Total Synthesis and Biological Profiling of Putative (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A
Abstract
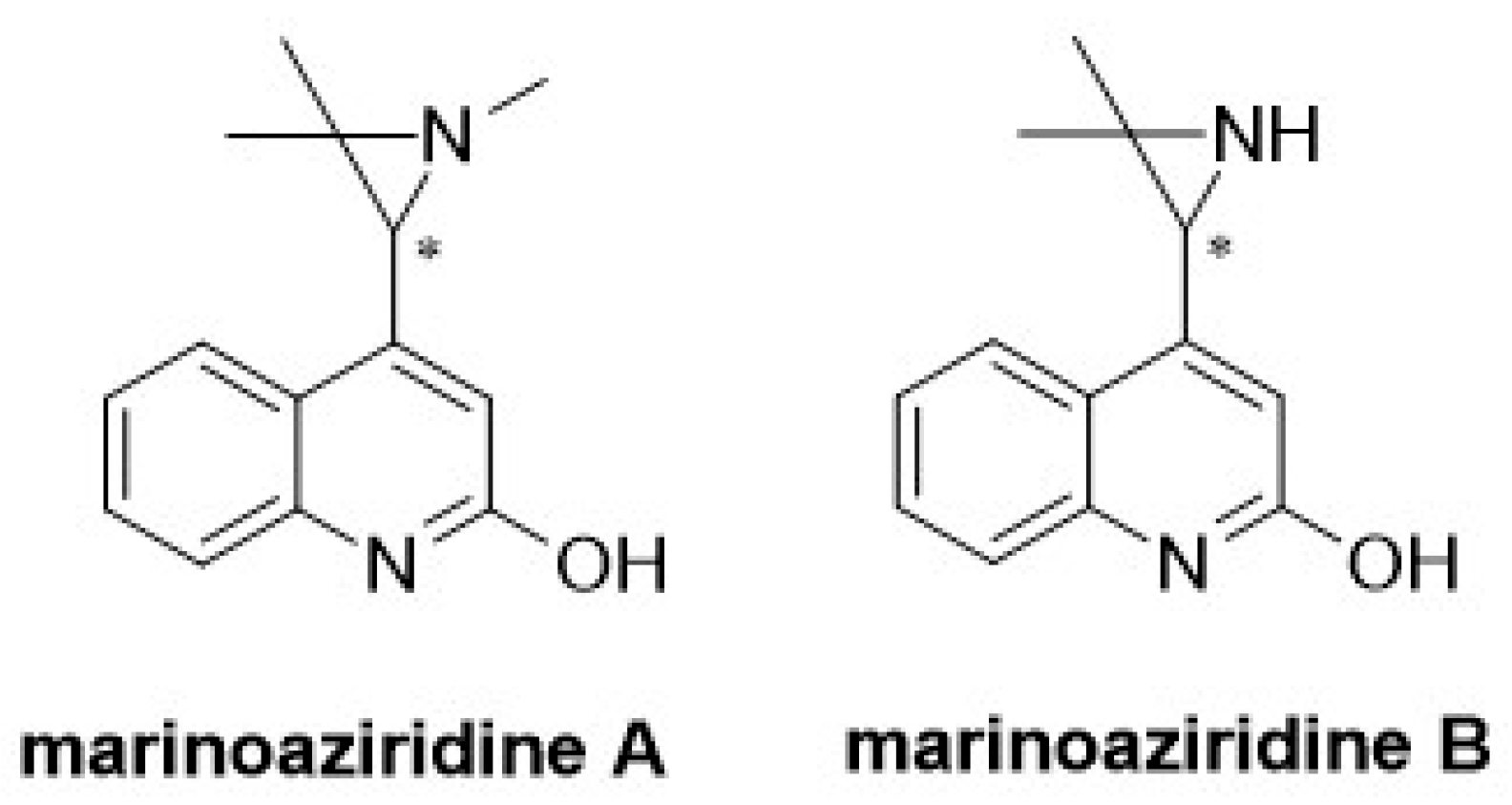
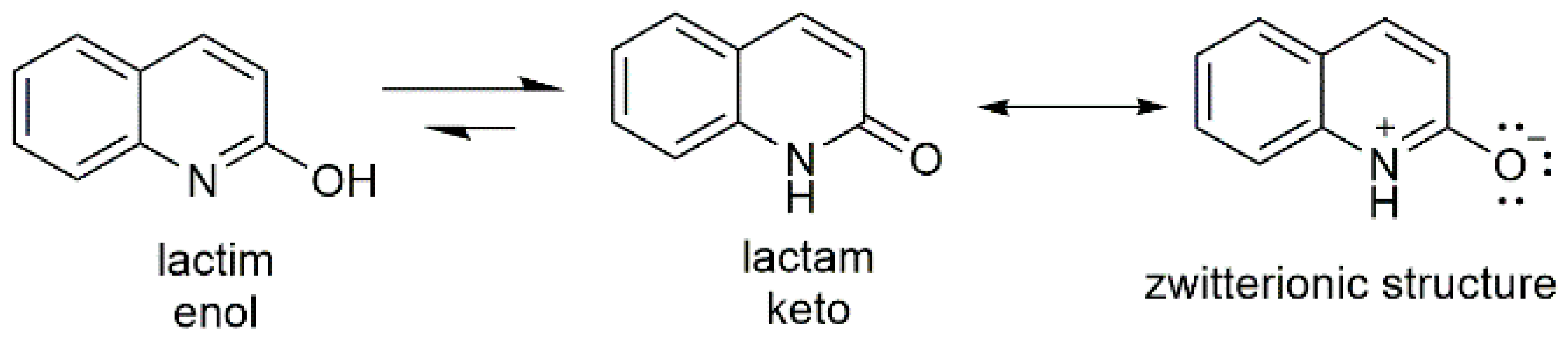
:1. Introduction
2. Results and Discussion
2.1. Total Synthesis of (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A

2.2. Biological Evaluation of Newly Synthesized Compounds: (±)-7 and (±)-8
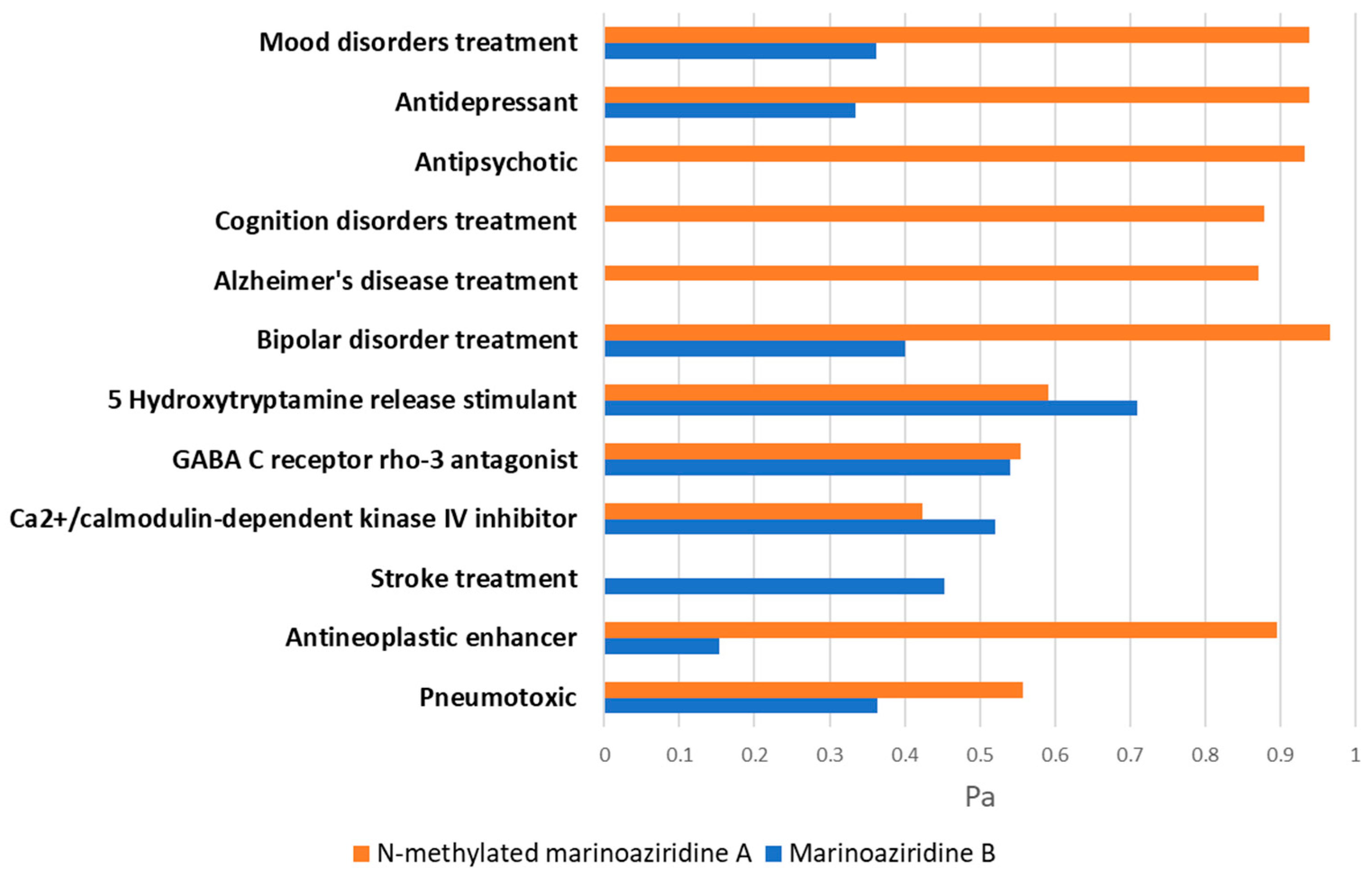
2.2.1. In Silico ADMET and Bioactivity Profiling
2.2.2. Antiproliferative and Cytotoxic Effect In Vitro
2.2.3. Antibacterial Activity In Vitro
2.2.4. Evaluation of Embryotoxicity Using Zebrafish (Danio rerio) In Vivo Model
3. Materials and Methods
3.1. Synthesis
3.2. Structure Characterization
3.3. Biological Activity
3.3.1. In Silico Physicochemical and Bioactivity Profiling
3.3.2. In Vitro Testing
Antiproliferative Activity
| Compound I.D. | Submitted MW a | Stock Solution/Solvent |
|---|---|---|
| (±)-7 | 214.27 | 4 × 10−2 M/DMSO |
| (±)-8 | 242.32 | 4 × 10−2 M/DMSO |
Cell Lines
Cell Culturing
Proliferation Assays
PG = 100 × (Atest − Atzero)/(Acont − Atzero)
PG = 100 × (Atest − Atzero)/Atzero
Antibacterial Activity
3.3.3. In Vivo Testing
Embryotoxicity Testing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karthikeyan, A.; Joseph, A.; Nair, B.G. Promising bioactive compounds from the marine environment and their potential effects on various diseases. J. Genet. Eng. Biotechnol. 2022, 20, 14. [Google Scholar] [CrossRef]
- Bergmann, W.; Feeney, R.J. The isolation of a new thymine pentoside from sponges. J. Am. Chem. Soc. 1950, 72, 2809–2810. [Google Scholar] [CrossRef]
- Gribble, G.W. Biological Activity of Recently Discovered Halogenated Marine Natural Products. Mar. Drugs 2015, 13, 4044–4136. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Parveen, S.; Tang, T.; Wei, J.; Huang, Z. Marine natural products in clinical use. Mar. Drugs 2022, 20, 528. [Google Scholar] [CrossRef] [PubMed]
- Ichiba, T.; Yoshida, W.Y.; Scheuer, P.J.; Higa, T.; Gravalos, D.G. Hennoxazoles, bioactive bisoxazoles from a marine sponge. J. Am. Chem. Soc. 1991, 113, 3173–3174. [Google Scholar] [CrossRef]
- Wipf, P.; Lim, S. Total synthesis of the enantiomer of the antiviral marine natural product hennoxazole A. J. Am. Chem. Soc. 1995, 117, 558–559. [Google Scholar] [CrossRef]
- Jiménez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [PubMed]
- Beattie, A.J.; Hay, M.; Magnusson, B.; de Nys, R.; Smeathers, J.; Vincent, J.F.V. Ecology and bioprospecting. Austral. Ecol. 2011, 3, 341–356. [Google Scholar] [CrossRef]
- Soejarto, D.D.; Farnsworth, N.R. Tropical rain forests: Potential source of new drugs. Persp. Biol. Med. 1989, 32, 244–246. [Google Scholar] [CrossRef]
- Sun, W.; Wu, W.; Liu, X.; Zaleta-Pinet, D.A.; Clark, B.R. Bioactive compounds isolated from marine-derived microbes in China: 2009–2018. Mar. Drugs 2019, 17, 339. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J.; Snader, K.M. Natural products in drug discovery and development. J. Nat. Prod. 1997, 60, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M.; Snader, K.M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod. 2003, 66, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Giddings, L.-A. Natural products as leads to antitumor drugs. Phytochem. Rev. 2014, 13, 123–137. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.J.; Nam, S.-J.; Paul, L.; Beatty, D.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Previously uncultured marine bacteria linked to novel alkaloid production. Chem. Biol. 2015, 22, 1270–1279. [Google Scholar] [CrossRef]
- Nimlos, M.R.; Krlley, D.F.; Bernstein, E.R. The spectroscopy and structure of 2-hydroxyquinoline. J. Phys. Chem. 1987, 91, 6610–6614. [Google Scholar] [CrossRef]
- Cook, M.J.; Katritzky, A.R.; Linda, P.; Tack, R.D. Aromaticity and tautomerism. Part II. The 4-pyridone, 2-quinolone, and 1-isoquinolone series. J. Chem. Soc., Perkin Trans. 1973, 2, 1080–1086. [Google Scholar] [CrossRef]
- Mason, S.F. The tautomerism of N-heteroaromatic hydroxy-compounds. Part III. Ionisation constants. J. Chem. Soc. 1958, 674–685. [Google Scholar] [CrossRef]
- Buljan, A.; Roje, M. Application of green chiral chromatography in enantioseparation of newly synthesized racemic marinoepoxides. Mar. Drugs 2022, 20, 530. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, J.B. Aziridines: Epoxides’ ugly cousins. Chem. Soc. Rev. 2002, 31, 247–258. [Google Scholar] [CrossRef]
- Mandal, S.; Bhuyan, S.; Jana, S.; Hossain, J.; Chhetri, K.; Roy, B.G. Efficient visible light mediated synthesis of quinolin 2(1H)-ones from quinoline N-oxides. Green Chem. 2021, 23, 5049–5055. [Google Scholar] [CrossRef]
- Simonetti, S.O.; Larghi, E.L.; Kaufman, T.S. The 3,4-dioxygenated 5-hydroxy-4-aryl-quinolin-2(1H)-one alkaloids. Results of 20 years of research, uncovering a new family of natural products. Nat. Prod. Rep. 2016, 33, 1425–1446. [Google Scholar] [CrossRef]
- Baraldi, P.G.; Budriesi, R.; Cacciari, B.; Chiarini, A.; Garuti, L.; Giovanninetti, G.; Leoni, A.; Roberti, M. Synthesis and calcium antagonist activity of dialkyl 1,4-dihydro-2,6-dimethyl-4-(nitrogenous heteroaryl)-3,5-pyridine dicarboxylates. Collect. Czech. Chem. Commun. 1992, 57, 169–178. [Google Scholar] [CrossRef]
- Durst, T.; Ben, R.N.; Roy, M.-N. e-EROS Encyclopedia of Reagents for Organic Synthesis; Wiley: New York, NY, USA, 2013; pp. 1–3. [Google Scholar]
- Parker, R.E.; Isaacs, N.S. Mechanisms of epoxide reactions. Chem. Rev. 1959, 59, 737–799. [Google Scholar] [CrossRef]
- Smith, J.G. Synthetically useful reactions of epoxides. Synthesis 1984, 8, 629–656. [Google Scholar] [CrossRef]
- Pocker, Y.; Ronald, B.P.; Anderson, K.W. Epoxides in vicinal diol dehydration. 7. A mechanistic characterization of the spontaneous ring opening process of epoxides in aqueous solution: Kinetic and product studies. J. Am. Chem. Soc. 1988, 110, 6492–6497. [Google Scholar] [CrossRef]
- Bonollo, S.; Lanari, D.; Vaccaro, L. Ring-Opening of Epoxides in water. Eur. J. Org. Chem. 2011, 2011, 2587–2598. [Google Scholar] [CrossRef]
- Amantini, D.; Fringuelli, F.; Piermatti, O.; Tortoioli, S.; Vaccaro, L. Nucleophilic ring opening of 1, 2-epoxides in aqueous medium. Arkivoc 2002, 11, 293–311. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Poroikov, V.V. Chemoinformatics Approaches to Virtual Screening; Varnek, A., Tropsha, A., Eds.; RSC: Cambridge, UK, 2008; pp. 182–216. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Sałaciak, K.; Koszałka, A.; Żmudzka, E.; Pytka, K. The calcium/calmodulin-dependent kinases II and IV as therapeutic targets in neurodegenerative and neuropsychiatric disorders. Int. J. Mol. Sci. 2021, 9, 4307. [Google Scholar] [CrossRef] [PubMed]
- Zakharov, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. Quantitative prediction of antitarget interaction profiles for chemical compounds. Chem. Res. Toxicol. 2012, 11, 2378–2385. [Google Scholar] [CrossRef]
- Lowless, M.S.; Waldman, M.; Franczkiewicz, R.; Clark, R.D. Using chemoinformatics in drug discovery. In New Approaches to Drug Discovery, Handbook of Experimental Pharmacology; Nielsch, U., Fuhrmann, U., Jaroch, S., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016; Volume 232, pp. 139–170. [Google Scholar]
- ChemAxon. Calculator Plugins Were Used for Structure Property Prediction and Calculation, Marvin 22.5.0. 2022. Available online: http://www.chemaxon.com (accessed on 26 April 2024).
- Patel, J.B.; Cockerill, F.R.; Bradford, P.A.; Eliopoulos, G.M.; Hindler, J.A.; Jenkins, S.G.; Zimmer, B.L. M07-A10 Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 10th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- OECD. Test No. 236: Fish Embryo Acute Toxicity (FET) Test. OECD Guidelines for the Testing of Chemicals, Section 2; Organization for Economic Cooperation and Development: Paris, France, 2013. [Google Scholar]
- Babić, S.; Čizmek, L.; Maršavelski, A.; Malev, O.; Pflieger, M.; Strunjak-Perović, I.; Topić Popović, N.; Čož-Rakovac, R.; Trebše, P. Utilization of the zebrafish model to unravel the harmful effects of biomass burning during Amazonian wildfires. Sci. Rep. 2021, 11, 2527. [Google Scholar] [CrossRef] [PubMed]
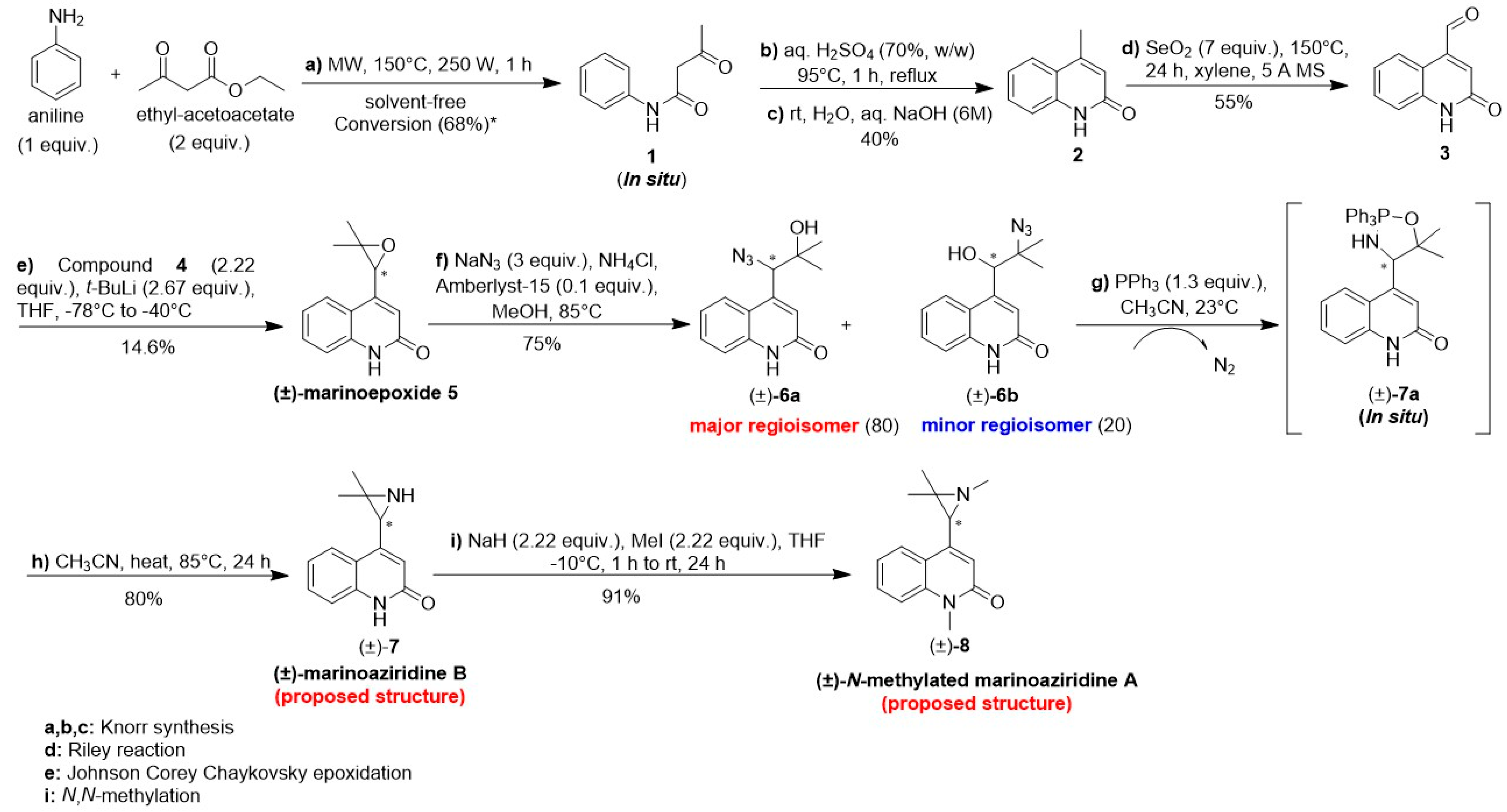
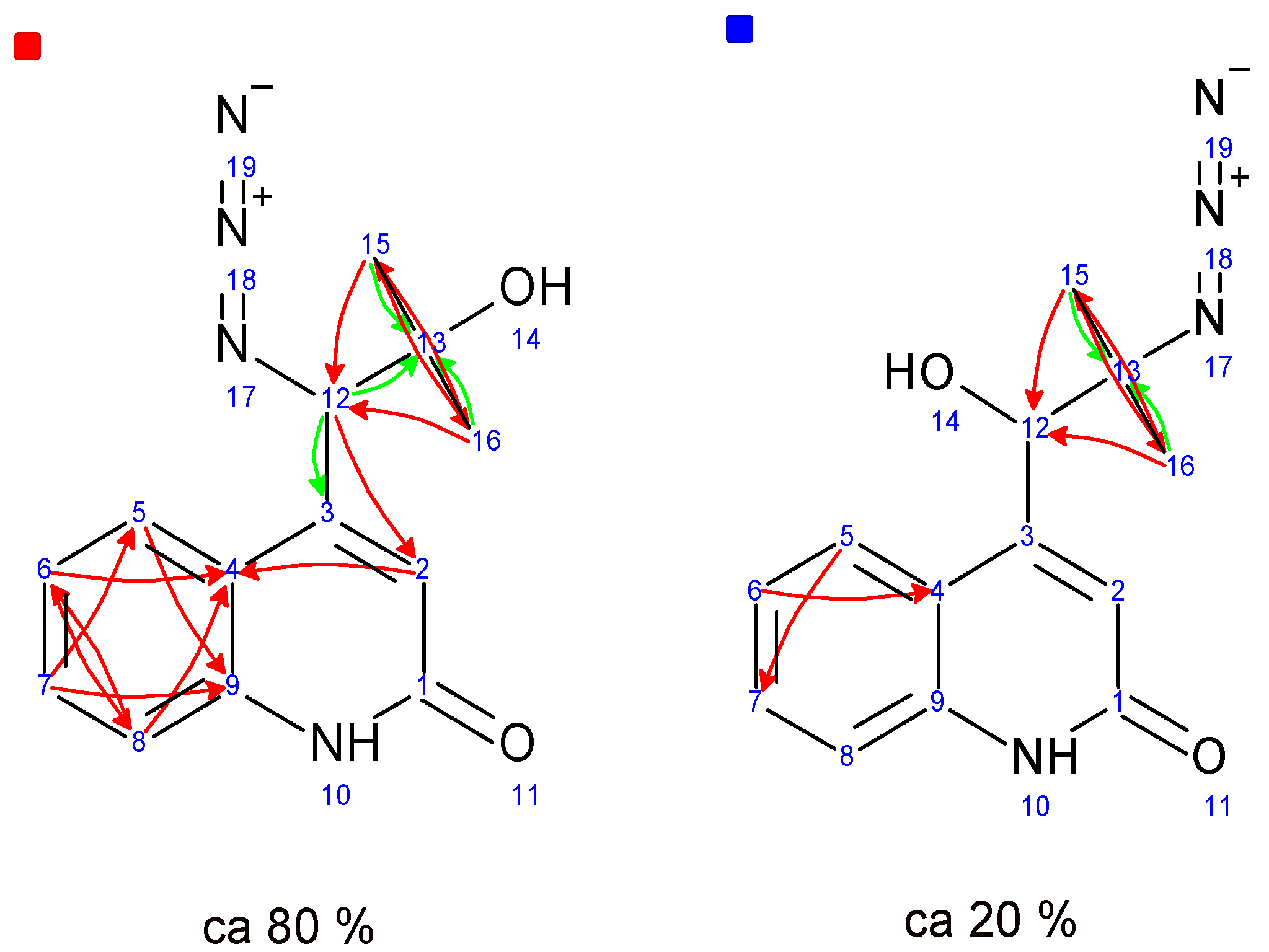
| Entry | Conditions | Yield ((±)-6a + (±)-6b)% | Ratio ((±)-6a/(±)-6b) a |
|---|---|---|---|
| i. | NaN3 (2 equiv.), NH4Cl (2 equiv.), 18-crown-6 (0.05 equiv.), MeOH, 65 °C, 72 h | 80 | 40/60 |
| ii. | NaN3 (2 equiv.), NH4Cl (2 equiv.), 18-crown-6 (0.05 equiv.), MeOH/H2O (1/1), 65 °C, 72 h | 77 | 42/58 |
| iii. | NaN3 (2 equiv.), NH4Cl (2 equiv.), Amberlyst® (0.1 equiv.) MeOH/H2O (1/1), 65 °C, 72 h | 50 | 52/48 |
| iv. | NaN3 (2 equiv.), NH4Cl (2 equiv.), Amberlyst® (0.1 equiv.), MeOH, 65 °C, 72 h | 75 | 80/20 |
| v. | Et2AlN3 (2 equiv.), toluene, −10 °C to RT, 24 h | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buljan, A.; Stepanić, V.; Čikoš, A.; Babić Brčić, S.; Bojanić, K.; Roje, M. Total Synthesis and Biological Profiling of Putative (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A. Mar. Drugs 2024, 22, 310. https://doi.org/10.3390/md22070310
Buljan A, Stepanić V, Čikoš A, Babić Brčić S, Bojanić K, Roje M. Total Synthesis and Biological Profiling of Putative (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A. Marine Drugs. 2024; 22(7):310. https://doi.org/10.3390/md22070310
Chicago/Turabian StyleBuljan, Anđela, Višnja Stepanić, Ana Čikoš, Sanja Babić Brčić, Krunoslav Bojanić, and Marin Roje. 2024. "Total Synthesis and Biological Profiling of Putative (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A" Marine Drugs 22, no. 7: 310. https://doi.org/10.3390/md22070310
APA StyleBuljan, A., Stepanić, V., Čikoš, A., Babić Brčić, S., Bojanić, K., & Roje, M. (2024). Total Synthesis and Biological Profiling of Putative (±)-Marinoaziridine B and (±)-N-Methyl Marinoaziridine A. Marine Drugs, 22(7), 310. https://doi.org/10.3390/md22070310






