Exploring Epigenetic Age in Response to Intensive Relaxing Training: A Pilot Study to Slow Down Biological Age
Abstract
:1. Background
2. Materials and Methods
2.1. Study Subjects
2.2. DNA Extraction
2.3. DNAmAge
2.4. LTL and Telomerase Expression
2.5. Sample Size Estimation
2.6. Statistical Analysis
3. Results
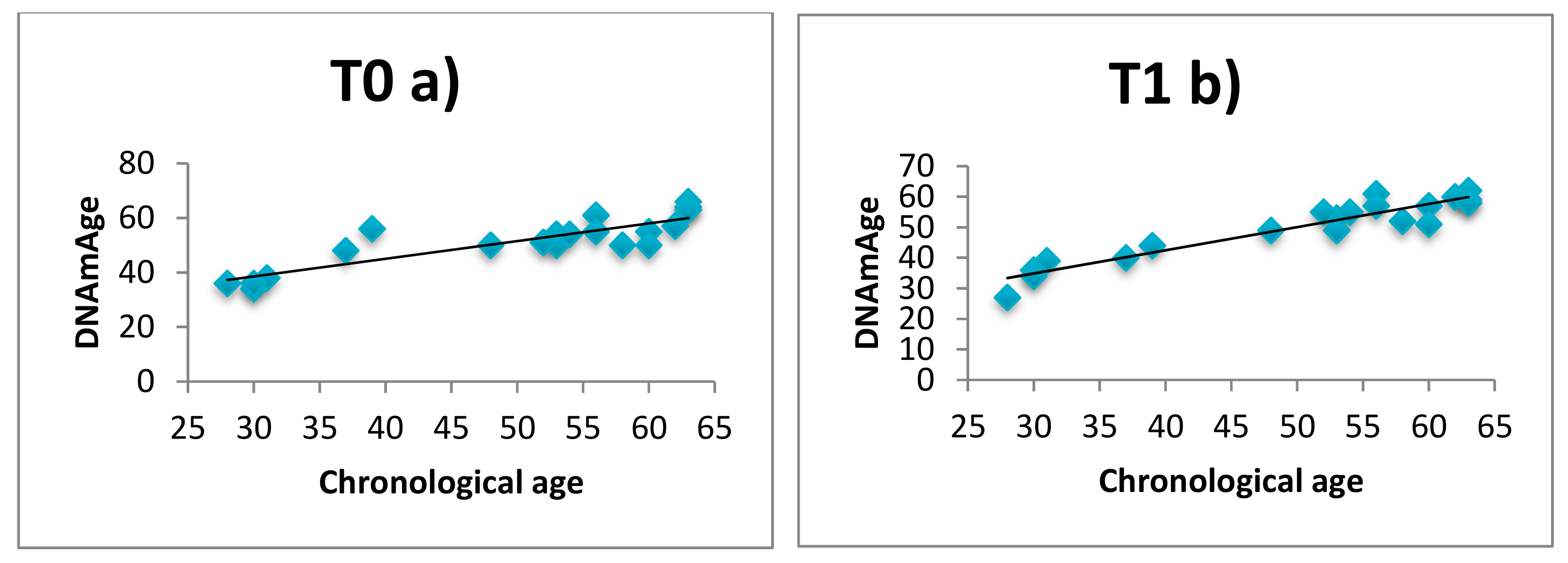
3.1. DNAmAge Correlation with Chronological Age
3.2. DNAmAge after 60 Days of Relaxing Practices
3.3. LTL, Telomerase and Relaxing Practices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Ż.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Goel, N.; Karir, P.; Garg, V.K. Role of DNA methylation in human age prediction. Mech. Ageing Dev. 2017, 166, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic predictor of age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef] [PubMed]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef] [PubMed]
- Perna, L.; Zhang, Y.; Mons, U.; Holleczek, B.; Saum, K.U.; Brenner, H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenet. 2016, 8, 64. [Google Scholar] [CrossRef]
- Horvath, S.; Gurven, M.; Levine, M.E.; Trumble, B.C.; Kaplan, H.; Allayee, H.; Ritz, B.R.; Chen, B.; Lu, A.T.; Rickabaugh, T.M.; et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016, 17, 171. [Google Scholar] [CrossRef]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Chen, B.H.; Colicino, E.; Harris, S.E.; Gibson, J.; Henders, A.K.; Redmond, P.; Cox, S.R.; et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015, 16, 25. [Google Scholar] [CrossRef]
- Fransquet, P.D.; Wrigglesworth, J.; Woods, R.L.; Ernst, M.E.; Ryan, J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin. Epigenet. 2019, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Simons, R.L.; Lei, M.K.; Beach, S.R.; Philibert, R.A.; Cutrona, C.E.; Gibbons, F.X.; Barr, A. Economic hardship and biological weathering: The epigenetics of aging in a U.S. sample of black women. Soc. Sci. Med. 2016, 150, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Quach, A.; Levine, M.E.; Tanaka, T.; Lu, A.T.; Chen, B.H.; Ferrucci, L.; Ritz, B.; Bandinelli, S.; Neuhouser, M.L.; Beasley, J.M.; et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 2017, 9, 419–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwanaji-Enwerem, J.C.; Colicino, E.; Dai, L.; Di, Q.; Just, A.C.; Hou, L.; Vokonas, P.; De Vivo, I.; Lemos, B.; Lu, Q.; et al. miRNA processing gene polymorphisms, blood DNA methylation age and long-term ambient PM2.5 exposure in elderly men. Epigenomics 2017, 9, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Ward-Caviness, C.K.; Nwanaji-Enwerem, J.C.; Wolf, K.; Wahl, S.; Colicino, E.; Trevisi, L.; Kloog, I.; Just, A.C.; Vokonas, P.; Cyrys, J.; et al. Long-term exposure to air pollution is associated with biological aging. Oncotarget 2016, 7, 74510–74525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorito, G.; McCrory, C.; Robinson, O.; Carmeli, C.; Rosales, C.O.; Zhang, Y.; Colicino, E.; Dugué, P.A.; Artaud, F.; McKay, G.J.; et al. Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: A multi-cohort analysis. Aging 2019, 11, 2045–2070. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Arloth, J.; Carrillo-Roa, T.; Iurato, S.; Röh, S.; Ressler, K.J.; Nemeroff, C.B.; Smith, A.K.; Bradley, B.; Heim, C.; et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: Relevance of glucocorticoid signaling. Genome Biol. 2015, 16, 266. [Google Scholar] [CrossRef] [PubMed]
- Gassen, N.C.; Chrousos, G.P.; Binder, E.B.; Zannas, A.S. Life stress, glucocorticoid signaling, and the aging epigenome: Implications for aging-related diseases. Neurosci. Biobehav. Rev. 2017, 74, 356–365. [Google Scholar] [CrossRef]
- Spólnicka, M.; Pośpiech, E.; Adamczyk, J.G.; Freire-Aradas, A.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Lareu, M.V.; Phillips, C.; et al. Modified aging of elite athletes revealed by analysis of epigenetic age markers. Aging 2018, 10, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Chaix, R.; Alvarez-López, M.J.; Fagny, M.; Lemee, L.; Regnault, B.; Davidson, R.J.; Lutz, A.; Kaliman, P. Epigenetic clock analysis in long-term meditators. Psychoneuroendocrinology 2017, 85, 210–214. [Google Scholar] [CrossRef] [Green Version]
- Levine, G.N.; Lange, R.A.; Bairey-Merz, C.N.; Davidson, R.J.; Jamerson, K.; Mehta, P.K.; Michos, E.D.; Norris, K.; Ray, I.B.; Saban, K.L.; et al. Meditation and cardiovascular risk reduction: A scientific statement from the American Heart association. J. Am. Heart Assoc. 2017, 6, e002218. [Google Scholar] [CrossRef] [PubMed]
- Lazar, S.W.; Bush, G.; Gollub, R.L.; Fricchione, G.L.; Khalsa, G.; Benson, H. Functional brain mapping of the relaxation response and meditation. Neuroreport 2000, 11, 1581–1585. [Google Scholar] [CrossRef] [PubMed]
- Paul-Labrador, M.; Polk, D.; Dwyer, J.H.; Velasquez, I.; Nidich, S.; Rainforth, M.; Schneider, R.; Merz, C.N. Effects of a randomized controlled trial of transcendental meditation on components of the metabolic syndrome in subjects with coronary heart disease. Arch. Intern. Med. 2006, 166, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Kaliman, P.; Alvarez-López, M.J.; Cosín-Tomás, M.; Rosenkranz, M.A.; Lutz, A.; Davidson, R.J. Rapid changes in histone deacetylases and inflammatory gene expression in expert meditators. Psychoneuroendocrinology 2014, 40, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Black, D.S.; Slavich, G.M. Mindfulness meditation and the immune system: A systematic review of randomized controlled trials. Ann. N. Y. Acad. Sci. 2016, 1373, 13–24. [Google Scholar] [CrossRef]
- Epel, E.S.; Lithgow, G.J. Stress biology and aging mechanisms: Toward understanding the deep connection between adaptation to stress and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. S1), S10–S16. [Google Scholar] [CrossRef]
- Jacobs, T.L.; Epel, E.S.; Lin, J.; Blackburn, E.H.; Wolkowitz, O.M.; Bridwell, D.A.; Zanesco, A.P.; Aichele, S.R.; Sahdra, B.K.; MacLean, K.A.; et al. Intensive meditation training, immune cell telomerase activity, and psychological mediators. Psychoneuroendocrinology 2011, 36, 664–681. [Google Scholar] [CrossRef]
- Schutte, N.S.; Malouff, J.M. A meta-analytic review of the effects of mindfulness meditation on telomerase activity. Psychoneuroendocrinology 2014, 42, 45–48. [Google Scholar] [CrossRef]
- Thimmapuram, J.; Pargament, R.; Sibliss, K.; Grim, R.; Risques, R.; Toorens, E. Effect of heartfulness meditation on burnout, emotional wellness, and telomere length in health care professionals. J. Community Hosp. Intern. Med. Perspect. 2017, 7, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Conklin, Q.A.; King, B.G.; Zanesco, A.P.; Lin, J.; Hamidi, A.B.; Pokorny, J.J.; Álvarez-López, M.J.; Cosín-Tomás, M.; Huang, C.; Kaliman, P.; et al. Insight meditation and telomere biology: The effects of intensive retreat and the moderating role of personality. Brain Behav. Immun. 2018, 70, 233–245. [Google Scholar] [CrossRef]
- Carlson, L.E.; Beattie, T.L.; Giese-Davis, J.; Faris, P.; Tamagawa, R.; Fick, L.J.; Degelman, E.S.; Speca, M. Mindfulness-based cancer recovery and supportive-expressive therapy maintain telomere length relative to controls in distressed breast cancer survivors. Cancer 2015, 121, 476–484. [Google Scholar] [CrossRef]
- Lengacher, C.A.; Reich, R.R.; Kip, K.E.; Barta, M.; Ramesar, S.; Paterson, C.L.; Moscoso, M.S.; Carranza, I.; Budhrani, P.H.; Kim, S.J.; et al. Influence of mindfulness-based stress reduction (MBSR) on telomerase activity in women with breast cancer (BC). Biol. Res. Nurs. 2014, 16, 438–447. [Google Scholar] [CrossRef]
- Wang, X.; Sundquist, K.; Hedelius, A.; Palmér, K.; Memon, A.A.; Sundquist, J. Leukocyte telomere length and depression, anxiety and stress and adjustment disorders in primary health care patients. BMC Psychiatr. 2017, 17, 148. [Google Scholar] [CrossRef]
- Dal Lin, C.; Marinova, M.; Rubino, G.; Gola, E.; Brocca, A.; Pantano, G.; Brugnolo, L.; Sarais, C.; Cucchini, U.; Volpe, B.; et al. Thoughts modulate the expression of inflammatory genes and may improve the coronary blood flow in patients after a myocardial infarction. J. Tradit. Complement. Med. 2017, 8, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Pavanello, S.; Stendardo, M.; Mastrangelo, G.; Bonci, M.; Bottazzi, B.; Campisi, M.; Nardini, M.; Leone, R.; Mantovani, A.; Boschetto, P. Inflammatory long pentraxin 3 is associated with leukocyte telomere length in night-shift workers. Front. Immunol. 2017, 8, 516. [Google Scholar] [CrossRef]
- Pavanello, S.; Angelici, L.; Hoxha, M.; Cantone, L.; Campisi, M.; Tirelli, A.S.; Vigna, L.; Pesatori, A.C.; Bollati, V. Sterol 27-hydroxylase polymorphism significantly associates with shorter telomere, higher cardiovascular and type-2 diabetes risk in obese subjects. Front. Endocrinol. 2018, 9, 309. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Klengel, T.; Mehta, D.; Anacker, C.; Rex-Haffner, M.; Pruessner, J.C.; Pariante, C.M.; Pace, T.W.; Mercer, K.B.; Mayberg, H.S.; Bradley, B.; et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013, 16, 33–41. [Google Scholar] [CrossRef]
- Bamberger, C.M.; Schulte, H.M.; Chrousos, G.P. Molecular determinants of glucocorticoid receptor function and tissue sensitivity to glucocorticoids. Endocr. Rev. 1996, 17, 245–261. [Google Scholar] [CrossRef]
- Thomassin, H.; Flavin, M.; Espinás, M.L.; Grange, T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001, 20, 1974–1983. [Google Scholar] [CrossRef]
- Zannas, A.S.; Chrousos, G.P. Epigenetic programming by stress and glucocorticoids along the human lifespan. Mol. Psychiatr. 2017, 22, 640–646. [Google Scholar] [CrossRef]
- Swamynathan, S.K. Krüppel-like factors: Three fingers in control. Hum. Genomics 2010, 4, 263–270. [Google Scholar] [CrossRef]
- Iwaya, C.; Kitajima, H.; Yamamoto, K.; Maeda, Y.; Sonoda, N.; Shibata, H.; Inoguchi, T. DNA methylation of the Klf14 gene region in whole blood cells provides prediction for the chronic inflammation in the adipose tissue. Biochem. Biophys. Res. Commun. 2018, 497, 908–915. [Google Scholar] [CrossRef]
- Small, K.S.; Hedman, A.K.; Grundberg, E.; Nica, A.C.; Thorleifsson, G.; Kong, A.; Thorsteindottir, U.; Shin, S.Y.; Richards, H.B.; Soranzo, N.; et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011, 43, 561–564. [Google Scholar] [CrossRef]
- Berdasco, M.; Esteller, M. Hot topics in epigenetic mechanisms of aging: 2011. Aging Cell 2012, 11, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Steegenga, W.T.; Boekschoten, M.V.; Lute, C.; Hooiveld, G.J.; De Groot, P.J.; Morris, T.J.; Teschendorff, A.E.; Butcher, L.M.; Beck, S.; Müller, M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age 2014, 36, 9648. [Google Scholar] [CrossRef]
- Epel, E.S.; Puterman, E.; Lin, J.; Blackburn, E.H.; Lum, P.Y.; Beckmann, N.D.; Zhu, J.; Lee, E.; Gilbert, A.; Rissman, R.A.; et al. Meditation and vacation effects have an impact on disease-associated molecular phenotypes. Transl. Psychiatr. 2016, 6, e880. [Google Scholar] [CrossRef]
- Tolahunase, M.; Sagar, R.; Dada, R. Impact of yoga and meditation on cellular aging in apparently healthy individuals: A prospective, open-label single-arm exploratory study. Oxid. Med. Cell Longev. 2017, 2017, 7928981. [Google Scholar] [CrossRef]
- Vyas, C.M.; Hazra, A.; Chang, S.C.; Qiu, W.; Reynolds, C.F., 3rd; Mischoulon, D.; Chang, G.; Manson, J.E.; De Vivo, I.; Okereke, O.I. Pilot study of DNA methylation, molecular aging markers and measures of health and well-being in aging. Transl. Psychiatr. 2019, 9, 118. [Google Scholar] [CrossRef]
- Marioni, R.E.; Harris, S.E.; Shah, S.; McRae, A.F.; Von Zglinicki, T.; Martin-Ruiz, C.; Wray, N.R.; Visscher, P.M.; Deary, I.J. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int. J. Epidemiol. 2018, 47, 356. [Google Scholar] [CrossRef]
- Belsky, D.W.; Moffitt, T.E.; Cohen, A.A.; Corcoran, D.L.; Levine, M.E.; Prinz, J.A.; Schaefer, J.; Sugden, K.; Williams, B.; Poulton, R.; et al. Eleven telomere, epigenetic clock, and biomarker-composite quantifications of Biological Aging: Do They Measure the Same Thing? Am. J. Epidemiol. 2018, 187, 1220–1230. [Google Scholar] [CrossRef]
- Lin, J.; Smith, D.L.; Esteves, K.; Drury, S. Telomere length measurement by qPCR—Summary of critical factors and recommendations for assay design. Psychoneuroendocrinology 2019, 99, 271–278. [Google Scholar] [CrossRef]
- Bischoff, C.; Petersen, H.C.; Graakjaer, J.; Andersen-Ranberg, K.; Vaupel, J.W.; Bohr, V.A.; Kølvraa, S.; Christensen, K. No association between telomere length and survival among the elderly and oldest old. Epidemiology 2006, 17, 190–194. [Google Scholar] [CrossRef]
- Cassidy, A.; De Vivo, I.; Liu, Y.; Han, J.; Prescott, J.; Hunter, D.J.; Rimm, E.B. Associations between diet, lifestyle factors, and telomere length in women. Am. J. Clin. Nutr. 2010, 91, 1273–1280. [Google Scholar] [CrossRef] [Green Version]
- Astuti, Y.; Wardhana, A.; Watkins, J.; Wulaningsih, W.; PILAR Research Network. Cigarette smoking and telomere length: A systematic review of 84 studies and meta-analysis. Environ. Res. 2017, 158, 480–489. [Google Scholar] [CrossRef]



| T0 | T1 | p § | ||
|---|---|---|---|---|
| DNAmAge | ΔT1 − T0 DNAmAge | |||
| Mean (SD) | ||||
| All subjects | 51.4 (9.37) | 49.9 (10.0) | 1.50 (4.36) | 0.143 |
| Patients | 55.7 (5.66) | 55.6 (4.29) | −0.14 (2.88) | 0.428 |
| Healthy subjects | 41.3 (8.73) | 36.7 (5.85) | −4.67 (5.78) | 0.053 |
| b | r | t | p | |
|---|---|---|---|---|
| Healthy subjects | 14.836 | 0.631 | 3.260 | 0.005 |
| Chronological age | −0.400 | −0.507 | 2.350 | 0.032 |
| Gender | −3.497 | −0.443 | 1.977 | 0.075 |
| T0 | T1 | p§ | |
|---|---|---|---|
| ELOVL2 % Met Mean (SD) | |||
| All subjects | 61.2 (4.46) | 61.9 (6.02) | 0.253 |
| Patients | 63.0 (2.66) | 64.6 (3.86) | 0.071 |
| Healthy subjects | 56.8 (5.04) | 55.7 (5.61) | 0.135 |
| C1orf132 % Met mean (SD) | |||
| All subjects | 49.7 (10.9) | 40.9 (8.52) | 0.554 |
| Patients | 45.4 (9.35) | 46.9 (6.55) | 0.517 |
| Healthy subjects | 59.8 (6.88) | 60.2 (3.97) | 0.935 |
| TRIM59 % Met mean (SD) | |||
| All subjects | 50.6 (6.57) | 51.9 (7.67) | 0.204 |
| Patients | 53.9 (4.26) | 56.1 (3.61) | 0.131 |
| Healthy subjects | 43.2 (4.62) | 42.2 (5.04) | 0.110 |
| KLF14 % Met mean (SD) | |||
| All subjects | 13.3 (3.21) | 11.5 (1.99) | 0.037 |
| Patients | 13.4 (2.95) | 12.5 (1.45) | 0.260 |
| Healthy subjects | 12.8 (4.02) | 9.2 (0.41) | 0.087 |
| FHL2 % Met mean (SD) | |||
| All subjects | 45.0 (8.09) | 45.7 (6.88) | 0.609 |
| Patients | 47.7 (8.06) | 48.6 (6.06) | 0.671 |
| Healthy subjects | 38.5 (2.95) | 39.0 (2.76) | 0.774 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavanello, S.; Campisi, M.; Tona, F.; Dal Lin, C.; Iliceto, S. Exploring Epigenetic Age in Response to Intensive Relaxing Training: A Pilot Study to Slow Down Biological Age. Int. J. Environ. Res. Public Health 2019, 16, 3074. https://doi.org/10.3390/ijerph16173074
Pavanello S, Campisi M, Tona F, Dal Lin C, Iliceto S. Exploring Epigenetic Age in Response to Intensive Relaxing Training: A Pilot Study to Slow Down Biological Age. International Journal of Environmental Research and Public Health. 2019; 16(17):3074. https://doi.org/10.3390/ijerph16173074
Chicago/Turabian StylePavanello, Sofia, Manuela Campisi, Francesco Tona, Carlo Dal Lin, and Sabino Iliceto. 2019. "Exploring Epigenetic Age in Response to Intensive Relaxing Training: A Pilot Study to Slow Down Biological Age" International Journal of Environmental Research and Public Health 16, no. 17: 3074. https://doi.org/10.3390/ijerph16173074
APA StylePavanello, S., Campisi, M., Tona, F., Dal Lin, C., & Iliceto, S. (2019). Exploring Epigenetic Age in Response to Intensive Relaxing Training: A Pilot Study to Slow Down Biological Age. International Journal of Environmental Research and Public Health, 16(17), 3074. https://doi.org/10.3390/ijerph16173074






