Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View
Abstract
:1. Introduction
2. Results and Discussion
2.1. Growth Kinetics
2.2. Differentially Expressed Genes Analysis
2.3. Targeted Analysis of Interesting Pathway from a Bioproduction Point of View
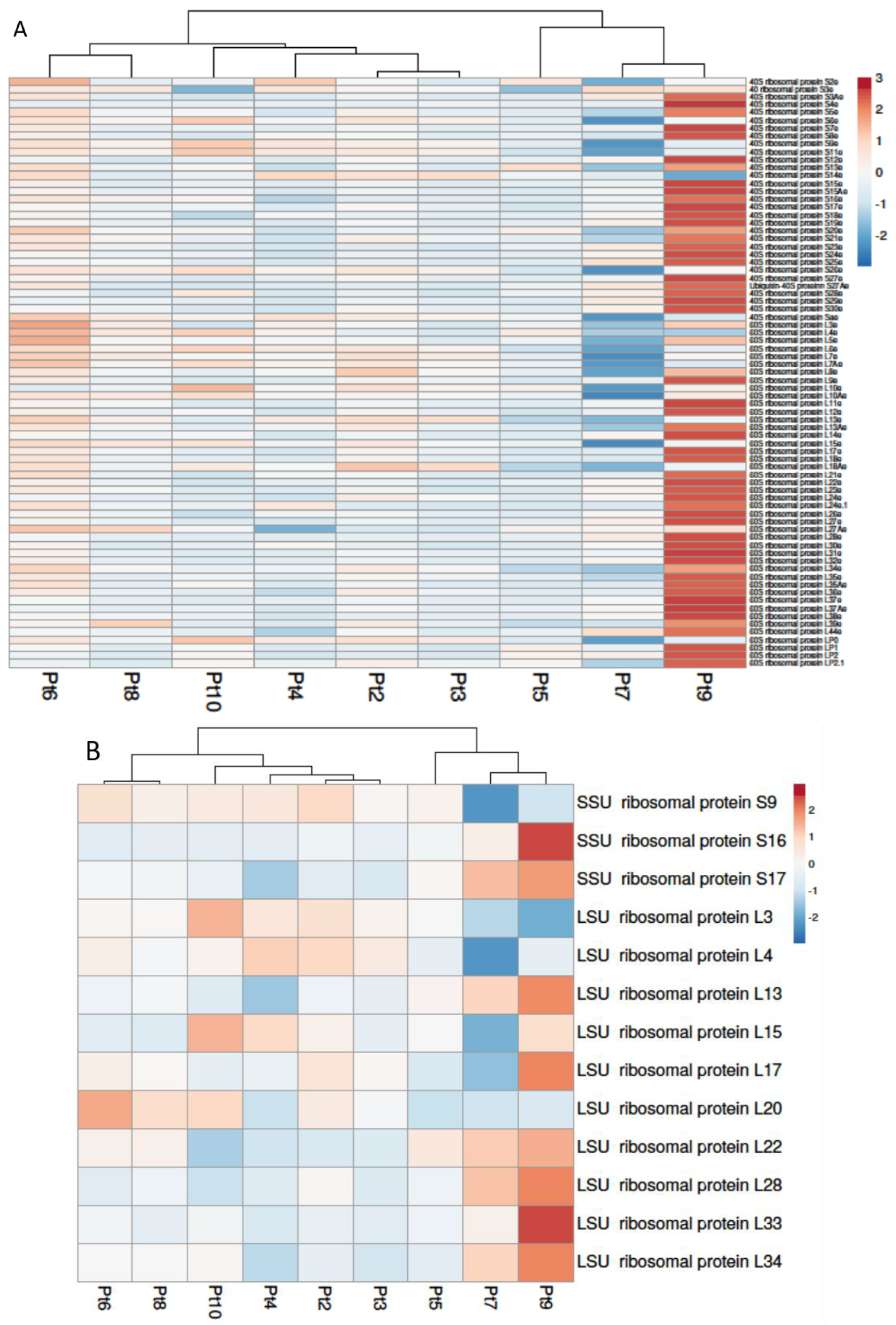
2.3.1. Protein Synthesis: Translation and Ribosome Biogenesis
2.3.2. Protein Export
2.3.3. N-Glycosylation
2.3.4. Quality Control and Proteasomes
2.3.5. Proteases
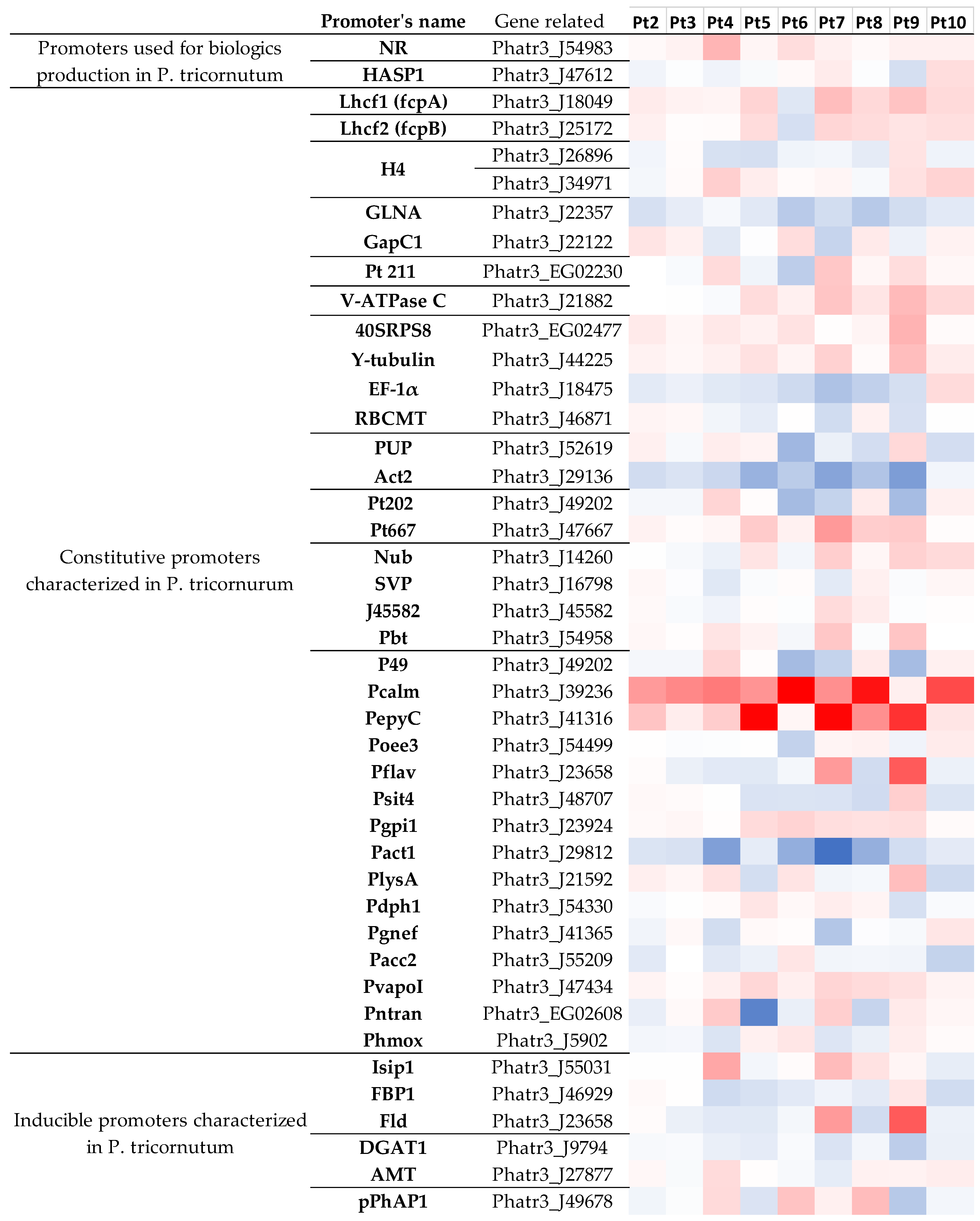
2.3.6. Genetic Tools
3. Materials and Methods
3.1. Cell Culture and Growth Conditions
3.2. Growth Curves
3.3. Preparation of RNA Samples
3.4. Generation of cDNA Libraries and Quality Control of Reads
3.5. Data Analysis
3.5.1. Trimming, Alignment, and Gene Count
3.5.2. Differential Gene Expression Analysis
3.5.3. Gene Ontology
3.6. Protease Activities Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lingg, N.; Zhang, P.; Song, Z.; Bardor, M. The Sweet Tooth of Biopharmaceuticals: Importance of Recombinant Protein Glycosylation Analysis. Biotechnol. J. 2012, 7, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Butler, M. Animal Cell Cultures: Recent Achievements and Perspectives in the Production of Biopharmaceuticals. Appl. Microbiol. Biotechnol. 2005, 68, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Ward, V. Production of Recombinant and Therapeutic Proteins in Microalgae. Curr. Opin. Biotechnol. 2022, 78, 102784. [Google Scholar] [CrossRef] [PubMed]
- Kornecki, M.; Mestmäcker, F.; Zobel-Roos, S.; Heikaus de Figueiredo, L.; Schlüter, H.; Strube, J. Host Cell Proteins in Biologics Manufacturing: The Good, the Bad, and the Ugly. Antibodies 2017, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Djemal, L.; Fournier, C.; von Hagen, J.; Kolmar, H.; Deparis, V. Review: High Temperature Short Time Treatment of Cell Culture Media and Feed Solutions to Mitigate Adventitious Viral Contamination in the Biopharmaceutical Industry. Biotechnol. Prog. 2021, 37, e3117. [Google Scholar] [CrossRef] [PubMed]
- Prade, E.; Zeck, A.; Stiefel, F.; Unsoeld, A.; Mentrup, D.; Arango Gutierrez, E.; Gorr, I.H. Cysteine in Cell Culture Media Induces Acidic IgG1 Species by Disrupting the Disulfide Bond Network. Biotechnol. Bioeng. 2021, 118, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.D.; de Oliveira, C.F.M.; Molino, J.V.D.; Ferreira-Camargo, L.S.; Matsudo, M.C.; de Carvalho, J.C.M. Production of Recombinant Biopharmaceuticals in Chlamydomonas Reinhardtii. Int. J. Plant Biol. 2023, 14, 39–52. [Google Scholar] [CrossRef]
- Daniell, H.; Streatfield, S.J.; Wycoff, K. Medical Molecular Farming: Production of Antibodies, Biopharmaceuticals and Edible Vaccines in Plants. Trends Plant Sci. 2001, 6, 219–226. [Google Scholar] [CrossRef]
- Taunt, H.N.; Stoffels, L.; Purton, S. Green Biologics: The Algal Chloroplast as a Platform for Making Biopharmaceuticals. Bioengineered 2018, 9, 48–54. [Google Scholar] [CrossRef]
- Moon, K.-B.; Park, J.-S.; Park, Y.-I.; Song, I.-J.; Lee, H.-J.; Cho, H.S.; Jeon, J.-H.; Kim, H.-S. Development of Systems for the Production of Plant-Derived Biopharmaceuticals. Plants 2020, 9, 30. [Google Scholar] [CrossRef]
- Barolo, L.; Abbriano, R.M.; Commault, A.S.; George, J.; Kahlke, T.; Fabris, M.; Padula, M.P.; Lopez, A.; Ralph, P.J.; Pernice, M. Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae. Cells 2020, 9, 633. [Google Scholar] [CrossRef]
- Gimpel, J.A.; Hyun, J.S.; Schoepp, N.G.; Mayfield, S.P. Production of Recombinant Proteins in Microalgae at Pilot Greenhouse Scale. Biotechnol. Bioeng. 2015, 112, 339–345. [Google Scholar] [CrossRef]
- Mathieu-Rivet, E.; Kiefer-Meyer, M.-C.; Vanier, G.; Ovide, C.; Burel, C.; Lerouge, P.; Bardor, M. Protein N-Glycosylation in Eukaryotic Microalgae and Its Impact on the Production of Nuclear Expressed Biopharmaceuticals. Front. Plant Sci. 2014, 5, 359. [Google Scholar] [CrossRef]
- Mathieu-Rivet, E.; Lerouge, P.; Bardor, M. Chlamydomonas Reinhardtii: Protein Glycosylation and Production of Biopharmaceuticals. In Chlamydomonas: Biotechnology and Biomedicine; Hippler, M., Ed.; Microbiology Monographs; Springer International Publishing: Cham, Switzerland, 2017; pp. 45–72. ISBN 978-3-319-66360-9. [Google Scholar]
- Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Mar. Drugs 2022, 20, 657. [Google Scholar] [CrossRef]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as Protein Factories: Expression of a Human Antibody and the Respective Antigen in the Diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424. [Google Scholar] [CrossRef]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From Hybridomas to a Robust Microalgal-Based Production Platform: Molecular Design of a Diatom Secreting Monoclonal Antibodies Directed against the Marburg Virus Nucleoprotein. Microb. Cell Factories 2017, 16, 131. [Google Scholar] [CrossRef]
- Hempel, F.; Maier, U.G. An Engineered Diatom Acting like a Plasma Cell Secreting Human IgG Antibodies with High Efficiency. Microb. Cell Factories 2012, 11, 126. [Google Scholar] [CrossRef]
- Slattery, S.S.; Giguere, D.J.; Stuckless, E.E.; Shrestha, A.; Briere, L.-A.K.; Galbraith, A.; Reaume, S.; Boyko, X.; Say, H.H.; Browne, T.S.; et al. Phosphate-Regulated Expression of the SARS-CoV-2 Receptor-Binding Domain in the Diatom Phaeodactylum tricornutum for Pandemic Diagnostics. Sci. Rep. 2022, 12, 7010. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Serif, M.; Lepetit, B.; Weißert, K.; Kroth, P.G.; Rio Bartulos, C. A Fast and Reliable Strategy to Generate TALEN-Mediated Gene Knockouts in the Diatom Phaeodactylum tricornutum. Algal Res. 2017, 23, 186–195. [Google Scholar] [CrossRef]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 System Adapted for Gene Editing in Marine algae. Sci. Rep. 2016, 6, 24951. [Google Scholar] [CrossRef] [PubMed]
- Stukenberg, D.; Zauner, S.; Dell’Aquila, G.; Maier, U.G. Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2018, 9, 740. [Google Scholar] [CrossRef]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J.; et al. An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef]
- Rastogi, A.; Vieira, F.R.J.; Deton-Cabanillas, A.-F.; Veluchamy, A.; Cantrel, C.; Wang, G.; Vanormelingen, P.; Bowler, C.; Piganeau, G.; Hu, H.; et al. A Genomics Approach Reveals the Global Genetic Polymorphism, Structure, and Functional Diversity of Ten Accessions of the Marine Model Diatom Phaeodactylum tricornutum. ISME J. 2020, 14, 347–363. [Google Scholar] [CrossRef]
- De Martino, A.; Meichenin, A.; Shi, J.; Pan, K.; Bowler, C. Genetic and Phenotypic Characterization of Phaeodactylum tricornutum (Bacillariophyceae) Accessions1. J. Phycol. 2007, 43, 992–1009. [Google Scholar] [CrossRef]
- Zhao, X.; Rastogi, A.; Deton Cabanillas, A.F.; Ait Mohamed, O.; Cantrel, C.; Lombard, B.; Murik, O.; Genovesio, A.; Bowler, C.; Bouyer, D.; et al. Genome Wide Natural Variation of H3K27me3 Selectively Marks Genes Predicted to Be Important for Cell Differentiation in Phaeodactylum tricornutum. New Phytol. 2021, 229, 3208–3220. [Google Scholar] [CrossRef]
- De Martino, A.; Bartual, A.; Willis, A.; Meichenin, A.; Villazán, B.; Maheswari, U.; Bowler, C. Physiological and Molecular Evidence That Environmental Changes Elicit Morphological Interconversion in the Model Diatom Phaeodactylum tricornutum. Protist 2011, 162, 462–481. [Google Scholar] [CrossRef]
- Chaumier, T.; Yang, F.; Manirakiza, E.; Ait-Mohamed, O.; Wu, Y.; Chandola, U.; Jesus, B.; Piganeau, G.; Groisillier, A.; Tirichine, L. Genome-Wide Assessment of Genetic Diversity and Transcript Variations in 17 Accessions of the Model Diatom Phaeodactylum tricornutum. ISME Commun. 2024, 4, ycad008. [Google Scholar] [CrossRef]
- Montsant, A.; Jabbari, K.; Maheswari, U.; Bowler, C. Comparative Genomics of the Pennate Diatom Phaeodactylum tricornutum. Plant Physiol. 2005, 137, 500–513. [Google Scholar] [CrossRef]
- Rastogi, A.; Maheswari, U.; Dorrell, R.G.; Vieira, F.R.J.; Maumus, F.; Kustka, A.; McCarthy, J.; Allen, A.E.; Kersey, P.; Bowler, C.; et al. Integrative Analysis of Large Scale Transcriptome Data Draws a Comprehensive Landscape of Phaeodactylum tricornutum Genome and Evolutionary Origin of Diatoms. Sci. Rep. 2018, 8, 4834. [Google Scholar] [CrossRef]
- Rogato, A.; Richard, H.; Sarazin, A.; Voss, B.; Cheminant Navarro, S.; Champeimont, R.; Navarro, L.; Carbone, A.; Hess, W.R.; Falciatore, A. The Diversity of Small Non-Coding RNAs in the Diatom Phaeodactylum tricornutum. BMC Genom. 2014, 15, 698. [Google Scholar] [CrossRef] [PubMed]
- Hoguin, A.; Rastogi, A.; Bowler, C.; Tirichine, L. Genome-Wide Analysis of Allele-Specific Expression of Genes in the Model Diatom Phaeodactylum tricornutum. Sci. Rep. 2021, 11, 2954. [Google Scholar] [CrossRef] [PubMed]
- Sapriel, G.; Quinet, M.; Heijde, M.; Jourdren, L.; Tanty, V.; Luo, G.; Crom, S.L.; Lopez, P.J. Genome-Wide Transcriptome Analyses of Silicon Metabolism in Phaeodactylum tricornutum Reveal the Multilevel Regulation of Silicic Acid Transporters. PLoS ONE 2009, 4, e7458. [Google Scholar] [CrossRef] [PubMed]
- Cruz de Carvalho, M.H.; Sun, H.-X.; Bowler, C.; Chua, N.-H. Noncoding and Coding Transcriptome Responses of a Marine Diatom to Phosphate Fluctuations. New Phytol. 2016, 210, 497–510. [Google Scholar] [CrossRef] [PubMed]
- König, S.; Eisenhut, M.; Bräutigam, A.; Kurz, S.; Weber, A.P.M.; Büchel, C. The Influence of a Cryptochrome on the Gene Expression Profile in the Diatom Phaeodactylum tricornutum under Blue Light and in Darkness. Plant Cell Physiol. 2017, 58, 1914–1923. [Google Scholar] [CrossRef]
- Li, S.; Ismar, S.M.H. Transcriptome, Biochemical and Growth Responses of the Marine Phytoplankter Phaeodactylum tricornutum Bohlin (Bacillariophyta) to Copepod Grazer Presence. CPB 2018, 46, 1091–1111. [Google Scholar] [CrossRef] [PubMed]
- Ovide, C.; Kiefer-Meyer, M.-C.; Bérard, C.; Vergne, N.; Lecroq, T.; Plasson, C.; Burel, C.; Bernard, S.; Driouich, A.; Lerouge, P.; et al. Comparative in Depth RNA Sequencing of P. Tricornutum’s Morphotypes Reveals Specific Features of the Oval Morphotype. Sci. Rep. 2018, 8, 14340. [Google Scholar] [CrossRef]
- Feng, T.-Y.; Yang, Z.-K.; Zheng, J.-W.; Xie, Y.; Li, D.-W.; Murugan, S.B.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Examination of Metabolic Responses to Phosphorus Limitation via Proteomic Analyses in the Marine Diatom Phaeodactylum tricornutum. Sci. Rep. 2015, 5, 10373. [Google Scholar] [CrossRef]
- Bai, X.; Song, H.; Lavoie, M.; Zhu, K.; Su, Y.; Ye, H.; Chen, S.; Fu, Z.; Qian, H. Proteomic Analyses Bring New Insights into the Effect of a Dark Stress on Lipid Biosynthesis in Phaeodactylum tricornutum. Sci. Rep. 2016, 6, 25494. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome Response of Phaeodactylum tricornutum, during Lipid Accumulation Induced by Nitrogen Depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Poirier, I.; Pallud, M.; Kuhn, L.; Hammann, P.; Demortière, A.; Jamali, A.; Chicher, J.; Caplat, C.; Gallon, R.K.; Bertrand, M. Toxicological Effects of CdSe Nanocrystals on the Marine Diatom Phaeodactylum tricornutum: The First Mass Spectrometry-Based Proteomic Approach. Ecotoxicol. Environ. Saf. 2018, 152, 78–90. [Google Scholar] [CrossRef]
- Chuberre, C.; Chan, P.; Walet-Balieu, M.-L.; Thiébert, F.; Burel, C.; Hardouin, J.; Gügi, B.; Bardor, M. Comparative Proteomic Analysis of Q12 the Diatom Phaeodactylum tricornutum Reveals New Insights Into Intra- and Extra-Cellular Protein Contents of Its Oval, Fusiform, and Triradiate Morphotypes. Front. Plant Sci. 2022, 13, 673113. [Google Scholar] [CrossRef]
- Yang, Z.-K.; Ma, Y.-H.; Zheng, J.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Proteomics to Reveal Metabolic Network Shifts towards Lipid Accumulation Following Nitrogen Deprivation in the Diatom Phaeodactylum tricornutum. J. Appl. Phycol. 2014, 26, 73–82. [Google Scholar] [CrossRef]
- Huang, A.; Li, Y.; Duan, J.; Guo, S.; Cai, X.; Zhang, X.; Long, H.; Ren, W.; Xie, Z. Metabolomic, Proteomic and Lactylated Proteomic Analyses Indicate Lactate Plays Important Roles in Maintaining Energy and C:N Homeostasis in Phaeodactylum tricornutum. Biotechnol. Biofuels Bioprod. 2022, 15, 61. [Google Scholar] [CrossRef]
- Duarte, B.; Feijão, E.; Cruz de Carvalho, R.; Duarte, I.A.; Marques, A.P.; Maia, M.; Hertzog, J.; Matos, A.R.; Cabrita, M.T.; Caçador, I.; et al. Untargeted Metabolomics Reveals Antidepressant Effects in a Marine Photosynthetic Organism: The Diatom Phaeodactylum tricornutum as a Case Study. Biology 2022, 11, 1770. [Google Scholar] [CrossRef]
- Stolfa, G.; Smonskey, M.T.; Boniface, R.; Hachmann, A.-B.; Gulde, P.; Joshi, A.D.; Pierce, A.P.; Jacobia, S.J.; Campbell, A. CHO-Omics Review: The Impact of Current and Emerging Technologies on Chinese Hamster Ovary Based Bioproduction. Biotechnol. J. 2018, 13, 1700227. [Google Scholar] [CrossRef]
- Lewis, A.M.; Abu-Absi, N.R.; Borys, M.C.; Li, Z.J. The Use of ’Omics Technology to Rationally Improve Industrial Mammalian Cell Line Performance. Biotechnol. Bioeng. 2016, 113, 26–38. [Google Scholar] [CrossRef]
- Boulogne, I.; Toustou, C.; Bardor, M. Meta-Analysis of RNA-Seq Datasets Helps to Better Understand P. tricornutum Cellular Biology, a Requirement to Improve the Production of Biologics. Université de Rouen Normandie, Rouen, France, 2024. submitted. [Google Scholar]
- Bohutskyi, P.; Kligerman, D.C.; Byers, N.; Nasr, L.K.; Cua, C.; Chow, S.; Su, C.; Tang, Y.; Betenbaugh, M.J.; Bouwer, E.J. Effects of Inoculum Size, Light Intensity, and Dose of Anaerobic Digestion Centrate on Growth and Productivity of Chlorella and Scenedesmus Microalgae and Their Poly-Culture in Primary and Secondary Wastewater. Algal Res. 2016, 19, 278–290. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Ma, Q.; Yang, J.; Li, X.; Yuan, Y.-J. Phospholipid Metabolism in an Industry Microalga Chlorella Sorokiniana: The Impact of Inoculum Sizes. PLoS ONE 2013, 8, e70827. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, J.; Lu, S.; Lv, Y.; Yuan, Y. Quantitative Proteomic Profiling Reveals Photosynthesis Responsible for Inoculum Size Dependent Variation in Chlorella Sorokiniana. Biotechnol. Bioeng. 2013, 110, 773–784. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and Gene Expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef]
- Veluchamy, A.; Lin, X.; Maumus, F.; Rivarola, M.; Bhavsar, J.; Creasy, T.; O’Brien, K.; Sengamalay, N.A.; Tallon, L.J.; Smith, A.D.; et al. Insights into the Role of DNA Methylation in Diatoms by Genome-Wide Profiling in Phaeodactylum tricornutum. Nat. Commun. 2013, 4, 2091. [Google Scholar] [CrossRef]
- Scarsini, M.; Thurotte, A.; Veidl, B.; Amiard, F.; Niepceron, F.; Badawi, M.; Lagarde, F.; Schoefs, B.; Marchand, J. Metabolite Quantification by Fourier Transform Infrared Spectroscopy in Diatoms: Proof of Concept on Phaeodactylum tricornutum. Front. Plant Sci. 2021, 12, 756421. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes, P.-E. An Overview of Pre-Ribosomal RNA Processing in Eukaryotes. WIREs RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Jüttner, M.; Ferreira-Cerca, S. A Comparative Perspective on Ribosome Biogenesis: Unity and Diversity Across the Tree of Life. In Ribosome Biogenesis: Methods and Protocols; Entian, K.-D., Ed.; Springer: New York, NY, USA, 2022; pp. 3–22. ISBN 978-1-07-162501-9. [Google Scholar]
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical Analysis of the Commercial Potential of Plants for the Production of Recombinant Proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef]
- Ves-Urai, P.; Krobthong, S.; Thongsuk, K.; Roytrakul, S.; Yokthongwattana, C. Comparative Secretome Analysis between Salinity-Tolerant and Control Chlamydomonas Reinhardtii Strains. Planta 2021, 253, 68. [Google Scholar] [CrossRef]
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to Facilitate Transgene Expression in Chlamydomonas Reinhardtii. Planta 2009, 229, 873–883. [Google Scholar] [CrossRef]
- Kiefer, A.M.; Niemeyer, J.; Probst, A.; Erkel, G.; Schroda, M. Production and Secretion of Functional SARS-CoV-2 Spike Protein in Chlamydomonas Reinhardtii. Front. Plant Sci. 2022, 13, 988870. [Google Scholar] [CrossRef]
- Galas, L.; Burel, C.; Schapman, D.; Ropitaux, M.; Bernard, S.; Bénard, M.; Bardor, M. Comparative Structural and Functional Analyses of the Fusiform, Oval, and Triradiate Morphotypes of Phaeodactylum tricornutum Pt3 Strain. Front. Plant Sci. 2021, 12, 638181. [Google Scholar] [CrossRef]
- van Beers, M.M.C.; Bardor, M. Minimizing Immunogenicity of Biopharmaceuticals by Controlling Critical Quality Attributes of Proteins. Biotechnol. J. 2012, 7, 1473–1484. [Google Scholar] [CrossRef]
- Colley, K.J.; Varki, A.; Kinoshita, T. Cellular Organization of Glycosylation. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Vanier, G.; Hempel, F.; Chan, P.; Rodamer, M.; Vaudry, D.; Maier, U.G.; Lerouge, P.; Bardor, M. Biochemical Characterization of Human Anti-Hepatitis B Monoclonal Antibody Produced in the Microalgae Phaeodactylum tricornutum. PLoS ONE 2015, 10, e0139282. [Google Scholar] [CrossRef] [PubMed]
- Baïet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.-C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-Glycans of Phaeodactylum tricornutum Diatom and Functional Characterization of Its N-Acetylglucosaminyltransferase I Enzyme. J. Biol. Chem. 2011, 286, 6152–6164. [Google Scholar] [CrossRef] [PubMed]
- Aebi, M. N-Linked Protein Glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef]
- Lucas, P.-L.; Dumontier, R.; Loutelier-Bourhis, C.; Mareck, A.; Afonso, C.; Lerouge, P.; Mati-Baouche, N.; Bardor, M. User-Friendly Extraction and Multistage Tandem Mass Spectrometry Based Analysis of Lipid-Linked Oligosaccharides in Microalgae. Plant Methods 2018, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Silberstein, S.; Gilmore, R. Biochemistry, Molecular Biology, and Genetics of the Oligosaccharyltransferase. FASEB J. 1996, 10, 849–858. [Google Scholar] [CrossRef]
- Breitling, J.; Aebi, M. N-Linked Protein Glycosylation in the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013359. [Google Scholar] [CrossRef]
- Toustou, C.; Walet-Balieu, M.-L.; Kiefer-Meyer, M.-C.; Houdou, M.; Lerouge, P.; Foulquier, F.; Bardor, M. Towards Understanding the Extensive Diversity of Protein N-Glycan Structures in Eukaryotes. Biol. Rev. 2022, 97, 732–748. [Google Scholar] [CrossRef]
- Stanley, P.; Taniguchi, N.; Aebi, M. N-Glycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Bardor, M.; Faveeuw, C.; Fitchette, A.-C.; Gilbert, D.; Galas, L.; Trottein, F.; Faye, L.; Lerouge, P. Immunoreactivity in Mammals of Two Typical Plant Glyco-Epitopes, Core Alpha(1,3)-Fucose and Core Xylose. Glycobiology 2003, 13, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Altmann, F.; Strasser, R.; Mach, L.; Schähs, M.; Kunert, R.; Rademacher, T.; Glössl, J.; Steinkellner, H. A Plant-Derived Human Monoclonal Antibody Induces an Anti-Carbohydrate Immune Response in Rabbits. Glycobiology 2008, 18, 235–241. [Google Scholar] [CrossRef]
- Zhang, P.; Burel, C.; Plasson, C.; Kiefer-Meyer, M.-C.; Ovide, C.; Gügi, B.; Wan, C.; Teo, G.; Mak, A.; Song, Z.; et al. Characterization of a GDP-Fucose Transporter and a Fucosyltransferase Involved in the Fucosylation of Glycoproteins in the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2019, 10, 610. [Google Scholar] [CrossRef]
- Hebert, D.N.; Molinari, M. In and out of the ER: Protein Folding, Quality Control, Degradation, and Related Human Diseases. Physiol. Rev. 2007, 87, 1377–1408. [Google Scholar] [CrossRef] [PubMed]
- Lamriben, L.; Graham, J.B.; Adams, B.M.; Hebert, D.N. N-Glycan-Based ER Molecular Chaperone and Protein Quality Control System: The Calnexin Binding Cycle. Traffic 2016, 17, 308–326. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like Protein Specialized for Protein Folding and Quality Control in the Endoplasmic Reticulum. Biochim. Biophys. Acta 2012, 1823, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Kyeong, M.; Lee, J.S. Endogenous BiP Reporter System for Simultaneous Identification of ER Stress and Antibody Production in Chinese Hamster Ovary Cells. Metab. Eng. 2022, 72, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Treiber, D.; McCoy, R.E.; Miller, A.K.; Han, M.; He, F.; Domnitz, S.; Heath, C.; Reddy, P. Non-Invasive UPR Monitoring System and Its Applications in CHO Production Cultures. Biotechnol. Bioeng. 2013, 110, 2184–2194. [Google Scholar] [CrossRef] [PubMed]
- Kober, L.; Zehe, C.; Bode, J. Development of a Novel ER Stress Based Selection System for the Isolation of Highly Productive Clones. Biotechnol. Bioeng. 2012, 109, 2599–2611. [Google Scholar] [CrossRef]
- Braakman, I.; Hebert, D.N. Protein Folding in the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013201. [Google Scholar] [CrossRef]
- Fornace, A.J., Jr.; Alamo, I., Jr.; Hollander, M.C.; Lamoreaux, E. Ubiquftin MRNA Is a Major Stress-Induced Transcript in Mammalian Cells. Nucleic Acids Res. 1989, 17, 1215–1230. [Google Scholar] [CrossRef]
- Sheng, X.; Xia, Z.; Yang, H.; Hu, R. The Ubiquitin Codes in Cellular Stress Responses. Protein Cell 2024, 15, 157. [Google Scholar] [CrossRef]
- Li, G.-Q.; Zang, X.-N.; Zhang, X.-C.; Lu, N.; Ding, Y.; Gong, L.; Chen, W.-C. Cloning of Ubiquitin-Activating Enzyme and Ubiquitin-Conjugating Enzyme Genes from Gracilaria lemaneiformis and Their Activity under Heat Shock. Gene 2014, 538, 155–163. [Google Scholar] [CrossRef]
- Li, W.; Schmidt, W. A Lysine-63-Linked Ubiquitin Chain-Forming Conjugase, UBC13, Promotes the Developmental Responses to Iron Deficiency in Arabidopsis Roots. Plant J. 2010, 62, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Sang, X.; Lu, S.; Dong, C.; Zhao, Q.; Chen, H.; Jia, L. Genome-Wide Identification, Phylogenetic and Expression Analyses of the Ubiquitin-Conjugating Enzyme Gene Family in Maize. PLoS ONE 2015, 10, e0143488. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Du, H.; Kazmi, S.S.U.H.; Chen, J.; Chen, W.; Fan, Y.; Liu, Z.; Luo, H.; Fang, H.; Wang, Z.; et al. UBC Gene Family and Their Potential Functions on the Cellular Homeostasis under the Elevated pCO2 Stress in the Diatom Phaeodactylum tricornutum. Ecol. Indic. 2023, 148, 110106. [Google Scholar] [CrossRef]
- Kunert, R.; Reinhart, D. Advances in Recombinant Antibody Manufacturing. Appl. Microbiol. Biotechnol. 2016, 100, 3451–3461. [Google Scholar] [CrossRef]
- Bond, J.S. Proteases: History, Discovery, and Roles in Health and Disease. J. Biol. Chem. 2019, 294, 1643–1651. [Google Scholar] [CrossRef]
- Gilgunn, S.; Bones, J. Challenges to Industrial MAb Bioprocessing—Removal of Host Cell Proteins in CHO Cell Bioprocesses. Curr. Opin. Chem. Eng. 2018, 22, 98–106. [Google Scholar] [CrossRef]
- Yang, B.; Li, W.; Zhao, H.; Wang, A.; Lei, Y.; Xie, Q.; Xiong, S. Discovery and Characterization of CHO Host Cell Protease-Induced Fragmentation of a Recombinant Monoclonal Antibody during Production Process Development. J. Chromatogr. B 2019, 1112, 1–10. [Google Scholar] [CrossRef]
- Jutras, P.V.; Marusic, C.; Lonoce, C.; Deflers, C.; Goulet, M.-C.; Benvenuto, E.; Michaud, D.; Donini, M. An Accessory Protease Inhibitor to Increase the Yield and Quality of a Tumour-Targeting MAb in Nicotiana Benthamiana Leaves. PLoS ONE 2016, 11, e0167086. [Google Scholar] [CrossRef]
- Gao, S.X.; Zhang, Y.; Stansberry-Perkins, K.; Buko, A.; Bai, S.; Nguyen, V.; Brader, M.L. Fragmentation of a Highly Purified Monoclonal Antibody Attributed to Residual CHO Cell Protease Activity. Biotechnol. Bioeng. 2011, 108, 977–982. [Google Scholar] [CrossRef]
- Dorrell, R.G.; Villain, A.; Perez-Lamarque, B.; Audren de Kerdrel, G.; McCallum, G.; Watson, A.K.; Ait-Mohamed, O.; Alberti, A.; Corre, E.; Frischkorn, K.R.; et al. Phylogenomic Fingerprinting of Tempo and Functions of Horizontal Gene Transfer within Ochrophytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2009974118. [Google Scholar] [CrossRef]
- Clincke, M.-F.; Guedon, E.; Yen, F.T.; Ogier, V.; Goergen, J.-L. Characterization of Metalloprotease and Serine Protease Activities in Batch CHO Cell Cultures: Control of Human Recombinant IFN-γ Proteolysis by Addition of Iron Citrate. BMC Proc. 2011, 5, P115. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Yu, B.; Byrne, G.; Wright, M.; O’Rourke, S.; Mesa, K.; Berman, P.W. Identification and CRISPR/Cas9 Inactivation of the C1s Protease Responsible for Proteolysis of Recombinant Proteins Produced in CHO Cells. Biotechnol. Bioeng. 2019, 116, 2130–2145. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, H.; Lütkemeyer, D.; Kuprin, S.; Wrangel, M.; Almstedt, A.; Persson, P.; Ek, V.; Mikaelsson, M. Mapping and Partial Characterization of Proteases Expressed by a CHO Production Cell Line. Biotechnol. Bioeng. 2006, 95, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Laux, H.; Romand, S.; Nuciforo, S.; Farady, C.J.; Tapparel, J.; Buechmann-Moeller, S.; Sommer, B.; Oakeley, E.J.; Bodendorf, U. Degradation of Recombinant Proteins by Chinese Hamster Ovary Host Cell Proteases Is Prevented by Matriptase-1 Knockout. Biotechnol. Bioeng. 2018, 115, 2530–2540. [Google Scholar] [CrossRef] [PubMed]
- Aboulaich, N.; Chung, W.K.; Thompson, J.H.; Larkin, C.; Robbins, D.; Zhu, M. A Novel Approach to Monitor Clearance of Host Cell Proteins Associated with Monoclonal Antibodies. Biotechnol. Prog. 2014, 30, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Goetze, A.M.; Cui, H.; Wylie, J.; Trimble, S.; Hewig, A.; Flynn, G.C. Comprehensive Tracking of Host Cell Proteins during Monoclonal Antibody Purifications Using Mass Spectrometry. mAbs 2014, 6, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Goetze, A.M.; Cui, H.; Wylie, J.; Tillotson, B.; Hewig, A.; Hall, M.P.; Flynn, G.C. Characterization of the Co-Elution of Host Cell Proteins with Monoclonal Antibodies during Protein A Purification. Biotechnol. Prog. 2016, 32, 708–717. [Google Scholar] [CrossRef]
- Ho, S.C.L.; Bardor, M.; Feng, H.; Mariati; Tong, Y.W.; Song, Z.; Yap, M.G.S.; Yang, Y. IRES-Mediated Tricistronic Vectors for Enhancing Generation of High Monoclonal Antibody Expressing CHO Cell Lines. J. Biotechnol. 2012, 157, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.C.L.; Bardor, M.; Li, B.; Lee, J.J.; Song, Z.; Tong, Y.W.; Goh, L.-T.; Yang, Y. Comparison of Internal Ribosome Entry Site (IRES) and Furin-2A (F2A) for Monoclonal Antibody Expression Level and Quality in CHO Cells. PLoS ONE 2013, 8, e63247. [Google Scholar] [CrossRef]
- Bayat, H.; Hoseinzadeh, S.; Pourmaleki, E.; Ahani, R.; Rahimpour, A. Evaluation of Different Vector Design Strategies for the Expression of Recombinant Monoclonal Antibody in CHO Cells. Prep. Biochem. Biotechnol. 2018, 48, 160–164. [Google Scholar] [CrossRef]
- Chng, J.; Wang, T.; Nian, R.; Lau, A.; Hoi, K.M.; Ho, S.C.L.; Gagnon, P.; Bi, X.; Yang, Y. Cleavage Efficient 2A Peptides for High Level Monoclonal Antibody Expression in CHO Cells. mAbs 2015, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.K.; Morse, N.J.; Wagner, J.M.; Tucker, S.K.; Alper, H.S. Design and Evaluation of Synthetic Terminators for Regulating Mammalian Cell Transgene Expression. ACS Synth. Biol. 2019, 8, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Windhagauer, M.; Abbriano, R.M.; Ashworth, J.; Barolo, L.; Jaramillo-Madrid, A.C.; Pernice, M.; Doblin, M.A. Characterisation of Novel Regulatory Sequences Compatible with Modular Assembly in the Diatom Phaeodactylum tricornutum. Algal Res. 2021, 53, 102159. [Google Scholar] [CrossRef]
- Garza, E.A.; Bielinski, V.A.; Espinoza, J.L.; Orlandi, K.; Alfaro, J.R.; Bolt, T.M.; Beeri, K.; Weyman, P.D.; Dupont, C.L. Validating a Promoter Library for Application in Plasmid-Based Diatom Genetic Engineering. ACS Synth. Biol. 2023, 12, 3215–3228. [Google Scholar] [CrossRef]
- Erdene-Ochir, E.; Shin, B.-K.; Kwon, B.; Jung, C.; Pan, C.-H. Identification and Characterisation of the Novel Endogenous Promoter HASP1 and Its Signal Peptide from Phaeodactylum tricornutum. Sci. Rep. 2019, 9, 9941. [Google Scholar] [CrossRef]
- Apt, K.E.; Kroth-Pancic, P.G.; Grossman, A.R. Stable Nuclear Transformation of the Diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 1996, 252, 572–579. [Google Scholar]
- Falciatore, A.; Casotti, R.; Leblanc, C.; Abrescia, C.; Bowler, C. Transformation of Nonselectable Reporter Genes in Marine Diatoms. Mar. Biotechnol. 1999, 1, 239–251. [Google Scholar] [CrossRef]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular Toolbox for Studying Diatom Biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef]
- Erdene-Ochir, E.; Shin, B.-K.; Huda, M.N.; Kim, D.H.; Lee, E.H.; Song, D.-G.; Kim, Y.-M.; Kim, S.M.; Pan, C.-H. Cloning of a Novel Endogenous Promoter for Foreign Gene Expression in Phaeodactylum tricornutum. Appl. Biol. Chem. 2016, 59, 861–867. [Google Scholar] [CrossRef]
- Zou, L.-G.; Chen, J.-W.; Zheng, D.-L.; Balamurugan, S.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. High-Efficiency Promoter-Driven Coordinated Regulation of Multiple Metabolic Nodes Elevates Lipid Accumulation in the Model Microalga Phaeodactylum tricornutum. Microb. Cell Factories 2018, 17, 54. [Google Scholar] [CrossRef]
- Watanabe, Y.; Kadono, T.; Kira, N.; Suzuki, K.; Iwata, O.; Ohnishi, K.; Yamaguchi, H.; Adachi, M. Development of Endogenous Promoters That Drive High-Level Expression of Introduced Genes in the Model Diatom Phaeodactylum tricornutum. Mar. Genom. 2018, 42, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Adler-Agnon (Shemesh), Z.; Leu, S.; Zarka, A.; Boussiba, S.; Khozin-Goldberg, I. Novel Promoters for Constitutive and Inducible Expression of Transgenes in the Diatom Phaeodactylum tricornutum under Varied Nitrate Availability. J. Appl. Phycol. 2018, 30, 2763–2772. [Google Scholar] [CrossRef]
- Zou, L.-G.; Balamurugan, S.; Zhou, T.-B.; Chen, J.-W.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Potentiation of Concurrent Expression of Lipogenic Genes by Novel Strong Promoters in the Oleaginous Microalga Phaeodactylum tricornutum. Biotechnol. Bioeng. 2019, 116, 3006–3015. [Google Scholar] [CrossRef]
- Yoshinaga, R.; Niwa-Kubota, M.; Matsui, H.; Matsuda, Y. Characterization of Iron-Responsive Promoters in the Marine Diatom Phaeodactylum tricornutum. Mar. Genom. 2014, 16, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, Z.; Leu, S.; Khozin-Goldberg, I.; Didi-Cohen, S.; Zarka, A.; Boussiba, S. Inducible Expression of Haematococcus Oil Globule Protein in the Diatom Phaeodactylum tricornutum: Association with Lipid Droplets and Enhancement of TAG Accumulation under Nitrogen Starvation. Algal Res. 2016, 18, 321–331. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Yen, S.-C.; Kuo, P.-C.; Chung, C.-Y.; Yeh, K.-L.; Huang, C.-H.; Chang, J.; Lin, H.-J. Alkaline Phosphatase Promoter as an Efficient Driving Element for Exogenic Recombinant in the Marine Diatom Phaeodactylum tricornutum. Algal Res. 2017, 23, 58–65. [Google Scholar] [CrossRef]
- Lupette, J.; Tardif, M.; Brugière, S.; Couté, Y.; Salvaing, J.; Maréchal, E. Quantitative Proteomic Analyses Reveal the Impact of Nitrogen Starvation on the Proteome of the Model Diatom Phaeodactylum tricornutum. Proteomics 2022, 22, 2200155. [Google Scholar] [CrossRef]
- Abida, H.; Dolch, L.-J.; Meï, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane Glycerolipid Remodeling Triggered by Nitrogen and Phosphorus Starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef]
- The Galaxy Community The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2022 Update. Nucleic Acids Res. 2022, 50, W345–W351. [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toustou, C.; Boulogne, I.; Gonzalez, A.-A.; Bardor, M. Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View. Mar. Drugs 2024, 22, 353. https://doi.org/10.3390/md22080353
Toustou C, Boulogne I, Gonzalez A-A, Bardor M. Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View. Marine Drugs. 2024; 22(8):353. https://doi.org/10.3390/md22080353
Chicago/Turabian StyleToustou, Charlotte, Isabelle Boulogne, Anne-Alicia Gonzalez, and Muriel Bardor. 2024. "Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View" Marine Drugs 22, no. 8: 353. https://doi.org/10.3390/md22080353
APA StyleToustou, C., Boulogne, I., Gonzalez, A.-A., & Bardor, M. (2024). Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View. Marine Drugs, 22(8), 353. https://doi.org/10.3390/md22080353





