Proteomics Analysis of the Protective Effect of Polydeoxyribonucleotide Extracted from Sea Cucumber (Apostichopus japonicus) Sperm in a Hydrogen Peroxide-Induced RAW264.7 Cell Injury Model
Abstract
:1. Introduction
2. Results
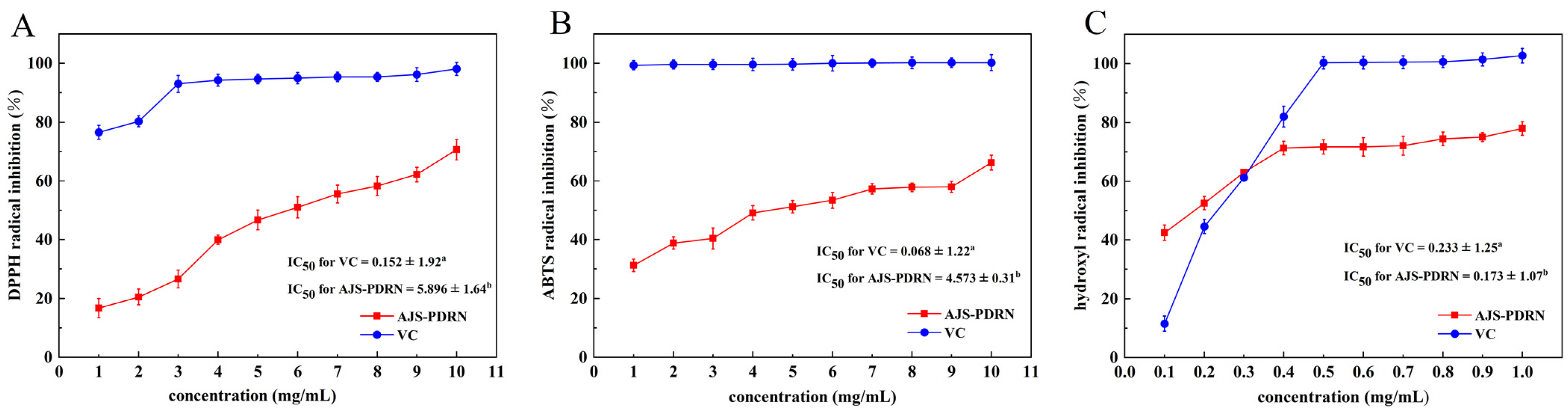
2.1. In Vitro Antioxidant Experiment of AJS-PDRN
2.2. Protective Effect of AJS-PDRN on H2O2-Induced RAW264.7 Cell Injury Model
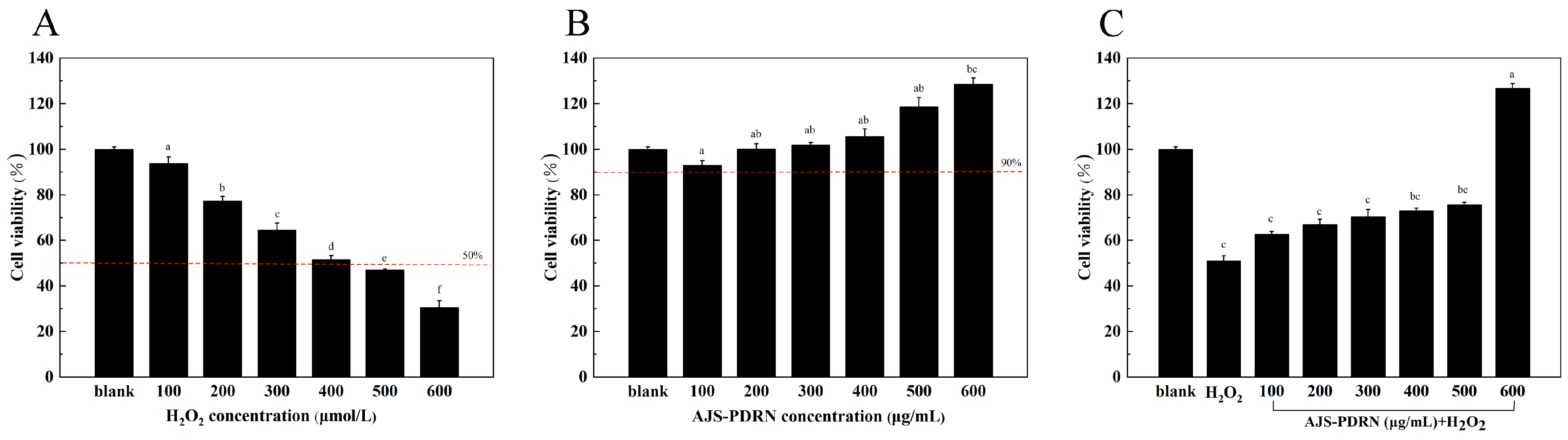
2.2.1. Effect of AJS-PDRN on RAW264.7 Cell Viability
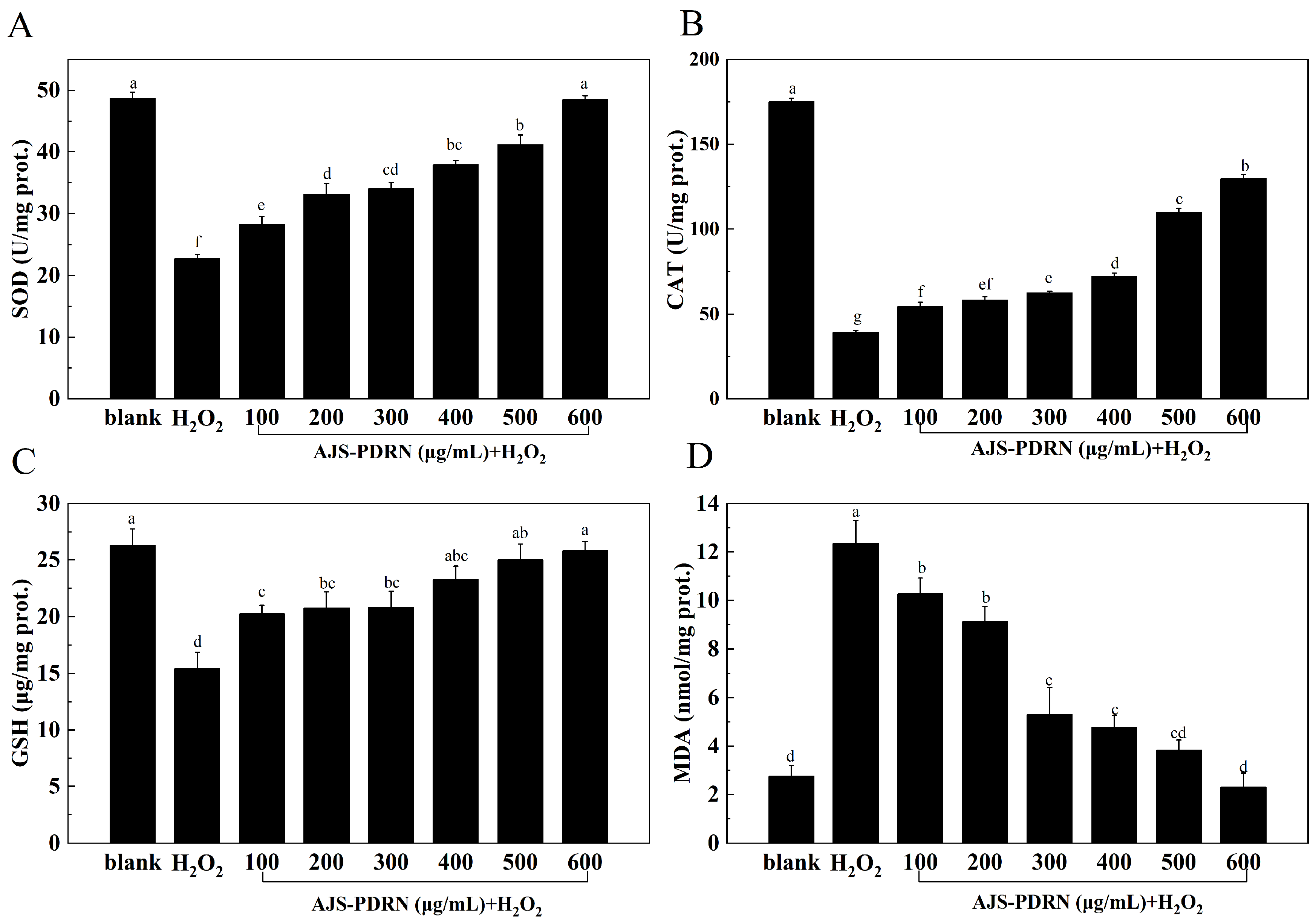
2.2.2. Oxidative Stress Biomarker Levels in RAW264.7 Cells
2.3. Effect of AJS-PDRN on Protein Expression Profiles in RAW264.7 Cells Induced by H2O2
2.3.1. Screening of Differentially Expressed Proteins
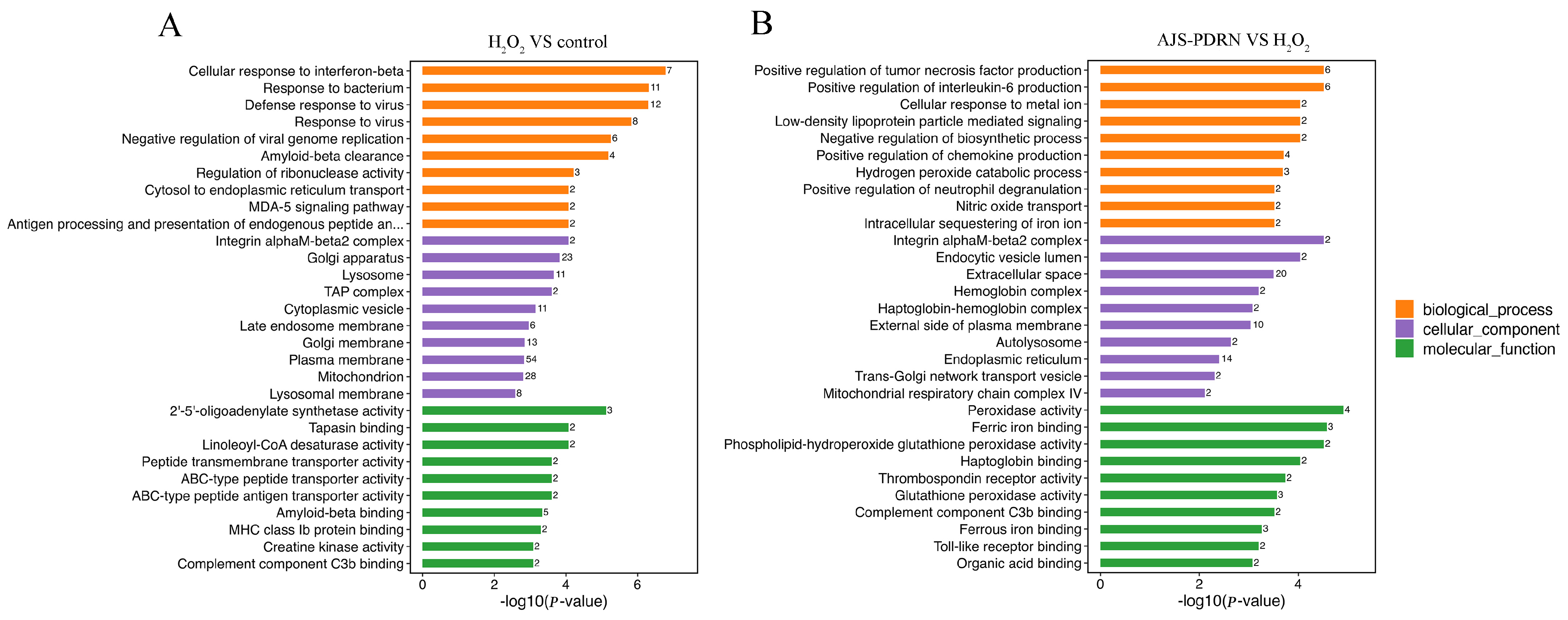
2.3.2. Gene Ontology (GO) Functional Annotation Enrichment Analysis of DEPs
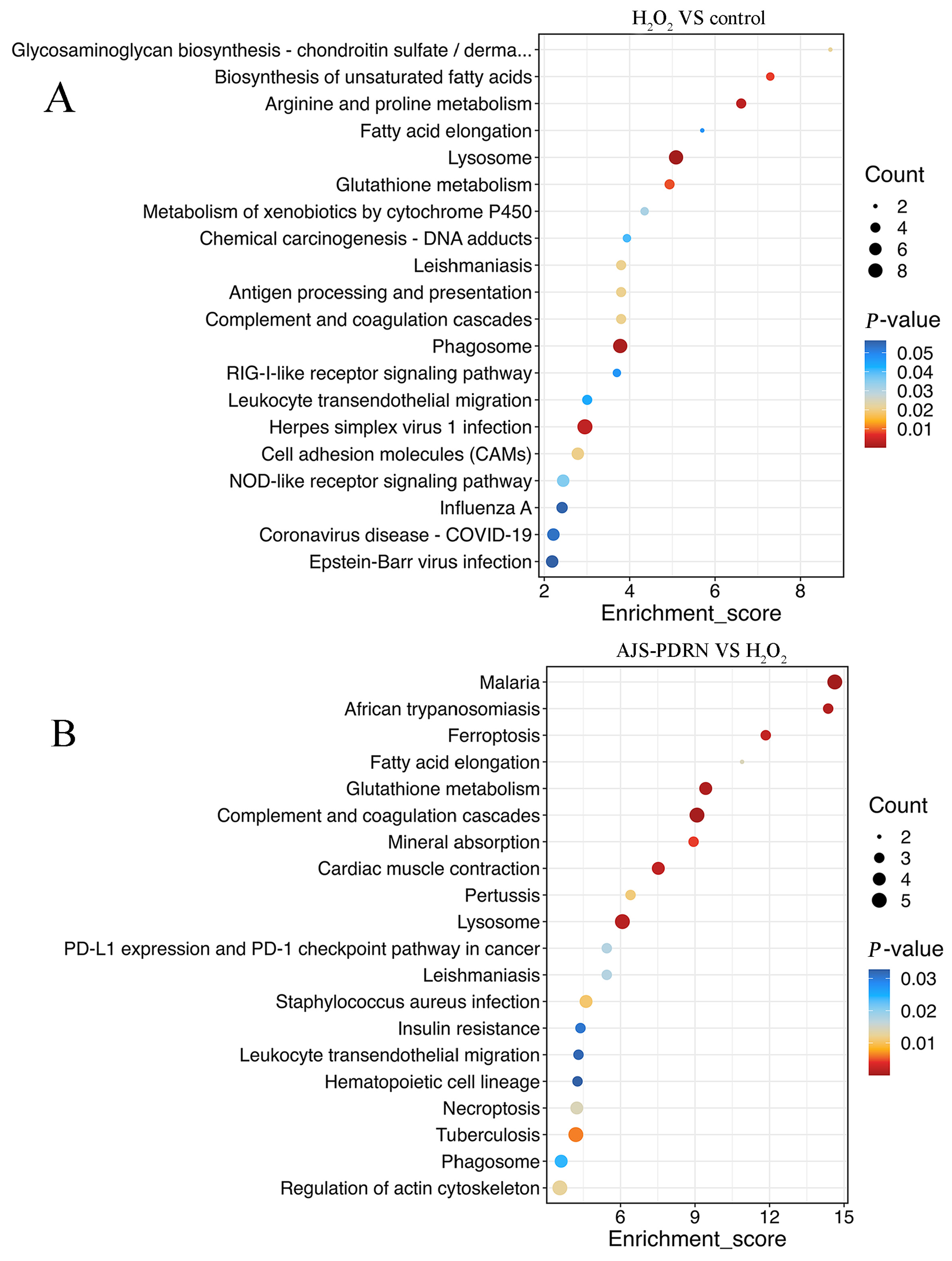
2.3.3. KEGG Pathway Enrichment Analysis of DEPs
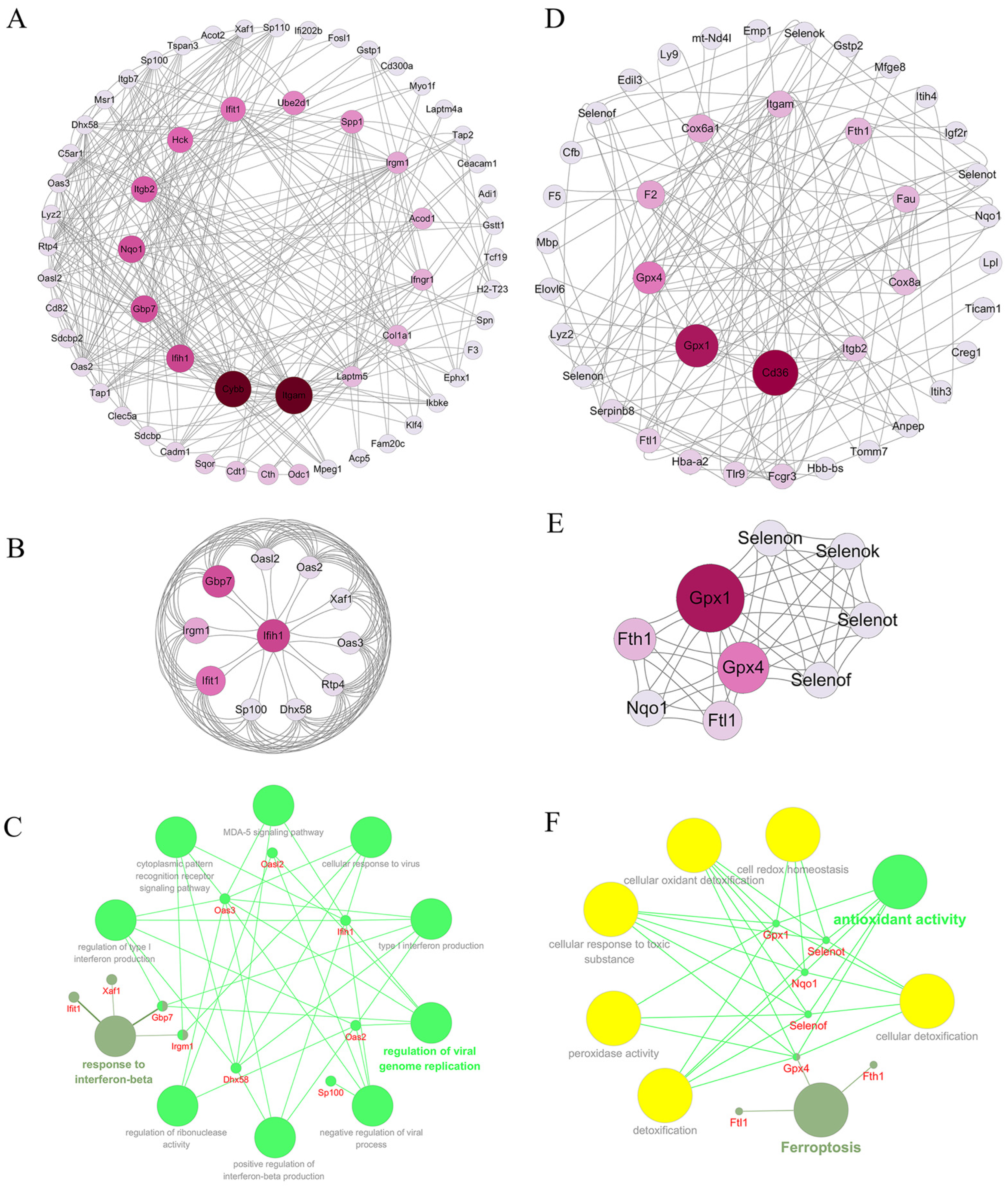
2.3.4. Analysis of Protein Interaction Network
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Extraction and Preparation of AJS-PDRN
4.3. DPPH Radical Scavenging Assay by AJS-PDRN
4.4. ABTS Radical Scavenging Assay by AJS-PDRN
4.5. Hydroxyl Radical Scavenging Activity
4.6. Cell Culture
4.7. Cell Viability Assay
4.8. Establishment of H2O2-Induced Oxidative Stress Model of RAW264.7 Cells
4.9. Toxic Effect of AJS-PDRN on RAW264.7 Cells
4.10. Protective Effect of AJS-PDRN on H2O2-Induced Cellular Oxidative Stress Model
4.11. CAT, SOD, GSH, and MDA in RAW264.7 Cells
4.12. Protein Preparation and iTRAQ Labelling
4.13. LC-MS/MS High-Resolution Mass Spectrometry Analysis
4.14. Protein Identification and Bioinformatic Analysis
4.15. Statistical Processing and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Pangestuti, R.; Arifin, Z. Medicinal and health benefit effects of functional sea cucumbers. J. Tradit. Complement. Med. 2018, 8, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-C.; Xue, C.-H.; Zhang, T.-T.; Wang, Y.-M. Saponins from Sea Cucumber and Their Biological Activities. J. Agric. Food Chem. 2018, 66, 7222–7237. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, Y. Pharmacological Potential of Sea Cucumbers. Int. J. Mol. Sci. 2018, 19, 1342. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, S.; Li, F.; Du, X.; Kong, B.; Wang, H.; Xia, X. Physicochemical properties and antioxidant activity of polysaccharides obtained from sea cucumber gonads via ultrasound-assisted enzymatic techniques. LWT 2022, 160, 113307. [Google Scholar] [CrossRef]
- Luo, L.; Wu, M.; Xu, L.; Lian, W.; Xiang, J.; Lu, F.; Gao, N.; Xiao, C.; Wang, S.; Zhao, J. Comparison of Physicochemical Characteristics and Anticoagulant Activities of Polysaccharides from Three Sea Cucumbers. Mar. Drugs 2013, 11, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.-X.; Wang, B.-K.; Wu, Y.-C.; Li, Q.-Y.; Qin, B.-W.; Li, H.-J. Release of antidiabetic peptides from Stichopus japonicas by simulated gastrointestinal digestion. Food Chem. 2020, 315, 126273. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Luo, W.; Yao, L.; Li, X.; Wang, Z.; Xu, H.; Li, H.; Hu, Z.; Bao, W.; Xu, X. Activation of murine RAW264.7 macrophages by oligopeptides from sea cucumber (Apostichopus japonicus) and its molecular mechanisms. J. Funct. Foods 2020, 75, 104229. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Z.; Chen, Y.; Wu, T.; Fersht, V.; Jin, Y.; Meng, J.; Zhang, M. Sea cucumber peptides inhibit the malignancy of NSCLC by regulating miR-378a-5p targeted TUSC2. Food Funct. 2021, 12, 12362–12371. [Google Scholar] [CrossRef] [PubMed]
- Cusimano, M.G.; Spinello, A.; Barone, G.; Schillaci, D.; Cascioferro, S.; Magistrato, A.; Parrino, B.; Arizza, V.; Vitale, M. A Synthetic Derivative of Antimicrobial Peptide Holothuroidin 2 from Mediterranean Sea Cucumber (Holothuria tubulosa) in the Control of Listeria monocytogenes. Mar. Drugs 2019, 17, 159. [Google Scholar] [CrossRef]
- Hoang, L.; Le Thi, V.; Tran Thi Hong, H.; Nguyen Van, T.; Nguyen Xuan, C.; Nguyen Hoai, N.; Do Cong, T.; Ivanchina, N.V.; Do Thi, T.; Dmitrenok, P.S.; et al. Triterpene glycosides from the Vietnamese sea cucumber Holothuria edulis. Nat. Prod. Res. 2020, 34, 1061–1067. [Google Scholar] [CrossRef]
- Malaiwong, N.; Chalorak, P.; Jattujan, P.; Manohong, P.; Niamnont, N.; Suphamungmee, W.; Sobhon, P.; Meemon, K. Anti-Parkinson activity of bioactive substances extracted from Holothuria leucospilota. Biomed. Pharmacother. 2019, 109, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Aminin, D.; Pislyagin, E.; Astashev, M.; Es’kov, A.; Kozhemyako, V.; Avilov, S.; Zelepuga, E.; Yurchenko, E.; Kaluzhskiy, L.; Kozlovskaya, E.; et al. Glycosides from edible sea cucumbers stimulate macrophages via purinergic receptors. Sci. Rep. 2016, 6, 39683. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014, 224, 164–175. [Google Scholar] [CrossRef]
- Zhao, X.-C.; Zhang, L.; Yu, H.-X.; Sun, Z.; Lin, X.-F.; Tan, C.; Lu, R.-R. Curcumin protects mouse neuroblastoma Neuro-2A cells against hydrogen-peroxide-induced oxidative stress. Food Chem. 2011, 129, 387–394. [Google Scholar] [CrossRef]
- Casas, A.I.; Nogales, C.; Mucke, H.A.M.; Petraina, A.; Cuadrado, A.; Rojo, A.I.; Ghezzi, P.; Jaquet, V.; Augsburger, F.; Dufrasne, F.; et al. On the Clinical Pharmacology of Reactive Oxygen Species. Pharmacol. Rev. 2020, 72, 801. [Google Scholar] [CrossRef] [PubMed]
- Siti, H.N.; Kamisah, Y.; Kamsiah, J. The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review). Vasc. Pharmacol. 2015, 71, 40–56. [Google Scholar] [CrossRef]
- Xu, B.; Dong, Q.; Yu, C.; Chen, H.; Zhao, Y.; Zhang, B.; Yu, P.; Chen, M. Advances in Research on the Activity Evaluation, Mechanism and Structure-Activity Relationships of Natural Antioxidant Peptides. Antioxidants 2024, 13, 479. [Google Scholar] [CrossRef]
- Zhou, H.; Li, X.; Shang, Y.; Chen, K. Radical Scavenging Activity of Puerarin: A Theoretical Study. Antioxidants 2019, 8, 590. [Google Scholar] [CrossRef]
- Colangelo, M.T.; Galli, C.; Guizzardi, S. The effects of polydeoxyribonucleotide on wound healing and tissue regeneration: A systematic review of the literature. Regener. Med. 2020, 15, 1801–1821. [Google Scholar] [CrossRef]
- Kim, S.-E.; Ko, I.-G.; Jin, J.-J.; Hwang, L.; Kim, C.-J.; Kim, S.-H.; Han, J.-H.; Jeon, J.W. Polydeoxyribonucleotide Exerts Therapeutic Effect by Increasing VEGF and Inhibiting Inflammatory Cytokines in Ischemic Colitis Rats. BioMed Res. Int. 2020, 2020, 2169083. [Google Scholar] [CrossRef]
- Kim, H.M.; Byun, K.-A.; Oh, S.; Yang, J.Y.; Park, H.J.; Chung, M.S.; Son, K.H.; Byun, K. A Mixture of Topical Forms of Polydeoxyribonucleotide, Vitamin C, and Niacinamide Attenuated Skin Pigmentation and Increased Skin Elasticity by Modulating Nuclear Factor Erythroid 2-like 2. Molecules 2022, 27, 1276. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Heo, S.-Y.; Oh, G.-W.; Heo, S.-J.; Jung, W.-K. Applications of Marine Organism-Derived Polydeoxyribonucleotide: Its Potential in Biomedical Engineering. Mar. Drugs 2021, 19, 296. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Liu, Y.; Liu, Y.; Wang, Q.; Ye, H.L.; Cheng, Y.G.; Kuang, H.X.; Jiang, X.C.; Yang, B.Y. Proteomics Research on the Protective Effect of Mangiferin on H9C2 Cell Injury Induced by H2O2. Molecules 2019, 24, 1911. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, M.-c.; Li, H.; Yang, Y.-g.; Su, N.; Wu, Y.-j.; Wang, H. Proteomics analysis of the protective effect of canola (Brassica campestris L.) bee pollen flavonoids on the tert butyl hydroperoxide-induced EA.hy926 cell injury model. J. Funct. Foods 2020, 75, 104223. [Google Scholar] [CrossRef]
- Lai, C.; Liang, Y.; Zhang, L.; Huang, J.; Kaliaperumal, K.; Jiang, Y.; Zhang, J. Variations of Bioactive Phytochemicals and Antioxidant Capacity of Navel Orange Peel in Response to Different Drying Methods. Antioxidants 2022, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.V.; Javadov, S.; Sommer, N. Cellular ROS and Antioxidants: Physiological and Pathological Role. Antioxidants 2024, 13, 602. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, R.; Cao, S.; Ismael, M.; Wang, X.; Lü, X. A galacturonic acid-rich polysaccharide from Diospyros kaki peel: Isolation, characterization, rheological properties and antioxidant activities in vitro. Food Chem. 2023, 416, 135781. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Kim, M.-J.; Kweon, D.-K.; Lim, S.-T.; Lee, S.-J. Polydeoxyribonucleotide Activates Mitochondrial Biogenesis but Reduces MMP-1 Activity and Melanin Biosynthesis in Cultured Skin Cells. Appl. Biochem. Biotechnol. 2020, 191, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-J.; Kim, Y.-J.; Kim, J.; Kim, S.M.; Lee, S.Y.; Bae, J.-W.; Hwang, K.-K.; Kim, D.-W.; Cho, M.-C. Protective Effects of Peroxiredoxin on Hydrogen Peroxide Induced Oxidative Stress and Apoptosis in Cardiomyocytes. Korean Circ. J. 2012, 42, 23–32. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, Q.; Zhang, X.; Zhu, X.; Dong, X.; Shen, L.; Zhang, S.; Niu, L.; Chen, L.; Zhang, M.; et al. l-Arginine Alleviates LPS-Induced Oxidative Stress and Apoptosis via Activating SIRT1-AKT-Nrf2 and SIRT1-FOXO3a Signaling Pathways in C2C12 Myotube Cells. Antioxidants 2021, 10, 1957. [Google Scholar] [CrossRef]
- Pan, Y.; Long, X.; Yi, R.; Zhao, X. Polyphenols in Liubao Tea Can Prevent CCl4-Induced Hepatic Damage in Mice through Its Antioxidant Capacities. Nutrients 2018, 10, 1280. [Google Scholar] [CrossRef] [PubMed]
- Petriccione, M.; Mastrobuoni, F.; Pasquariello, M.S.; Zampella, L.; Nobis, E.; Capriolo, G.; Scortichini, M. Effect of Chitosan Coating on the Postharvest Quality and Antioxidant Enzyme System Response of Strawberry Fruit during Cold Storage. Foods 2015, 4, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, M.; Cao, F.; Chen, Y.; Zhang, L.; Li, H.; Cao, J.; Song, J.; Ma, Y.; Mi, W.; et al. The Ferroptosis Inhibitor Liproxstatin-1 Ameliorates LPS-Induced Cognitive Impairment in Mice. Nutrients 2022, 14, 4599. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Cardoso, C.; Pestana, C. Measurement of malondialdehyde in fish: A comparison study between HPLC methods and the traditional spectrophotometric test. Food Chem. 2009, 112, 1038–1045. [Google Scholar] [CrossRef]
- Li, S.; Sampson, C.; Liu, C.; Piao, H.-l.; Liu, H.-X. Integrin signaling in cancer: Bidirectional mechanisms and therapeutic opportunities. Cell Commun. Signal. 2023, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Cheng, X.; Liu, X.; Sun, H.; Guo, Z.; Li, J.; Tang, X.; Wang, Z.; Sun, W.; et al. LC-MS-Based Plasma Metabolomics and Lipidomics Analyses for Differential Diagnosis of Bladder Cancer and Renal Cell Carcinoma. Front. Oncol. 2020, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Coevolution of the coagulation and immune systems. Inflamm. Res. 2019, 68, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Val, A.; Bekker-Jensen, D.B.; Steigerwald, S.; Koenig, C.; Østergaard, O.; Mehta, A.; Tran, T.; Sikorski, K.; Torres-Vega, E.; Kwasniewicz, E.; et al. Spatial-proteomics reveals phospho-signaling dynamics at subcellular resolution. Nat. Commun. 2021, 12, 7113. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Song, G.-L. Roles of Selenoproteins in Brain Function and the Potential Mechanism of Selenium in Alzheimer’s Disease. Front. Neurosci. 2021, 15, 646518. [Google Scholar] [CrossRef]
- Barrett, C.W.; Short, S.P.; Williams, C.S. Selenoproteins and oxidative stress-induced inflammatory tumorigenesis in the gut. Cell. Mol. Life Sci. 2017, 74, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Varone, E.; Pozzer, D.; Di Modica, S.; Chernorudskiy, A.; Nogara, L.; Baraldo, M.; Cinquanta, M.; Fumagalli, S.; Villar-Quiles, R.N.; De Simoni, M.-G.; et al. SELENON (SEPN1) protects skeletal muscle from saturated fatty acid-induced ER stress and insulin resistance. Redox Biol. 2019, 24, 101176. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chang, Y.; Wu, F.; Xu, X.; Xue, C. Fucosylated chondroitin sulfate is covalently associated with collagen fibrils in sea cucumber Apostichopus japonicus body wall. Carbohydr. Polym. 2018, 186, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dave, D.; Trenholm, S.; Ramakrishnan, V.V.; Murphy, W. Effect of Drying on Nutritional Composition of Atlantic Sea Cucumber (Cucumaria frondosa) Viscera Derived from Newfoundland Fisheries. Processes 2021, 9, 4. [Google Scholar] [CrossRef]
- Mamelona, J.; Saint-Louis, R.; Pelletier, É. Proximate composition and nutritional profile of by-products from green urchin and Atlantic sea cucumber processing plants. Int. J. Food Sci. Technol. 2010, 45, 2119–2126. [Google Scholar] [CrossRef]
- Hossain, A.; Yeo, J.; Dave, D.; Shahidi, F. Phenolic Compounds and Antioxidant Capacity of Sea Cucumber (Cucumaria frondosa) Processing Discards as Affected by High-Pressure Processing (HPP). Antioxidants 2022, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Sun, L.-C.; Yan, L.-J.; Lin, Y.-C.; Liu, G.-M.; Cao, M.-J. Production, optimisation and characterisation of angiotensin converting enzyme inhibitory peptides from sea cucumber (Stichopus japonicus) gonad. Food Funct. 2018, 9, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, F.; Bitto, A.; Irrera, N.; Pizzino, G.; Pallio, G.; Minutoli, L.; Altavilla, D. Pharmacological Activity and Clinical Use of PDRN. Front. Pharmacol. 2017, 8, 224. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Zhang, J.; Cheng, W.; Wang, C.; Zhao, D. Ethanolic Extract from Seed Residues of Sea Buckthorn (Hippophae rhamnoides L.) Ameliorates Oxidative Stress Damage and Prevents Apoptosis in Murine Cell and Aging Animal Models. Foods 2023, 12, 3322. [Google Scholar] [CrossRef]
- Čakar, U.; Čolović, M.; Milenković, D.; Medić, B.; Krstić, D.; Petrović, A.; Đorđević, B. Protective Effects of Fruit Wines against Hydrogen Peroxide—Induced Oxidative Stress in Rat Synaptosomes. Agronomy 2021, 11, 1414. [Google Scholar] [CrossRef]
- Chiabrando, D.; Vinchi, F.; Fiorito, V.; Mercurio, S.; Tolosano, E. Heme in pathophysiology: A matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 2014, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Siegel, D. Functions of NQO1 in Cellular Protection and CoQ10 Metabolism and its Potential Role as a Redox Sensitive Molecular Switch. Front. Physiol. 2017, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xiao, J.-H. The Keap1-Nrf2 System: A Mediator between Oxidative Stress and Aging. Oxid. Med. Cell. Longev. 2021, 2021, 6635460. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.O.; Sforca, M.L.; Heleno Batista, F.A.; Migliorini Figueira, A.C.; Benedetti, C.E. The MAF1 Phosphoregulatory Region Controls MAF1 Interaction with the RNA Polymerase III C34 Subunit and Transcriptional Repression in Plants. Plant Cell 2020, 32, 3019–3035. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.N.; Darouich, S.; Masmoudi, A.; Gordon, D.; Zender, G.; Han, Z.; Fitzgerald-Butt, S.; White, P.; McBride, K.L.; Kharrat, M.; et al. Novel frameshift variant in MYL2 reveals molecular differences between dominant and recessive forms of hypertrophic cardiomyopathy. PLoS Genet. 2020, 16, e1008639. [Google Scholar] [CrossRef] [PubMed]
- Hsia, D.A.; Tepper, C.G.; Pochampalli, M.R.; Hsia, E.Y.C.; Izumiya, C.; Huerta, S.B.; Wright, M.E.; Chen, H.-W.; Kung, H.-J.; Izumiya, Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. USA 2010, 107, 9671–9676. [Google Scholar] [CrossRef] [PubMed]
- Anijs, M.; Devanna, P.; Vernes, S.C. ARHGEF39, a Gene Implicated in Developmental Language Disorder, Activates RHOA and Is Involved in Cell De-Adhesion and Neural Progenitor Cell Proliferation. Front. Mol. Neurosci. 2022, 15, 941494. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, K.; Kudin, A.P.; Zsurka, G.; Kornblum, C.; Reimann, J.; Stüve, B.; Waltz, S.; Hattingen, E.; Thiele, H.; Nürnberg, P.; et al. Loss of the smallest subunit of cytochrome c oxidase, COX8A, causes Leigh-like syndrome and epilepsy. Brain 2016, 139, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Mi, T.; Xiao, X.; Zhang, H.; Dong, Y.; Huang, N.; Gao, P.; Lee, J.; Guelakis, M.; Gu, X. Topical glutathione amino acid precursors protect skin against environmental and oxidative stress. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 3–11. [Google Scholar] [CrossRef]
- Zhang, Y.; Roh, Y.J.; Han, S.-J.; Park, I.; Lee, H.M.; Ok, Y.S.; Lee, B.C.; Lee, S.-R. Role of Selenoproteins in Redox Regulation of Signaling and the Antioxidant System: A Review. Antioxidants 2020, 9, 383. [Google Scholar] [CrossRef]
- Yu, M.; Yu, J.; Yi, Y.; Chen, T.; Yu, L.; Zeng, W.; Ouyang, X.-k.; Huang, C.; Sun, S.; Wang, Y.; et al. Oxidative stress-amplified nanomedicine for intensified ferroptosis-apoptosis combined tumor therapy. J. Control. Release 2022, 347, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, D.-M.; Steinman, H.A.; Witthuhn, R.C. Comparative study of different methods for the extraction of DNA from fish species commercially available in South Africa. Food Control 2011, 22, 231–244. [Google Scholar] [CrossRef]
- Shao, P.; Chen, X.; Sun, P. Improvement of antioxidant and moisture-preserving activities of Sargassum horneri polysaccharide enzymatic hydrolyzates. Int. J. Biol. Macromol. 2015, 74, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, A.; Zheng, Y.; Wang, H.; Zheng, X.; Jin, Z.; Liu, D.; Wang, N.; Kan, Y. Influences of superfine-grinding and enzymolysis separately assisted with carboxymethylation and acetylation on the in vitro hypoglycemic and antioxidant activities of oil palm kernel expeller fibre. Food Chem. 2024, 449, 139192. [Google Scholar] [CrossRef]
- Souza, B.W.S.; Cerqueira, M.A.; Bourbon, A.I.; Pinheiro, A.C.; Martins, J.T.; Teixeira, J.A.; Coimbra, M.A.; Vicente, A.A. Chemical characterization and antioxidant activity of sulfated polysaccharide from the red seaweed Gracilaria birdiae. Food Hydrocoll. 2012, 27, 287–292. [Google Scholar] [CrossRef]

| Accession | Protein Description | Gene Symbol | H2O2/Control a | PDRN/H2O2 b |
|---|---|---|---|---|
| P01942 | Hemoglobin subunit alpha | Hba | 1.718 | 0.607 |
| Q64669 | NAD(P)H dehydrogenase [quinone] 1 | Nqo1 | 1.641 | 0.515 |
| Q9D0U6 | Repressor of RNA polymerase III transcription MAF1 homolog | Maf1 | 2.329 | 0.035 |
| P51667 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | Myl2 | 5.929 | 0.453 |
| Q9CXT6 | Bifunctional peptidase and arginyl-hydroxylase JMJD5 | Kdm8 | 1.551 | 0.485 |
| Q66JY6 | Rho guanine nucleotide exchange factor 39 | Arhgef39 | 1.546 | 0.578 |
| Q64445 | Cytochrome c oxidase subunit 8A, mitochondrial | Cox8a | 0.069 | 1.979 |
| P08905 | Lysozyme C-2 | Lyz2 | 0.647 | 0.581 |
| Q05117 | Tartrate-resistant acid phosphatase type 5 | Acp5 | 0.404 | 0.463 |
| O88199 | Carbohydrate sulfotransferase 3 | Chst3 | 45.981 | 67.64 |
| Q9D722 | Oxidative stress-responsive serine-rich protein 1 | Oser1 | 10.169 | 16.966 |
| P54987 | Cis-aconitate decarboxylase | Acod1 | 0.392 | 0.571 |
| O70167 | Phosphatidylinositol 3-kinase C2 domain-containing subunit gamma | Pik3c2g | 0.168 | 0.069 |
| P05555 | Integrin alpha-M | Itgam | 0.638 | 0.642 |
| Q9CPU9 | Probable low-affinity copper uptake protein 2 | Slc31a2 | 0.6 | 0.661 |
| P11835 | Integrin beta-2 | Itgb2 | 0.663 | 0.661 |
| Q61704 | Inter-alpha-trypsin inhibitor heavy chain H3 | Itih3 | 1.933 | 2.489 |
| P54830 | Tyrosine protein phosphatase non-receptor type 5 | Ptpn5 | 0.408 | 0.464 |
| O88531 | Palmitoyl protein thioesterase 1 | Ppt1 | 0.653 | 0.628 |
| P31809 | Carcinoembryonic antigen-related cell adhesion molecule 1 | Ceacam1 | 0.543 | 0.538 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, Z.; Ji, Y.; Liu, F.; Jing, Y.; Jiao, C.; Li, Y.; Zhao, Y.; Wang, G.; Zhang, J. Proteomics Analysis of the Protective Effect of Polydeoxyribonucleotide Extracted from Sea Cucumber (Apostichopus japonicus) Sperm in a Hydrogen Peroxide-Induced RAW264.7 Cell Injury Model. Mar. Drugs 2024, 22, 325. https://doi.org/10.3390/md22070325
Shu Z, Ji Y, Liu F, Jing Y, Jiao C, Li Y, Zhao Y, Wang G, Zhang J. Proteomics Analysis of the Protective Effect of Polydeoxyribonucleotide Extracted from Sea Cucumber (Apostichopus japonicus) Sperm in a Hydrogen Peroxide-Induced RAW264.7 Cell Injury Model. Marine Drugs. 2024; 22(7):325. https://doi.org/10.3390/md22070325
Chicago/Turabian StyleShu, Zhiqiang, Yizhi Ji, Fang Liu, Yuexin Jing, Chunna Jiao, Yue Li, Yunping Zhao, Gongming Wang, and Jian Zhang. 2024. "Proteomics Analysis of the Protective Effect of Polydeoxyribonucleotide Extracted from Sea Cucumber (Apostichopus japonicus) Sperm in a Hydrogen Peroxide-Induced RAW264.7 Cell Injury Model" Marine Drugs 22, no. 7: 325. https://doi.org/10.3390/md22070325





