.; Wehrl, M.; Moitinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlik, J.R.; Lindquist, N.L.; Erpenbeck, D.; Wörheide, G.; Hentschel, U. The HMA-LMA dichotomy revisited: An electron microscopical survey of 56 sponge species. Biol. Bull. 2014, 227, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Schmidt, E.W. Parallel lives of symbionts and hosts: Chemical mutualism in marine animals. Nat. Prod. Rep. 2018, 35, 357–378. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.J.; Freeman, C.J.; Agarwal, V. Chemical ecology of marine sponges: New opportunities through “-omics”. Integr. Comp. Biol. 2019, 59, 765–776. [Google Scholar] [CrossRef]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Lin, Z.; Mohanty, I.; Garg, N.; Schmidt, E.W.; Agarwal, V. An obligate peptidyl brominase underlies the discovery of highly distributed biosynthetic gene clusters in marine sponge microbiomes. J. Am. Chem. Soc. 2021, 143, 10221–10231. [Google Scholar] [CrossRef]
- Cahn, J.K.B.; Piel, J. Opening up the single-cell toolbox for microbial natural products research. Angew. Chem. Int. Ed. 2021, 60, 18412–18428. [Google Scholar] [CrossRef]
- Simmons, T.L.; Coates, R.C.; Clark, B.R.; Engene, N.; Gonzalez, D.; Esquenazi, E.; Dorrestein, P.C.; Gerwick, W.H. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc. Natl. Acad. Sci. USA 2008, 105, 4587–4594. [Google Scholar] [CrossRef]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.R.; Easson, C.; Astudillo-García, C.; Olson, J.B.; Erwin, P.M.; López-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef]
- Bruck, L.B.; Towns, M.; Bretz, S.L. Faculty Perspectives of Undergraduate Chemistry Laboratory: Goals and Obstacles to Success. J. Chem. Educ. 2010, 87, 1416–1424. [Google Scholar] [CrossRef]
- Zwickl, B.M.; Finkelstein, N.; Lewandowski, H.J. The process of transforming an advanced lab course: Goals, curriculum, and assessments. Am. J. Phys. 2012, 81, 63–70. [Google Scholar] [CrossRef]
- Kerr, M.A.; Yan, F. Incorporating Course-Based Undergraduate Research Experiences into Analytical Chemistry Laboratory Curricula. J. Chem. Educ. 2016, 93, 658–662. [Google Scholar] [CrossRef]
- Velasco, J.B.; Knedeisen, A.; Xue, D.; Vickrey, T.L.; Abebe, M.; Stains, M. Characterizing Instructional Practices in the Laboratory: The Laboratory Observation Protocol for Undergraduate STEM. J. Chem. Educ. 2016, 93, 1191–1203. [Google Scholar] [CrossRef]
- Qiang, Z.; Obando, A.G.; Chen, Y.; Ye, C. Revisiting distance learning resources for undergraduate research and lab activities during COVID-19 pandemic. J. Chem. Educ. 2020, 97, 3446–3449. [Google Scholar] [CrossRef]
- Elmer, S.J.; Durocher, J.J. Moving student research forward during the COVID-19 pandemic. Adv. Physiol. Educ. 2020, 44, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Sunasee, R. Challenges of teaching organic chemistry during COVID-19 pandemic at a primarily undergraduate institution. J. Chem. Educ. 2020, 97, 3176–3181. [Google Scholar] [CrossRef]
- Grineski, S.E.; Morales, D.X.; Collins, T.W.; Nadybal, S.; Trego, S. Anxiety and depression among US college students engaging in undergraduate research during the COVID-19 pandemic. J. Am. Coll. Health 2021, 1–11. [Google Scholar] [CrossRef]
- Cantrell, T.P.; Freeman, C.J.; Paul, V.J.; Agarwal, V.; Garg, N. Mass spectrometry-based integration and expansion of the chemical diversity harbored within a marine sponge. J. Am. Soc. Mass Spectrom. 2019, 30, 1373–1384. [Google Scholar] [CrossRef]
- Mohanty, I.; Tapadar, S.; Moore, S.G.; Biggs, J.S.; Freeman, C.J.; Gaul, D.A.; Garg, N.; Agarwal, V. Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures. mSystems 2021, 6, e01387. [Google Scholar] [CrossRef]
- Moitinho-Silva, L.; Steinert, G.; Nielsen, S.; Hardoim, C.C.P.; Wu, Y.-C.; McCormack, G.P.; López-Legentil, S.; Marchant, R.; Webster, N.; Thomas, T.; et al. Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 2017, 8, 752. [Google Scholar] [CrossRef]
- Bayer, K.; Jahn, M.T.; Slaby, B.M.; Moitinho-Silva, L.; Hentschel, U. Marine sponges as Chloroflexi hot spots: Genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade. mSystems 2018, 3, e00150-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.X.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, J.; Hamann, M.T. The marine bromotyrosine derivatives. Alkaloids Chem. Biol. 2005, 61, 59–262. [Google Scholar] [PubMed]
- Bialonska, D.; Zjawiony, K.J. Aplysinopsins—marine indole alkaloids: Chemistry, bioactivity and ecological significance. Mar. Drugs 2009, 7, 166–183. [Google Scholar] [CrossRef]
- Teta, R.; Irollo, E.; Della Sala, G.; Pirozzi, G.; Mangoni, A.; Costantino, V. Smenamides A and B, chlorinated peptide/polyketide hybrids containing a dolapyrrolidinone unit from the Caribbean sponge Smenospongia aurea. Evaluation of their role as leads in antitumor drug research. Mar. Drugs 2013, 11, 4451–4463. [Google Scholar] [CrossRef]
- Via, C.W.; Glukhov, E.; Costa, S.; Zimba, P.V.; Moeller, P.D.R.; Gerwick, W.H.; Bertin, M.J. The metabolome of a cyanobacterial bloom visualized by MS/MS-based molecular networking reveals new neurotoxic smenamide analogs (C, D, and E). Front. Chem. 2018, 6, 316. [Google Scholar] [CrossRef]
- Djura, P.; Stierle, D.B.; Sullivan, B.; Faulkner, D.J.; Arnold, E.V.; Clardy, J. Some metabolites of the marine sponges Smenospongia aurea and Smenospongia (.ident.Polyfibrospongia) echina. J. Org. Chem. 1980, 45, 1435–1441. [Google Scholar] [CrossRef]
- Hu, J.F.; Schetz, J.A.; Kelly, M.; Peng, J.N.; Ang, K.K.H.; Flotow, H.; Leong, C.Y.; Ng, S.B.; Buss, A.D.; Wilkins, S.P.; et al. New antiinfective and human 5-HT2 receptor binding natural and semisynthetic compounds from the Jamaican sponge Smenospongia aurea. J. Nat. Prod. 2002, 65, 476–480. [Google Scholar] [CrossRef]
- Mohanty, I.; Moore, S.G.; Yi, D.; Biggs, J.S.; Gaul, D.A.; Garg, N.; Agarwal, V. Precursor-guided mining of marine sponge metabolomes lends insight into biosynthesis of pyrrole–imidazole alkaloids. ACS Chem. Biol. 2020, 15, 2185–2194. [Google Scholar] [CrossRef]
- Mohanty, I.; Podell, S.; Biggs, S.J.; Garg, N.; Allen, E.E.; Agarwal, V. Multi-omic profiling of Melophlus sponges reveals diverse metabolomic and microbiome architectures that are non-overlapping with ecological neighbors. Mar. Drugs 2020, 18, 124. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Bewley, C.A.; Faulkner, D.J. Lithistid sponges: Star performers or hosts to the stars. Angew. Chem. Int. Ed. 1998, 37, 2162–2178. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Mitova, M.; Popov, S.; De Rosa, S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod. 2004, 67, 1178–1181. [Google Scholar] [CrossRef] [PubMed]
- Quévrain, E.; Domart-Coulon, I.; Pernice, M.; Bourguet-Kondracki, M.-L. Novel natural parabens produced by a Microbulbifer bacterium in its calcareous sponge host Leuconia nivea. Environ. Microbiol. 2009, 11, 1527–1539. [Google Scholar] [CrossRef]
- Machado, H.; Sonnenschein, E.C.; Melchiorsen, J.; Gram, L. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genom. 2015, 16, 158. [Google Scholar] [CrossRef]
- Naughton, L.M.; Romano, S.; O’Gara, F.; Dobson, A.D.W. Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front. Microbiol. 2017, 8, 1494. [Google Scholar] [CrossRef]
- Romano, S. Ecology and biotechnological potential of bacteria belonging to the genus Pseudovibrio. Appl. Environ. Microbiol. 2018, 84, e02516-17. [Google Scholar] [CrossRef]
- Jayanetti, D.R.; Braun, D.R.; Barns, K.J.; Rajski, S.R.; Bugni, T.S. Bulbiferates A and B: Antibacterial acetamidohydroxybenzoates from a marine proteobacterium, Microbulbifer sp. J. Nat. Prod. 2019, 82, 1930–1934. [Google Scholar] [CrossRef]
- Karim, M.R.U.; Harunari, E.; Oku, N.; Akasaka, K.; Igarashi, Y. Bulbimidazoles A–C, antimicrobial and cytotoxic alkanoyl imidazoles from a marine gammaproteobacterium Microbulbifer species. J. Nat. Prod. 2020, 83, 1295–1299. [Google Scholar] [CrossRef]
- Ióca, L.P.; Dai, Y.; Kunakom, S.; Diaz-Espinosa, J.; Krunic, A.; Crnkovic, C.M.; Orjala, J.; Sanchez, L.M.; Ferreira, A.G.; Berlinck, R.G.S.; et al. A family of nonribosomal peptides modulate collective behavior in Pseudovibrio bacteria isolated from marine sponges. Angew. Chem. Int. Ed. 2021, 60, 15891–15898. [Google Scholar] [CrossRef]
- Nicacio, K.J.; Ióca, L.P.; Fróes, A.M.; Leomil, L.; Appolinario, L.R.; Thompson, C.C.; Thompson, F.L.; Ferreira, A.G.; Williams, D.E.; Andersen, R.J.; et al. Cultures of the marine bacterium Pseudovibrio denitrificans Ab134 produce bromotyrosine-derived alkaloids previously only isolated from marine sponges. J. Nat. Prod. 2017, 80, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Sipkema, D.; Schippers, K.; Maalcke Wouter, J.; Yang, Y.; Salim, S.; Blanch Harvey, W. Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl. Environ. Microbiol. 2011, 77, 2130–2140. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.I.S.; Amer, N.; Nguyen, M.; Thomas, T. Sample processing impacts the viability and cultivability of the sponge microbiome. Front. Microbiol. 2016, 7, 499. [Google Scholar] [CrossRef]
- Dat, T.T.H.; Steinert, G.; Cuc, N.T.K.; Smidt, H.; Sipkema, D. Bacteria cultivated from sponges and bacteria not yet cultivated from sponges—A review. Front. Microbiol. 2021, 12, 3427. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, N.F.; Davis, J.; Vicente, J.; Pittiglio, R.; Ravel, J.; Hill, R.T. Integration of culture-based and molecular analysis of a complex sponge-associated bacterial community. PLoS ONE 2014, 9, e90517. [Google Scholar] [CrossRef] [PubMed]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Mohanty, I.; Nguyen, N.A.; Moore, S.G.; Biggs, J.S.; Gaul, D.A.; Garg, N.; Agarwal, V. Enzymatic synthesis assisted discovery of proline-rich macrocyclic peptides in marine sponges. Chembiochem 2021, 22, 2614–2618. [Google Scholar] [CrossRef] [PubMed]
- Breci, L.A.; Tabb, D.L.; Yates, J.R.; Wysocki, V.H. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal. Chem. 2003, 75, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Kapp, E.A.; Schütz, F.; Reid, G.E.; Eddes, J.S.; Moritz, R.L.; O’Hair, R.A.J.; Speed, T.P.; Simpson, R.J. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal. Chem. 2003, 75, 6251–6264. [Google Scholar] [CrossRef] [PubMed]
- Petras, D.; Nothias, L.F.; Quinn, R.A.; Alexandrov, T.; Bandeira, N.; Bouslimani, A.; Castro-Falcon, G.; Chen, L.; Dang, T.; Floros, D.J.; et al. Mass spectrometry-based visualization of molecules associated with human habitats. Anal. Chem. 2016, 88, 10775–10784. [Google Scholar] [CrossRef] [PubMed]
- Betts, T.A.; Palkendo, J.A. Teaching undergraduates LC–MS/MS theory and operation via multiple reaction monitoring (MRM) method development. J. Chem. Educ. 2018, 95, 1035–1039. [Google Scholar] [CrossRef]
- Boyce, M.C.; Lawler, N.G.; Tu, Y.; Reinke, S.N. Introducing undergraduate students to metabolomics using liquid chromatography–high resolution mass spectrometry analysis of horse blood. J. Chem. Educ. 2019, 96, 745–750. [Google Scholar] [CrossRef]
- Kirk, R.D.; Carro, M.A.; Wu, C.; Aldine, M.J.; Wharton, A.M.; Goldstein, D.G.; Rosario, M.E.; Gallucci, G.M.; Zhao, Y.; Leibovitz, E.; et al. Integrating natural product chemistry workflows into medicinal chemistry laboratory training: Building the PRISM library and cultivating independent research. J. Chem. Educ. 2021, 98, 410–415. [Google Scholar] [CrossRef]
- Alker Amanda, T.; Gode Bhumika, S.; Aspiras Alpher, E.; Jones Jeffrey, E.; Michael Sama, R.; Aguilar, D.; Cain Audrea, D.; Candib Alec, M.; Cizmic Julian, M.; Clark Elise, A.; et al. Draft genome sequences of 10 bacteria from the marine Pseudoalteromonas group. Microbiol. Resour. Announc. 2021, 10, e00404-21. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Witting, M.; Tugizimana, F.; Dunn, W.B.; Reinke, S.N.; the Metabolomics Society Education and Training Committee. Providing metabolomics education and training: Pedagogy and considerations. Metabolomics 2022, 18, 106. [Google Scholar] [CrossRef]
- Deutsch, J.M.; Mandelare-Ruiz, P.; Yang, Y.; Foster, G.; Routhu, A.; Houk, J.; De La Flor, Y.T.; Ushijima, B.; Meyer, J.L.; Paul, V.J.; et al. Metabolomics Approaches to Dereplicate Natural Products from Coral-Derived Bioactive Bacteria. J. Nat. Prod. 2022, 85, 462–478. [Google Scholar] [CrossRef]
- Deutsch, J.M.; Jaiyesimi, O.A.; Pitts, K.A.; Houk, J.; Ushijima, B.; Walker, B.K.; Paul, V.J.; Garg, N. Metabolomics of Healthy and Stony Coral Tissue Loss Disease Affected Montastraea cavernosa Corals. Front. Mar. Sci. 2021, 8, 714778. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Shannon, P.; Markie, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deutsch, J.M.; Green, M.O.; Akavaram, P.; Davis, A.C.; Diskalkar, S.S.; Du Plessis, I.A.; Fallon, H.A.; Grason, E.M.; Kauf, E.G.; Kim, Z.M.; et al. Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech. Mar. Drugs 2023, 21, 53. https://doi.org/10.3390/md21010053
Deutsch JM, Green MO, Akavaram P, Davis AC, Diskalkar SS, Du Plessis IA, Fallon HA, Grason EM, Kauf EG, Kim ZM, et al. Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech. Marine Drugs. 2023; 21(1):53. https://doi.org/10.3390/md21010053
Chicago/Turabian StyleDeutsch, Jessica M., Madison O. Green, Priyanka Akavaram, Ashleigh C. Davis, Sarth S. Diskalkar, Isabelle A. Du Plessis, Hannah A. Fallon, Emma M. Grason, Emma G. Kauf, Zoe M. Kim, and et al. 2023. "Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech" Marine Drugs 21, no. 1: 53. https://doi.org/10.3390/md21010053
APA StyleDeutsch, J. M., Green, M. O., Akavaram, P., Davis, A. C., Diskalkar, S. S., Du Plessis, I. A., Fallon, H. A., Grason, E. M., Kauf, E. G., Kim, Z. M., Miller, J. R., II, Neal, A. L., Riera, T., Stroeva, S.-E., Tran, J., Tran, V., Coronado, A. V., Coronado, V. V., Wall, B. T., ... Agarwal, V. (2023). Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech. Marine Drugs, 21(1), 53. https://doi.org/10.3390/md21010053





