Salinity Stress Acclimation Strategies in Chlamydomonas sp. Revealed by Physiological, Morphological and Transcriptomic Approaches
Abstract
1. Introduction
2. Results
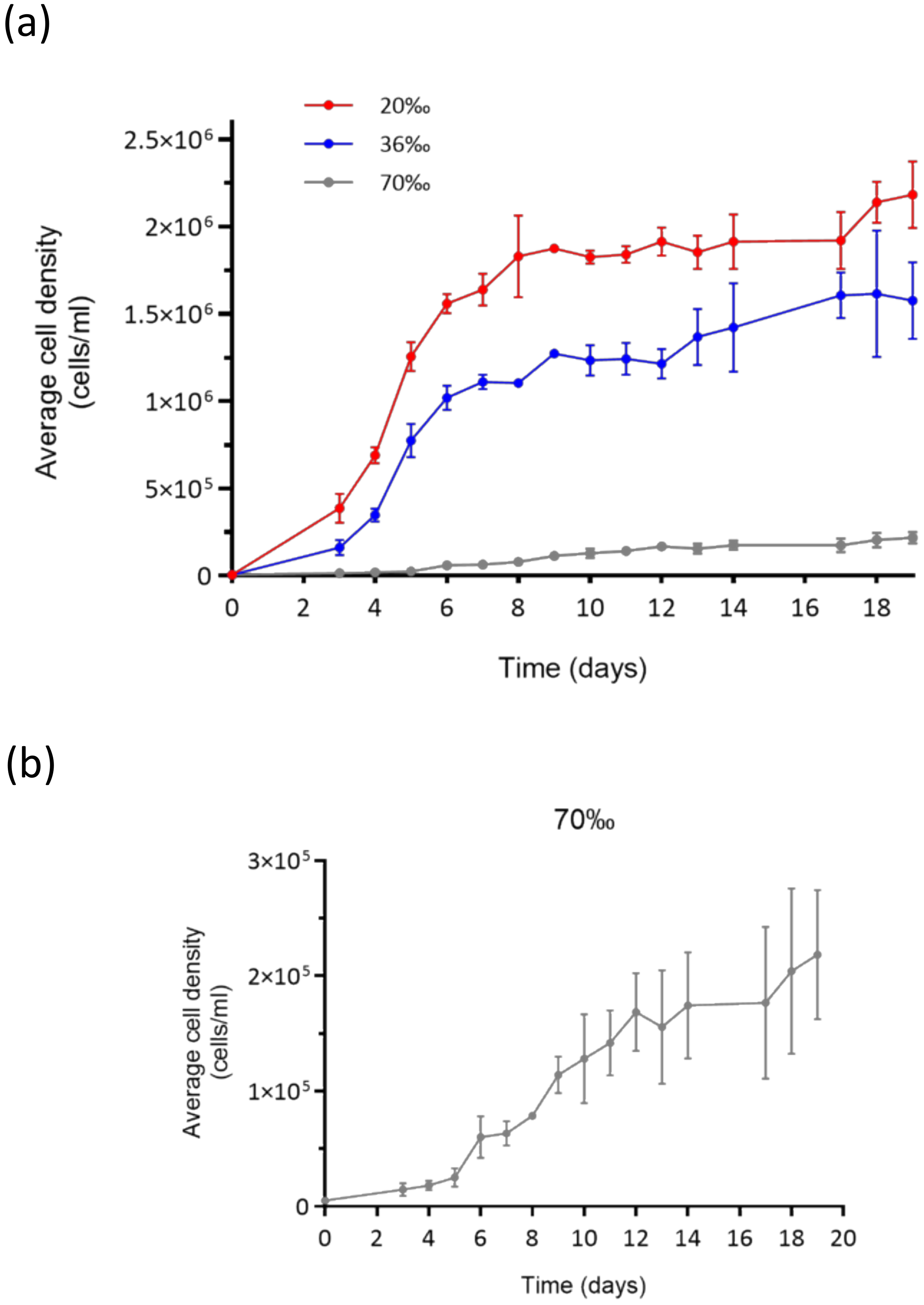
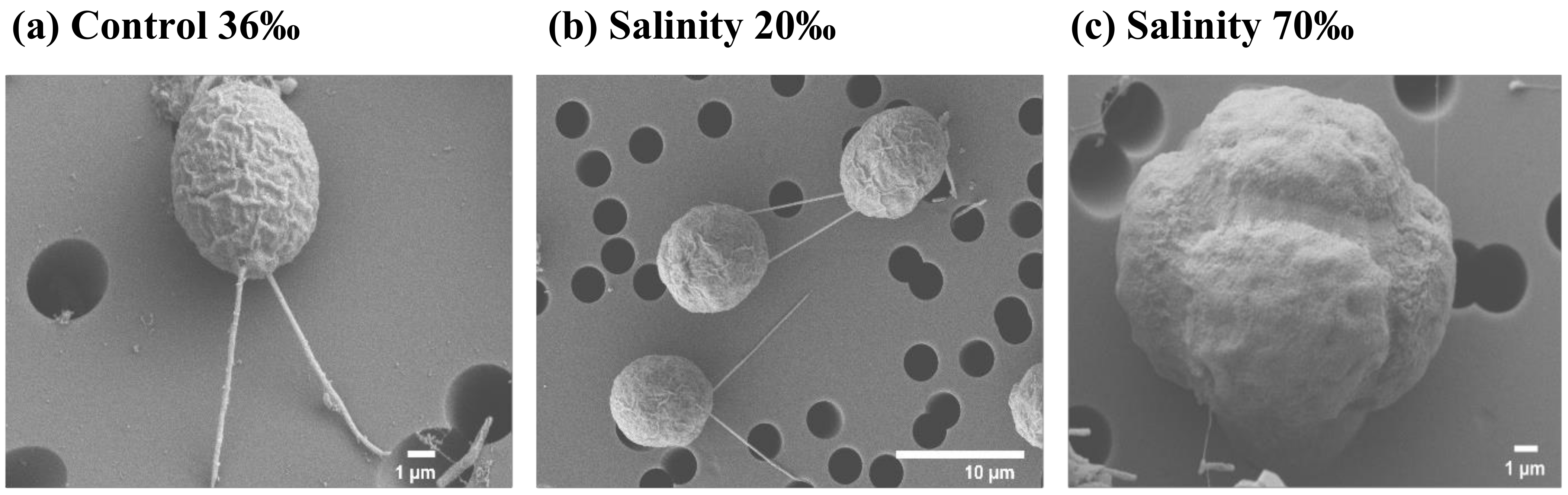
2.1. Growth Curves and Morphological Observations
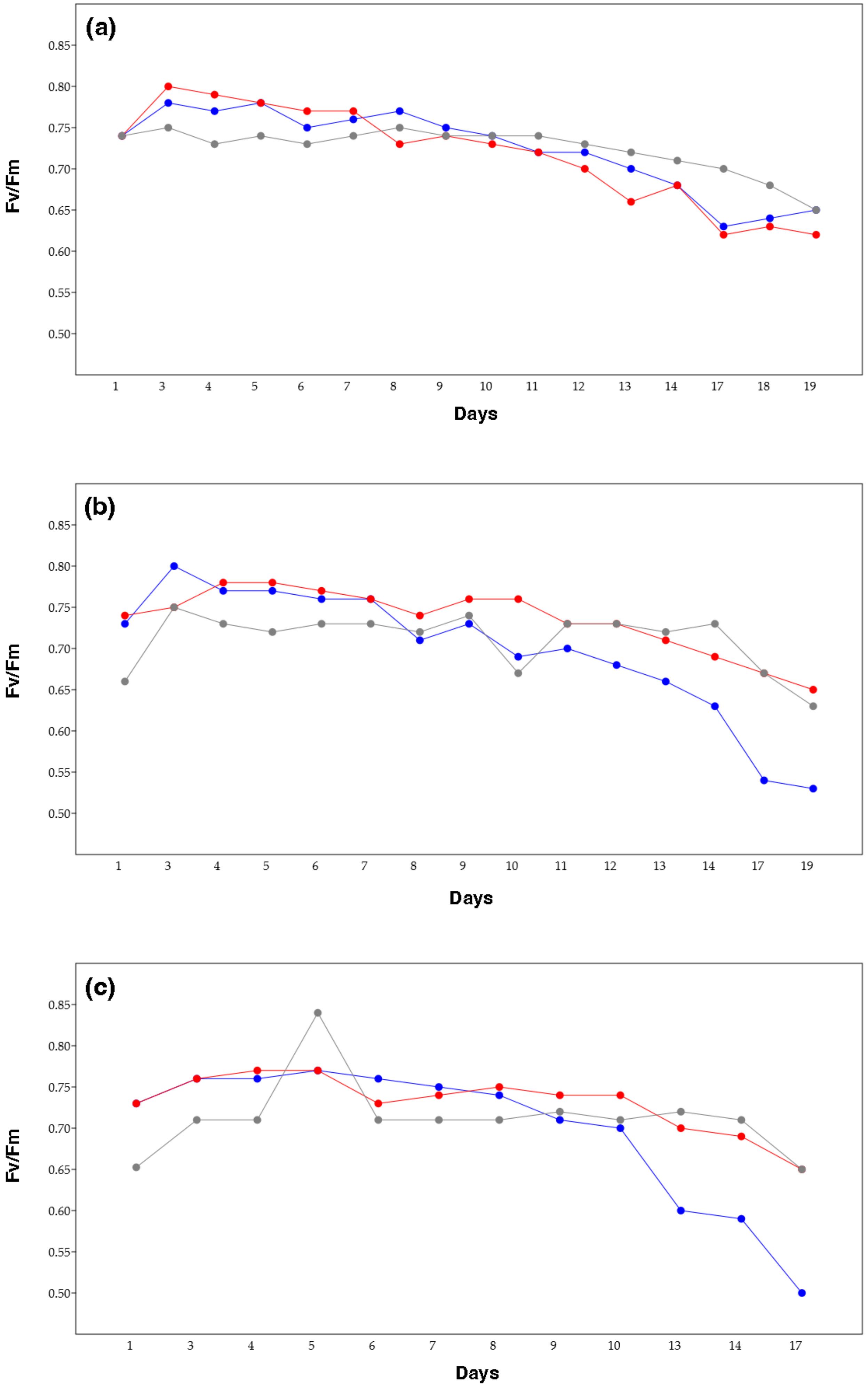
2.2. Photosynthetic Efficiency
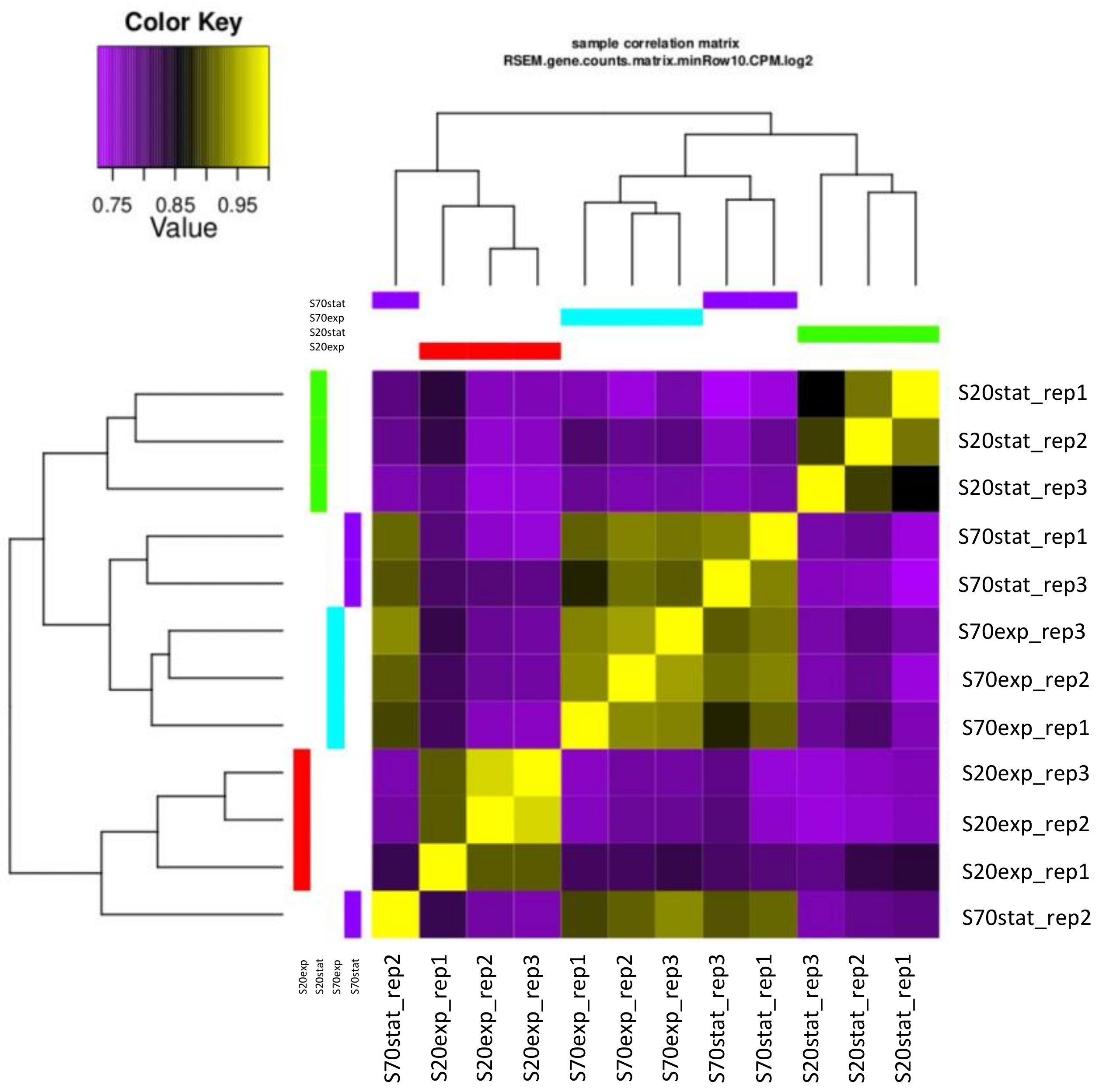
2.3. Sequencing
2.4. Differential Gene Expression Analysis
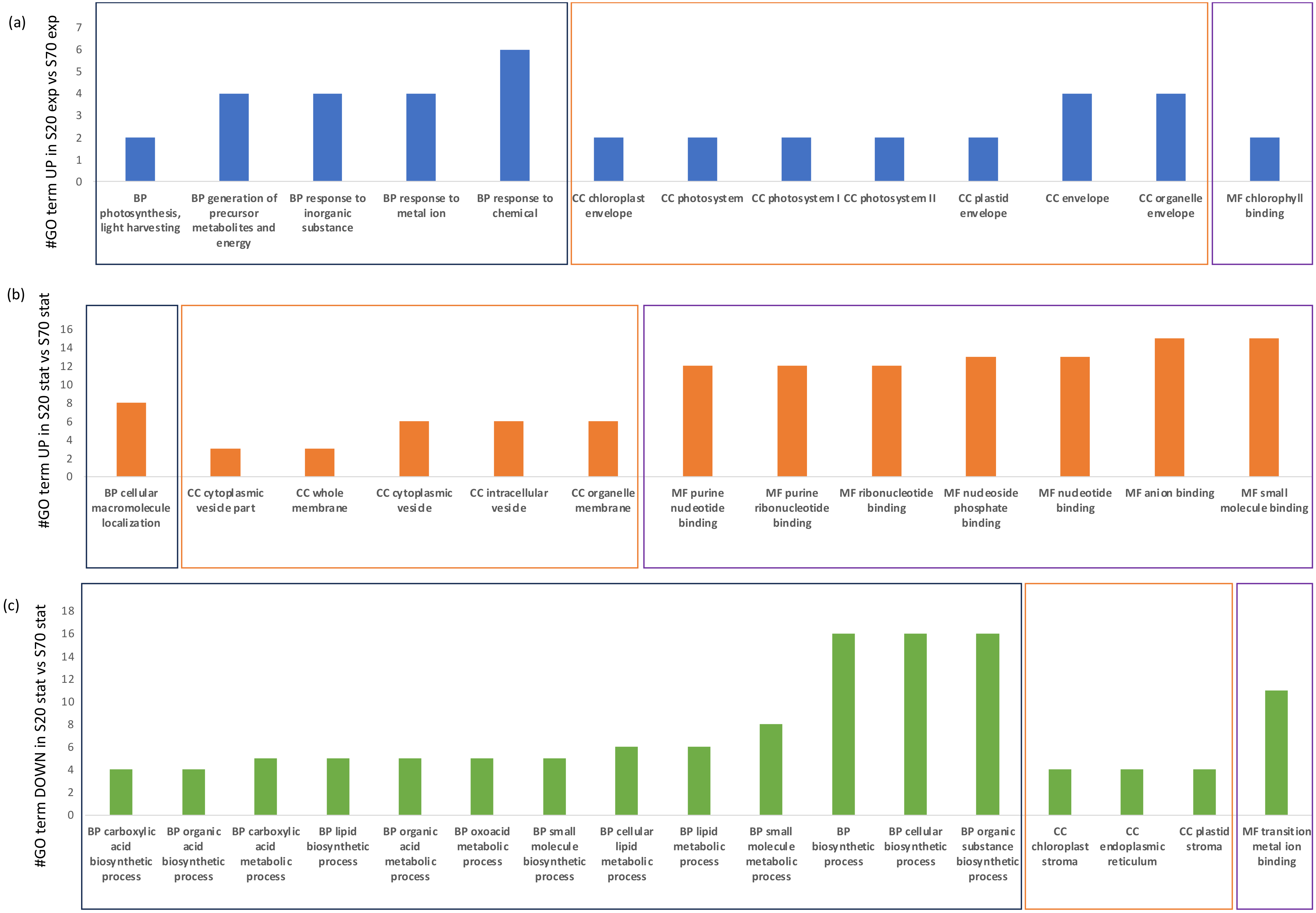
2.5. Functional Annotation
2.6. Genes Most Up- or Down-Regulated in S20_Exp vs. S20_Stat
2.7. Genes Most Up- or Down-Regulated in S20_Exp vs. S70_Exp
2.8. Genes Most Up- or Down-Regulated in S20_Stat vs. S70_Stat Comparison
2.9. Genes Most Up-Regulated in Exp vs. Stat Comparison
2.10. Reverse-Transcription–Quantitative PCR (RT-qPCR) Analyses on Selected Genes
3. Discussion
4. Materials and Methods
4.1. Microalgal Culturing
4.2. RNA Extraction and Quality Check
4.3. RNA Sequencing, De Novo Assembly, and Differential Expression Analysis
4.4. cDNA Synthesis and Reverse-Transcriptase–Quantitative PCR (RT-qPCR) Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durack, P.J. Ocean Salinity and the Global Water Cycle. Oceanography 2015, 28, 20–31. Available online: http://www.jstor.org/stable/24861839 (accessed on 28 May 2024). [CrossRef]
- Beardall, J.; Raven, J.A. The Potential Effects of Global Climate Change on Microalgal Photosynthesis, Growth and Ecology. Phycologia 2004, 43, 26–40. [Google Scholar] [CrossRef]
- Lauritano, C.; Ianora, A. Chemical Defense in Marine Organisms. Mar. Drugs 2020, 18, 518. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Coppola, D. Biodiversity, Adaptation Strategies, and Opportunities in Extreme Marine Environments. J. Mar. Sci. Eng. 2023, 11, 2367. [Google Scholar] [CrossRef]
- Lauritano, C.; Coppola, D. Biotechnological Applications of Products Released by Marine Microorganisms for Cold Adaptation Strategies: Polyunsaturated Fatty Acids, Antioxidants, and Antifreeze Proteins. J. Mar. Sci. Eng. 2023, 11, 1399. [Google Scholar] [CrossRef]
- Lauritano, C.; Rizzo, C.; Lo Giudice, A.; Saggiomo, M. Physiological and Molecular Responses to Main Environmental Stressors of Microalgae and Bacteria in Polar Marine Environments. Microorganisms 2020, 8, 1957. [Google Scholar] [CrossRef] [PubMed]
- Bazzani, E.; Lauritano, C.; Mangoni, O.; Bolinesi, F.; Saggiomo, M. Chlamydomonas Responses to Salinity Stress and Possible Biotechnological Exploitation. J. Mar. Sci. Eng. 2021, 9, 1242. [Google Scholar] [CrossRef]
- Shetty, P.; Gitau, M.M.; Maróti, G. Salinity Stress Responses and Adaptation Mechanisms in Eukaryotic Green Microalgae. Cells 2019, 8, 1657. [Google Scholar] [CrossRef] [PubMed]
- De Carpentier, F.; Lemaire, S.D.; Danon, A. When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells 2019, 8, 1307. [Google Scholar] [CrossRef] [PubMed]
- Saggiomo, M.; Escalera, L.; Saggiomo, V.; Bolinesi, F.; Mangoni, O. Phytoplankton Blooms below the Antarctic Landfast Ice during the Melt Season between Late Spring and Early Summer. J. Phycol. 2021, 57, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Grulois, D.; Quadrana, L.; Chevin, L.-M. Phenotypic Plasticity Evolves at Multiple Biological Levels in Response to Environmental Predictability in a Long-Term Experiment with a Halotolerant Microalga. PLoS Biol. 2023, 21, e3001895. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Grulois, D.; Chevin, L. Plasticity across Levels: Relating Epigenomic, Transcriptomic, and Phenotypic Responses to Osmotic Stress in a Halotolerant Microalga. Mol. Ecol. 2022, 31, 4672–4687. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jiang, J. Osmotic Responses of Dunaliella to the Changes of Salinity. J. Cell. Physiol. 2009, 219, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Gorelova, O.; Baulina, O.; Ismagulova, T.; Kokabi, K.; Lobakova, E.; Selyakh, I.; Semenova, L.; Chivkunova, O.; Karpova, O.; Scherbakov, P.; et al. Stress-Induced Changes in the Ultrastructure of the Photosynthetic Apparatus of Green Microalgae. Protoplasma 2019, 256, 261–277. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Lucker, B.F.; Zienkiewicz, K.; Miller, T.E.; Zienkiewicz, A.; Sears, B.B.; Kramer, D.M.; Benning, C. Galactoglycerolipid Lipase PGD1 Is Involved in Thylakoid Membrane Remodeling in Response to Adverse Environmental Conditions in Chlamydomonas. Plant Cell 2018, 30, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.; Croft, M.; Gladyshev, V.; Merchant, S.; Posewitz, M.; Prochnik, S.; Spalding, M. Novel Metabolism in Chlamydomonas through the Lens of Genomics. Curr. Opin. Plant Biol. 2007, 10, 190–198. [Google Scholar] [CrossRef]
- Cock, J.M.; Coelho, S.M. Algal Models in Plant Biology. J. Exp. Bot. 2011, 62, 2425–2430. [Google Scholar] [CrossRef]
- Salomé, P.A.; Merchant, S.S. A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism. Plant Cell 2019, 31, 1682–1707. [Google Scholar] [CrossRef]
- Falciatore, A.; Jaubert, M.; Bouly, J.-P.; Bailleul, B.; Mock, T. Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity. Plant Cell 2020, 32, 547–572. [Google Scholar] [CrossRef]
- Chang, C.; Bowman, J.L.; Meyerowitz, E.M. Field Guide to Plant Model Systems. Cell 2016, 167, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Carrasco Flores, D.; Fricke, M.; Wesp, V.; Desirò, D.; Kniewasser, A.; Hölzer, M.; Marz, M.; Mittag, M. A Marine Chlamydomonas sp. Emerging as an Algal Model. J. Phycol. 2021, 57, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Stallings, W.C.; Abdel-Meguid, S.S.; Lim, L.W.; Shieh, H.S.; Dayringer, H.E.; Leimgruber, N.K.; Stegeman, R.A.; Anderson, K.S.; Sikorski, J.A.; Padgette, S.R.; et al. Structure and Topological Symmetry of the Glyphosate Target 5-Enolpyruvylshikimate-3-Phosphate Synthase: A Distinctive Protein Fold. Proc. Natl. Acad. Sci. USA 1991, 88, 5046–5050. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Zuo, L.; Xu, H.; Li, C.; Qiao, G.; Guo, M.; Lin, X. Alkaloids as Anticancer Agents: A Review of Chinese Patents in Recent 5 Years. Pat. Anti-Cancer Drug Discov. 2020, 15, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.; Gandhi, A.; Fimognari, C.; Atanasov, A.G.; Bishayee, A. Alkaloids for Cancer Prevention and Therapy: Current Progress and Future Perspectives. Eur. J. Pharmacol. 2019, 858, 172472. [Google Scholar] [CrossRef] [PubMed]
- Wicks, C.; Hudlicky, T.; Rinner, U. Morphine Alkaloids: History, Biology, and Synthesis. In The Alkaloids: Chemistry and Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 86, pp. 145–342. ISBN 978-0-12-824618-4. [Google Scholar]
- Barbuti, A.; Chen, Z.-S. Paclitaxel through the Ages of Anticancer Therapy: Exploring Its Role in Chemoresistance and Radiation Therapy. Cancers 2015, 7, 2360–2371. [Google Scholar] [CrossRef] [PubMed]
- Seca, A.; Pinto, D. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; O’Brien, S.; Cortes, J. Homoharringtonine/Omacetaxine Mepesuccinate: The Long and Winding Road to Food and Drug Administration Approval. Clin. Lymphoma Myeloma Leuk. 2013, 13, 530–533. [Google Scholar] [CrossRef]
- D’Incalci, M.; Badri, N.; Galmarini, C.M.; Allavena, P. Trabectedin, a Drug Acting on Both Cancer Cells and the Tumour Microenvironment. Br. J. Cancer 2014, 111, 646–650. [Google Scholar] [CrossRef]
- Dybdal-Hargreaves, N.F.; Risinger, A.L.; Mooberry, S.L. Eribulin Mesylate: Mechanism of Action of a Unique Microtubule-Targeting Agent. Clin. Cancer Res. 2015, 21, 2445–2452. [Google Scholar] [CrossRef]
- Singh, I.P.; Shah, P. Tetrahydroisoquinolines in Therapeutics: A Patent Review (2010–2015). Expert Opin. Ther. Pat. 2017, 27, 17–36. [Google Scholar] [CrossRef]
- Baumann, T.W.; Frischknecht, P.M. Purines. In Phytochemicals in Plant Cell Cultures; Elsevier: Amsterdam, The Netherlands, 1988; pp. 403–417. ISBN 978-0-12-715005-5. [Google Scholar]
- Morrison, H. Carbonic Anhydrase II. In Enzyme Active Sites and Their Reaction Mechanisms; Elsevier: Amsterdam, The Netherlands, 2021; pp. 31–35. ISBN 978-0-12-821067-3. [Google Scholar]
- Ohbayashi, I.; Huang, S.; Fukaki, H.; Song, X.; Sun, S.; Morita, M.T.; Tasaka, M.; Millar, A.H.; Furutani, M. Mitochondrial Pyruvate Dehydrogenase Contributes to Auxin-Regulated Organ Development. Plant Physiol. 2019, 180, 896–909. [Google Scholar] [CrossRef]
- Khannpnavar, B.; Mehta, V.; Qi, C.; Korkhov, V. Structure and Function of Adenylyl Cyclases, Key Enzymes in Cellular Signaling. Curr. Opin. Struct. Biol. 2020, 63, 34–41. [Google Scholar] [CrossRef]
- Kimura, Y.; Ohno, A. A New Peptide-N4-(Acetyl-β-Glucosaminyl) Asparagine Amidase from Soybean (Glycine max) Seeds: Purification and Substrate Specificity. Biosci. Biotechnol. Biochem. 1998, 62, 412–418. [Google Scholar] [CrossRef]
- Miao, X.; Wu, J.; Chen, H.; Lu, G. Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation. Nutrients 2022, 14, 1690. [Google Scholar] [CrossRef]
- Brautigan, D.L.; Shenolikar, S. Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu. Rev. Biochem. 2018, 87, 921–964. [Google Scholar] [CrossRef]
- Green, B.R.; Pichersky, E.; Kloppstech, K. Chlorophyll a/b-Binding Proteins: An Extended Family. Trends Biochem. Sci. 1991, 16, 181–186. [Google Scholar] [CrossRef]
- Simonds, W.F. G Protein Regulation of Adenylate Cyclase. Trends Pharmacol. Sci. 1999, 20, 66–73. [Google Scholar] [CrossRef]
- Kozminski, K.G.; Johnson, K.A.; Forscher, P.; Rosenbaum, J.L. A Motility in the Eukaryotic Flagellum Unrelated to Flagellar Beating. Proc. Natl. Acad. Sci. USA 1993, 90, 5519–5523. [Google Scholar] [CrossRef]
- Khona, D.K.; Shirolikar, S.M.; Gawde, K.K.; Hom, E.; Deodhar, M.A.; D’Souza, J.S. Characterization of Salt Stress-Induced Palmelloids in the Green Alga, Chlamydomonas reinhardtii. Algal Res. 2016, 16, 434–448. [Google Scholar] [CrossRef]
- Lefebvre, P.A. Chapter 1—Flagellar Amputation and Regeneration in Chlamydomonas. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 1995; Volume 47, pp. 3–7. ISBN 978-0-12-564148-7. [Google Scholar]
- Suwannachuen, N.; Leetanasaksakul, K.; Roytrakul, S.; Phaonakrop, N.; Thaisakun, S.; Roongsattham, P.; Jantasuriyarat, C.; Sanevas, N.; Sirikhachornkit, A. Palmelloid Formation and Cell Aggregation Are Essential Mechanisms for High Light Tolerance in a Natural Strain of Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2023, 24, 8374. [Google Scholar] [CrossRef]
- Tanaka, S.; Ikeda, K.; Miyasaka, H.; Shioi, Y.; Suzuki, Y.; Tamoi, M.; Takeda, T.; Shigeoka, S.; Harada, K.; Hirata, K. Comparison of Three Chlamydomonas Strains Which Show Distinctive Oxidative Stress Tolerance. J. Biosci. Bioeng. 2011, 112, 462–468. [Google Scholar] [CrossRef]
- Zhang, Z.; An, M.; Miao, J.; Gu, Z.; Liu, C.; Zhong, B. The Antarctic Sea Ice Alga Chlamydomonas sp. ICE-L Provides Insights into Adaptive Patterns of Chloroplast Evolution. BMC Plant Biol. 2018, 18, 53. [Google Scholar] [CrossRef]
- Hounslow, E.; Evans, C.A.; Pandhal, J.; Sydney, T.; Couto, N.; Pham, T.K.; Gilmour, D.J.; Wright, P.C. Quantitative Proteomic Comparison of Salt Stress in Chlamydomonas reinhardtii and the Snow Alga Chlamydomonas nivalis Reveals Mechanisms for Salt-Triggered Fatty Acid Accumulation via Reallocation of Carbon Resources. Biotechnol. Biofuels 2021, 14, 121. [Google Scholar] [CrossRef]
- Lu, N.; Wei, D.; Jiang, X.-L.; Chen, F.; Yang, S.-T. Regulation of Lipid Metabolism in the Snow Alga Chlamydomonas nivalis in Response to NaCl Stress: An Integrated Analysis by Cytomic and Lipidomic Approaches. Process Biochem. 2012, 47, 1163–1170. [Google Scholar] [CrossRef]
- Lu, N.; Wei, D.; Jiang, X.-L.; Chen, F.; Yang, S.-T. Fatty Acids Profiling and Biomarker Identification in Snow Alga Chlamydomonas nivalis by NaCl Stress Using GC/MS and Multivariate Statistical Analysis. Anal. Lett. 2012, 45, 1172–1183. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, P.; Cong, B.; Liu, C.; Lin, X.; Shen, J.; Huang, X. Molecular Cloning and Expression Analysis of a Cytosolic Hsp70 Gene from Antarctic Ice Algae Chlamydomonas sp. ICE-L. Extremophiles 2010, 14, 329–337. [Google Scholar] [CrossRef]
- Lindskog, S. Structure and Mechanism of Carbonic Anhydrase. Pharmacol. Ther. 1997, 74, 1–20. [Google Scholar] [CrossRef]
- Cross, T.G.; Scheel-Toellner, D.; Henriquez, N.V.; Deacon, E.; Salmon, M.; Lord, J.M. Serine/Threonine Protein Kinases and Apoptosis. Exp. Cell Res. 2000, 256, 34–41. [Google Scholar] [CrossRef]
- Giladi, M.; Edri, I.; Goldenberg, M.; Newman, H.; Strulovich, R.; Khananshvili, D.; Haitin, Y.; Loewenstein, A. Purification and Characterization of Human Dehydrodolychil Diphosphate Synthase (DHDDS) Overexpressed in E. coli. Protein Expr. Purif. 2017, 132, 138–142. [Google Scholar] [CrossRef]
- Dittami, S.M.; Michel, G.; Collén, J.; Boyen, C.; Tonon, T. Chlorophyll-Binding Proteins Revisited—A Multigenic Family of Light-Harvesting and Stress Proteins from a Brown Algal Perspective. BMC Evol. Biol. 2010, 10, 365. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, X.; Gao, G.; Beardall, J.; Inaba, K.; Hall-Spencer, J.M.; Xu, D.; Zhang, X.; Han, W.; McMinn, A.; et al. Decreased Motility of Flagellated Microalgae Long-Term Acclimated to CO2-Induced Acidified Waters. Nat. Clim. Change 2020, 10, 561–567. [Google Scholar] [CrossRef]
- Silva, D.A.; Huang, X.; Behal, R.H.; Cole, D.G.; Qin, H. The RABL5 Homolog IFT22 Regulates the Cellular Pool Size and the Amount of IFT Particles Partitioned to the Flagellar Compartment in Chlamydomonas reinhardtii. Cytoskeleton 2012, 69, 33–48. [Google Scholar] [CrossRef]
- Elkahoui, S.; Smaoui, A.; Zarrouk, M.; Ghrir, R.; Limam, F. Salt-Induced Lipid Changes in Catharanthus Roseus Cultured Cell Suspensions. Phytochemistry 2004, 65, 1911–1917. [Google Scholar] [CrossRef]
- Ji, C.; Mao, X.; Hao, J.; Wang, X.; Xue, J.; Cui, H.; Li, R. Analysis of bZIP Transcription Factor Family and Their Expressions under Salt Stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 2800. [Google Scholar] [CrossRef]
- Maathuis, F.J.M. The Role of Monovalent Cation Transporters in Plant Responses to Salinity. J. Exp. Bot. 2006, 57, 1137–1147. [Google Scholar] [CrossRef]
- Uzlasir, T.; Selli, S.; Kelebek, H. Effect of Salt Stress on the Phenolic Compounds, Antioxidant Capacity, Microbial Load, and In Vitro Bioaccessibility of Two Microalgae Species (Phaeodactylum tricornutum and Spirulina platensis). Foods 2023, 12, 3185. [Google Scholar] [CrossRef]
- Nezafatian, E.; Farhadian, O.; Yegdaneh, A.; Safavi, M.; Daneshvar, E.; Bhatnagar, A. Enhanced Production of Bioactive Compounds from Marine Microalgae Tetraselmis tetrathele under Salinity and Light Stresses: A Two-Stage Cultivation Strategy. Bioresour. Technol. 2023, 376, 128899. [Google Scholar] [CrossRef]
- Saide, A.; Riccio, G.; Ianora, A.; Lauritano, C. The Diatom Cylindrotheca closterium and the Chlorophyll Breakdown Product Pheophorbide a for Photodynamic Therapy Applications. Appl. Sci. 2023, 13, 2590. [Google Scholar] [CrossRef]
- Dennis, R.K.; Stein, M.R. K2 of Discrete Valuation Rings. Adv. Math. 1975, 18, 182–238. [Google Scholar] [CrossRef]
- Lauritano, C.; Orefice, I.; Procaccini, G.; Romano, G.; Ianora, A. Key Genes as Stress Indicators in the Ubiquitous Diatom Skeletonema marinoi. BMC Genom. 2015, 16, 411. [Google Scholar] [CrossRef]
- Zar, J.H. Biostatistical Analysis, 4th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 1999; ISBN 978-0-13-081542-2. [Google Scholar]
- Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 9 December 2022).
- Available online: https://github.com/TransDecoder/TransDecoder (accessed on 9 December 2022).
- Available online: https://github.com/Trinotate/Trinotate (accessed on 9 December 2022).
- Colina, F.; Amaral, J.; Carbó, M.; Pinto, G.; Soares, A.; Cañal, M.J.; Valledor, L. Genome-Wide Identification and Characterization of CKIN/SnRK Gene Family in Chlamydomonas reinhardtii. Sci. Rep. 2019, 9, 350. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
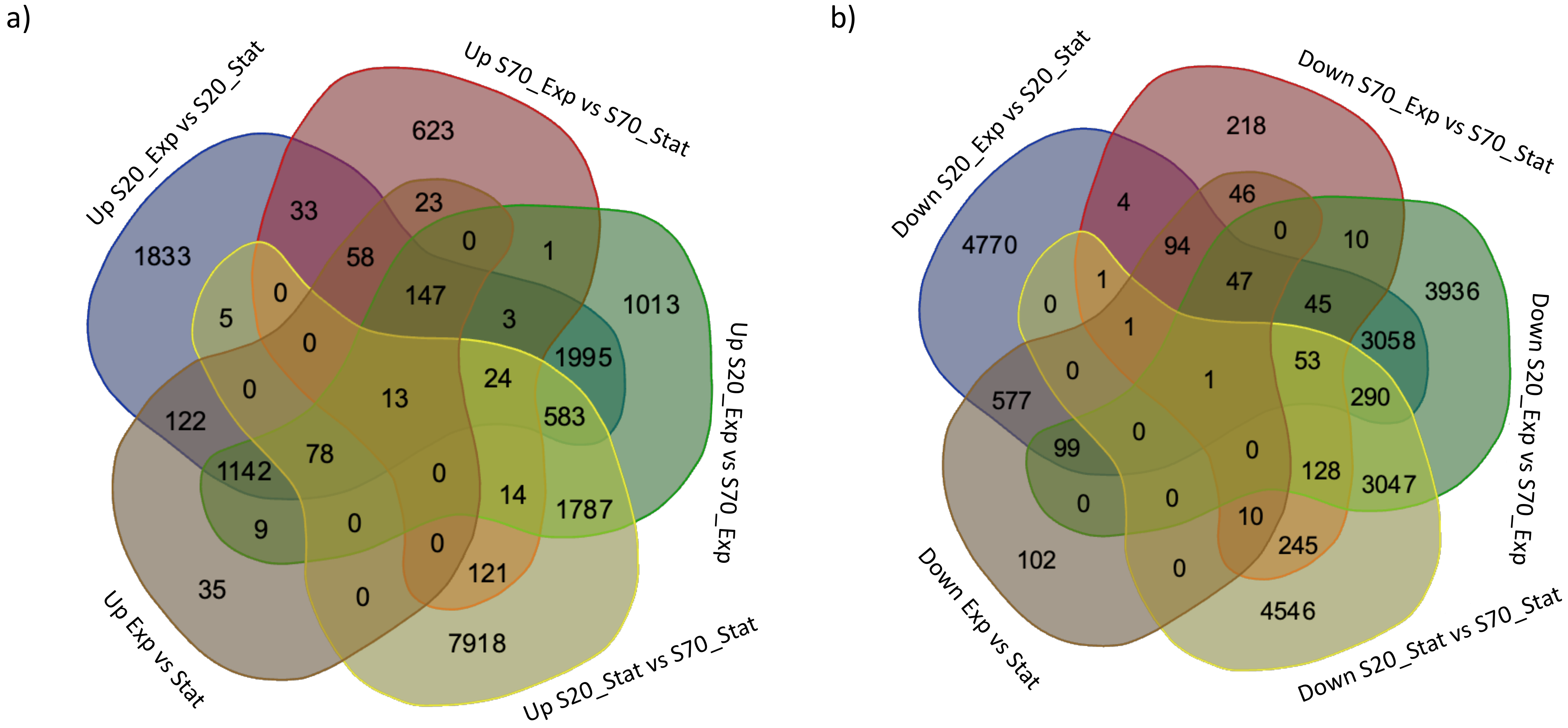

| Sample_Name | #Genes | #Isoforms |
|---|---|---|
| S20exp_rep1 | 47293 | 61173 |
| S20exp_rep2 | 38171 | 49096 |
| S20exp_rep3 | 42641 | 54014 |
| S20stat_rep1 | 73803 | 97149 |
| S20stat_rep2 | 76236 | 102698 |
| S20stat_rep3 | 66162 | 91150 |
| S70exp_rep1 | 74735 | 97753 |
| S70exp_rep2 | 73391 | 96712 |
| S70exp_rep3 | 71644 | 91918 |
| S70stat_rep1 | 64904 | 85302 |
| S70stat_rep2 | 68462 | 88701 |
| S70stat_rep3 | 59995 | 79730 |
| Treated_vs._CTRL | FDR ≤ 0.05 | FC ≥ 1.5 & FDR ≤ 0.05 | FC ≤ −1.5 & FDR ≤ 0.05 |
|---|---|---|---|
| S20_Exp vs. S20_Stat | 15094 | 6036 | 9040 |
| S70_Exp vs. S70_Stat | 2036 | 1060 | 903 |
| S20_Exp vs. S70_Exp | 17623 | 6809 | 10704 |
| S20_Stat vs. S70_Stat | 18926 | 10543 | 8322 |
| Exp vs. Stat | 2610 | 1627 | 977 |
| Treated_vs._CTRL | #GO Terms Enriched | |
|---|---|---|
| Up DEGs | Down DEGs | |
| S20_Exp vs. S20_Stat | 33 | 1 |
| S70_Exp vs. S70_Stat | 0 | 0 |
| S20_Exp vs. S70_Exp | 13 | 0 |
| S20_Stat vs. S70_Stat | 13 | 17 |
| Exp vs. Stat | 35 | 0 |
| Up-Regulated Genes | Fold Change | Down-Regulated Genes | Fold Changes |
|---|---|---|---|
| 3-phosphoshikimate1carboxyvinyltransferase | +56.70 | Adenylate cyclase | −6.54 |
| Carbonic anhydrase 2 | +51.19 | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase | −6.22 |
| UPF0225 protein VV1_2912 | +48.34 | Adenylate cyclase | −3.83 |
| Pyruvate dehydrogenase E1 component subunit beta-3 | +28.16 | Ubiquitin carboxyl-terminal hydrolase 24 | −2.83 |
| Chlorophyll a-b binding protein CP29.2, chloroplastic | +16.72 | Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit C | −2.78 |
| Up-Regulated Genes | Fold Change | Down-Regulated Genes | Fold Change |
|---|---|---|---|
| Chlorophyll a-b binding protein CP29.2, chloroplastic | +21.97 | Serine/threonine-protein kinase fray1 | −120.888 |
| Chlorophyll a-b binding protein type member F3, chloroplastic | +15.02 | Serine/threonine-protein kinase OSR1 | −120.888 |
| Carbonic anhydrase 2 | +11.84 | Isoprenyl transferase | −57.222 |
| UPF0225 protein VV1_2912 | +9.78 | Dehydrodolichyl diphosphate synthase 2 | −57.222 |
| 3-phosphoshikimate 1-carboxyvinyltransferase, chloroplastic | +8.05 | Nitric oxide synthase | −31,74 |
| Up-Regulated | Fold Change | Down-Regulated | Fold Change |
|---|---|---|---|
| Probable inactive leucine-rich repeat receptor kinase XIAO | +217.457 | Serine/threonine-protein kinase fray1 | −34.91 |
| Adenylate cyclase | +21.778 | Serine/threonine-protein kinase OSR1 | −34.91 |
| Adenylate cyclase | +10.119 | Isoprenyl transferase | −8.48 |
| Cys-loop ligand-gated ion channel | +9.003 | Dehydrodolichyl diphosphate synthase 2 | −8.48 |
| Intraflagellar transport protein 22 | +6.971 | Beta-carotene 3-hydroxylase, chloroplastic | −5.44 |
| Up-Regulated | Fold Change |
|---|---|
| Carbonic Anhydrase 2 | +33.138 |
| 3-phosphoshikimate 1-carboxyvinyltransferase, chloroplastic | +11.175 |
| Chlorophyll a-b binding protein CP29.2, chloroplastic | +9.657 |
| UPF0225 protein VV1_2912 | +9.569 |
| Pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic | +6.533 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauritano, C.; Bazzani, E.; Montuori, E.; Bolinesi, F.; Mangoni, O.; Riccio, G.; Buondonno, A.; Saggiomo, M. Salinity Stress Acclimation Strategies in Chlamydomonas sp. Revealed by Physiological, Morphological and Transcriptomic Approaches. Mar. Drugs 2024, 22, 351. https://doi.org/10.3390/md22080351
Lauritano C, Bazzani E, Montuori E, Bolinesi F, Mangoni O, Riccio G, Buondonno A, Saggiomo M. Salinity Stress Acclimation Strategies in Chlamydomonas sp. Revealed by Physiological, Morphological and Transcriptomic Approaches. Marine Drugs. 2024; 22(8):351. https://doi.org/10.3390/md22080351
Chicago/Turabian StyleLauritano, Chiara, Emma Bazzani, Eleonora Montuori, Francesco Bolinesi, Olga Mangoni, Gennaro Riccio, Angela Buondonno, and Maria Saggiomo. 2024. "Salinity Stress Acclimation Strategies in Chlamydomonas sp. Revealed by Physiological, Morphological and Transcriptomic Approaches" Marine Drugs 22, no. 8: 351. https://doi.org/10.3390/md22080351
APA StyleLauritano, C., Bazzani, E., Montuori, E., Bolinesi, F., Mangoni, O., Riccio, G., Buondonno, A., & Saggiomo, M. (2024). Salinity Stress Acclimation Strategies in Chlamydomonas sp. Revealed by Physiological, Morphological and Transcriptomic Approaches. Marine Drugs, 22(8), 351. https://doi.org/10.3390/md22080351










