Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole
Abstract
:1. Introduction
Highlights
- The evolution of antibiotic resistance was studied in an anoxic-aerobic system.
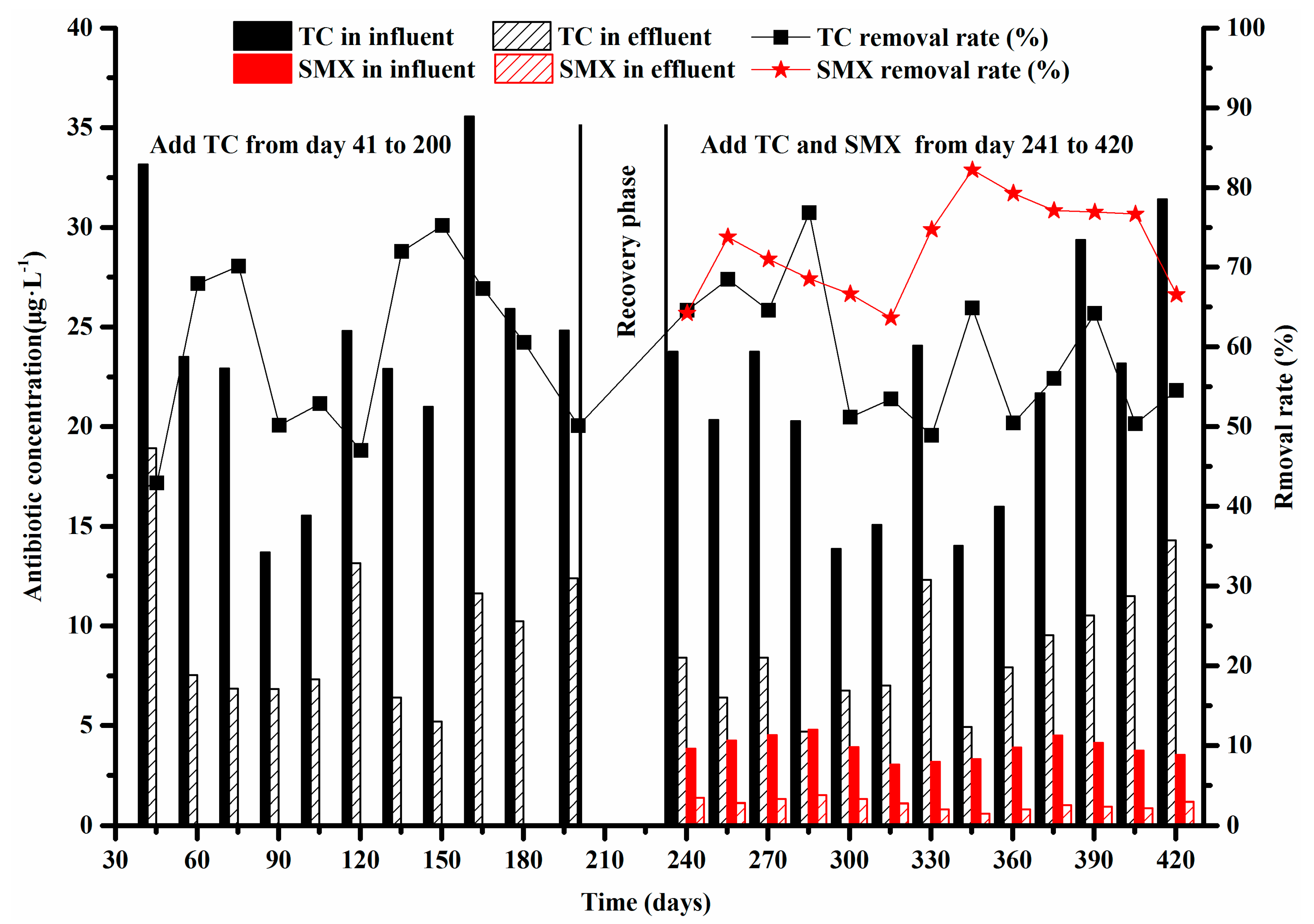
- The average removal rates of TC and SMX were about 59% and 72%, respectively.
- The ARGs abundance for TC and SMX increased significantly with antibiotic exposure.
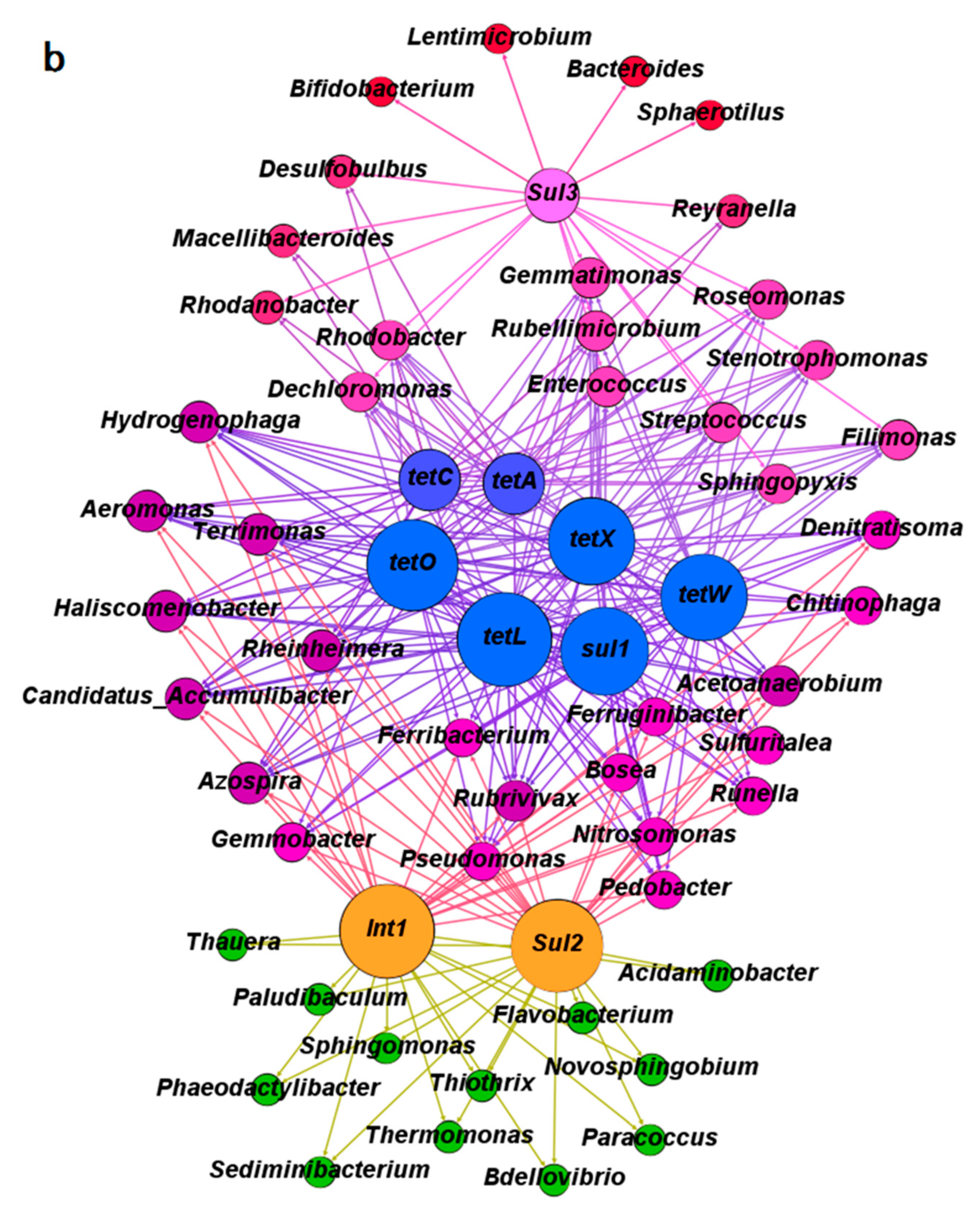
- The ARGs were possibly related to 20 main bacterial genera under antibiotic pressure.
- Gene intI1 played important roles in horizontal transfer of various ARGs except sul3.
2. Materials and Methods
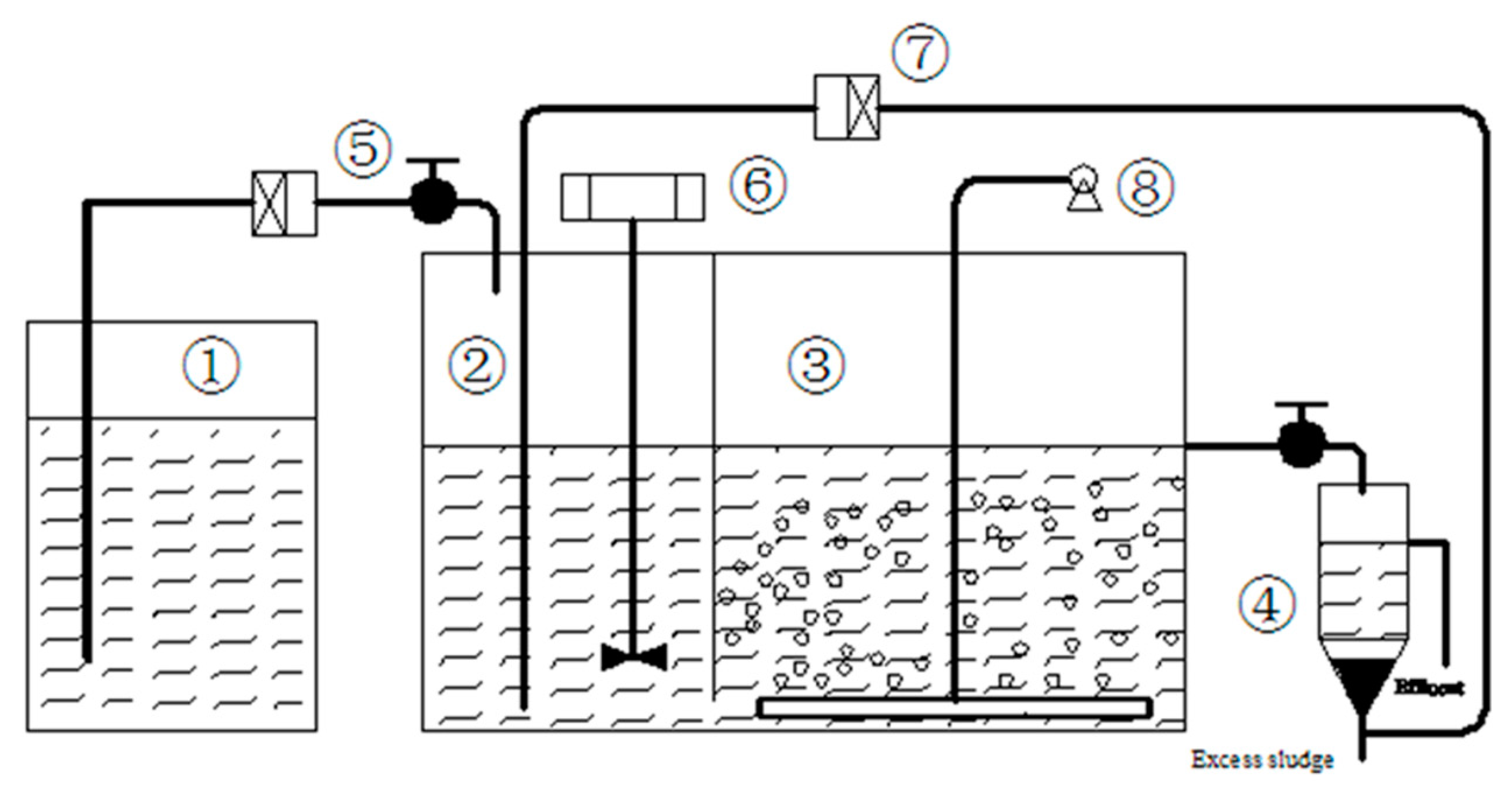
2.1. Design of the Experimental Approach
2.2. Total DNA Extraction and Bacterial Compositions Analysis
2.3. Concentration Determination of Tetracycline and Sulfamethoxazole
2.4. Quantification of Antibiotic Resistance Genes
2.5. Statistical Analysis
3. Results and Discussion
3.1. Efficiency of Antibiotics Removal in the Anoxic-Aerobic Wastewater Treatment System
3.2. Evolution of Antibiotic Resistance Genes under Long-Term Exposure to Antibiotics in the Anoxic-Aerobic Wastewater Treatment System
3.3. The Co-Occurrence of Microbial Communities and Their Corresponding Antibiotic Resistance Genes under the Pressure of Tetracycline and Sulfamethoxazole in the Anoxic-Aerobic Wastewater Treatment System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TC | tetracycline |
| SMX | sulfamethoxazole |
| ARG | antibiotic resistance gene |
| A-O | anoxic-aerobic |
| qPCR | quantitative polymerase chain reaction |
| HPLC | high performance liquid chromatography |
References
- Kor-Bicakci, G.; Pala-Ozkok, I.; Ural, A.; Jonas, D.; Orhon, D.; Ubay-Cokgor, E. Is the chronic impact of sulfamethoxazole different for slow growing culture? The effect of culture history. Bioresour. Technol. 2016, 206, 65–76. [Google Scholar] [CrossRef]
- Lu, Z.; Na, G.; Gao, H.; Wang, L.; Bao, C.; Yao, Z. Fate of sulfonamide resistance genes in estuary environment and effect of anthropogenic activities. Sci. Total Environ. 2015, 527–528, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Geng, J.; Ma, H.; Ren, H.; Xu, K.; Ding, L. Characterization of microbial community and antibiotic resistance genes in activated sludge under tetracycline and sulfamethoxazole selection pressure. Sci. Total Environ. 2016, 571, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Xu, C.; Huang, Y.; Nie, H.; Wang, J. Tetracyclines, sulfonamides and quinolones and their corresponding resistance genes in the Three Gorges Reservoir, China. Sci. Total Environ. 2018, 631–632, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Yang, Q.; Sun, L.; Yang, X.; Zhou, M.; Deng, R.; Bi, L. Plant growth, antibiotic uptake, and prevalence of antibiotic resistance in an endophytic system of pakchoi under antibiotic exposure. Int. J. Environ. Res. Public Health 2017, 14, 1336. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, J.; Yuan, X.; Chen, Y.; Yang, H.; Fei, X. Distribution characteristics and ecological risk assessment of tetracyclines pollution in the Weihe river, China. Int. J. Environ. Res. Public Health 2018, 15, 1803. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.L.; Andrei, A.S.; Rudi, K.; Balcázar, J.L.; Dragos, N.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 2017, 225, 304–315. [Google Scholar] [CrossRef]
- He, X.; Xu, Y.; Chen, J.; Ling, J.; Li, Y.; Huang, L.; Zhou, X.; Zheng, L.; Xie, G. Evolution of corresponding resistance genes in the water of fish tanks with multiple stresses of antibiotics and heavy metals. Water Res. 2017, 124, 39–48. [Google Scholar] [CrossRef]
- Berendonk, T.; Manaia, C.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 2355–2364. [Google Scholar] [CrossRef]
- Huang, K.; Xia, H.; Wu, Y.; Chen, J.; Cui, G.; Li, F.; Chen, Y.; Wu, N. Effects of earthworms on the fate of tetracycline and fluoroquinolone resistance genes of sewage sludge during vermicomposting. Bioresour. Technol. 2018, 259, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Song, H.; Yang, X.; Zhang, S.; Yang, Y.; Zhang, L.; Xu, H.; Wang, Y. A continuous flow MFC-CW coupled with a biofilm electrode reactor to simultaneously attenuate sulfamethoxazole and its corresponding resistance genes. Sci. Total Environ. 2018, 637–638, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Zhou, S.; Jiang, X.; Ma, X.; Liu, C. Robust performance of a membrane bioreactor for removing antibiotic resistance genes exposed to antibiotics: Role of membrane foulants. Water Res. 2018, 130, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Shan, J.; Yang, P.; Shang, X.; Xia, Y.; Yan, X. Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 2018, 240, 368–377. [Google Scholar] [CrossRef]
- Aydin, S.; Ince, B.; Ince, O. Development of antibiotic resistance genes in microbial communities during long-term operation of anaerobic reactors in the treatment of pharmaceutical wastewater. Water Res. 2015, 83, 337–344. [Google Scholar] [CrossRef]
- Bai, Y.; Xu, R.; Wang, Q.; Zhang, Y.; Yang, Z. Sludge anaerobic digestion with high concentrations of tetracyclines and sulfonamides: Dynamics of microbial communities and change of antibiotic resistance genes. Bioresour. Technol. 2019, 276, 51–59. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Sun, H.; Zhao, L.; Liu, Y. Effect of tetracycline on microbial community structure associated with enhanced biological N&P removal in sequencing batch reactor. Bioresour. Technol. 2018, 256, 414–420. [Google Scholar] [CrossRef]
- Zhao, R.; Feng, J.; Liu, J.; Fu, W.; Li, X.; Li, B. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar] [CrossRef]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.H.; Xu, H. Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef]
- Johnsen, P.; Townsend, J.; Bøhn, T.; Simonsen, G.; Sundsfjord, A.; Nielsen, K. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect. Dis. 2009, 9, 357–364. [Google Scholar] [CrossRef]
- Zhang, Q.; Tian, G.; Jin, R. The occurrence, maintenance, and proliferation of antibiotic resistance genes (ARGs) in the environment: Influencing factors, mechanisms, and elimination strategies. Appl. Microbiol. Biotechnol. 2018, 102, 8261–8274. [Google Scholar] [CrossRef] [PubMed]
- Jutkina, J.; Marathe, N.P.; Flach, C.F.; Larsson, D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Cetecioglu, Z.; Ince, B.; Gros, M.; Rodriguez-Mozaz, S.; Barceló, D.; Orhon, D.; Ince, O. Chronic impact of tetracycline on the biodegradation of an organic substrate mixture under anaerobic conditions. Water Res. 2013, 47, 2959–2969. [Google Scholar] [CrossRef] [PubMed]
- Collado, N.; Buttiglieri, G.; Marti, E.; Ferrando-Climent, L.; Rodriguez-Mozaz, S.; Barceló, D.; Comas, J.; Rodriguez-Roda, I. Effects on activated sludge bacterial community exposed to sulfamethoxazole. Chemosphere 2013, 93, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Wang, R.; Yang, Q.; Hu, H.; Li, X.; Duan, X. Impact of tetracycline on the performance and abundance of functional bacteria of a lab-scale anaerobic-aerobic wastewater treatment system. Biochem. Eng. J. 2018, 138, 98–105. [Google Scholar] [CrossRef]
- Kong, Q.; He, X.; Feng, Y.; Miao, M.; Wang, Q.; Du, Y.; Xu, F. Pollutant removal and microorganism evolution of activated sludge under ofloxacin selection pressure. Bioresour. Technol. 2017, 241, 849–856. [Google Scholar] [CrossRef]
- Yu, N.; Zhao, C.; Ma, B.; Li, S.; She, Z.; Guo, L.; Zhang, Q.; Zhao, Y.; Jin, C.; Gao, M. Impact of ampicillin on the nitrogen removal, microbial community and enzymatic activity of activated sludge. Bioresour. Technol. 2019, 272, 337–345. [Google Scholar] [CrossRef]
- Pham, V.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef]
- Vartoukian, S.; Palmer, R.; Wade, W. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Zhang, Q.; Ying, G.; Pan, C.; Liu, Y.; Zhao, J. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef]
- Du, B.; Yang, Q.; Li, X.; Yuan, W.; Chen, Y.; Wang, R. Impacts of long-term exposure to tetracycline and sulfamethoxazole on the sludge granules in an anoxic-aerobic wastewater treatment system. Sci. Total Environ. 2019, 684, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Ince, B.; Cetecioglu, Z.; Arikan, O.; Ozbayram, E.; Shahi, A.; Ince, O. Combined effect of erythromycin, tetracycline and sulfamethoxazole on performance of anaerobic sequencing batch reactors. Bioresour. Technol. 2015, 186, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Bui, X.; Luu, V.; Nguyen, P.; Guo, W.; Ngo, H. Removal of antibiotics in sponge membrane bioreactors treating hospital wastewater: Comparison between hollow fiber and flat sheet membrane systems. Bioresour. Technol. 2017, 240, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Wang, Y.; Liu, W.; Wang, Y.; Jiang, H. Enhanced adsorption performance of tetracycline in aqueous solutions by methanol-modified biochar. Chem. Eng. J. 2014, 248, 168–174. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Yang, Q. Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 2017, 621, 990–999. [Google Scholar] [CrossRef]
- Khan, M.; Bae, H.; Jung, J. Tetracycline degradation by ozonation in the aqueous phase: Proposed degradation intermediates and pathway. J. Hazard. Mater. 2010, 181, 659–665. [Google Scholar] [CrossRef]
- Yang, C.; Hsiao, W.; Chang, B. Biodegradation of sulfonamide antibiotics in sludge. Chemosphere 2016, 150, 559–565. [Google Scholar] [CrossRef]
- Yu, S.; Wang, Y.; Sun, F.; Rong, W.; Yan, Z. Novel mpg-C3N4 /TiO2 nanocomposite photocatalytic membrane reactor for sulfamethoxazole photodegradation. Chem. Eng. J. 2017, 337, 183–192. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, J.; Wang, H.; Chen, X.; Ren, S.; Li, X.; Xu, Y.; Zhang, H.; Li, X. Evolution of the microbial community in a full-scale printing and dyeing wastewater treatment system. Bioresour. Technol. 2012, 117, 155–163. [Google Scholar] [CrossRef]
- Pei, R.; Kim, S.; Carlson, K.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Li, B.; Zhang, T. Biodegradation and adsorption of antibiotics in the activated sludge process. Environ. Sci. Technol. 2011, 44, 3468–3473. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lin, C.; Yuchen Lin, A.; Andy Hong, P. Sorption and biodegradation of sulfonamide antibiotics by activated sludge: Experimental assessment using batch data obtained under aerobic conditions. Water Res. 2011, 45, 3389–3397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Jiang, X.; Zhou, S.; Wu, M.; Pan, M.; Chen, H. Microbial community compositional analysis for membrane bioreactor treating antibiotics containing wastewater. Chem. Eng. J. 2017, 325, 300–309. [Google Scholar] [CrossRef]
- Zhang, S.; Song, H.; Yang, X.; Yang, K.; Wang, X. Effect of electrical stimulation on the fate of sulfamethoxazole and tetracycline with their corresponding resistance genes in three-dimensional biofilm-electrode reactors. Chemosphere 2016, 164, 113–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harnisz, M.; Korzeniewska, E.; Ciesielski, S.; Gołaś, I. tet genes as indicators of changes in the water environment: Relationships between culture-dependent and culture-independent approaches. Sci. Total Environ. 2015, 505, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, X.; Yang, Q.; Chen, Y.; Du, B. Evolution of microbial community and drug resistance during enrichment of tetracycline-degrading bacteria. Ecotoxicol. Environ. Saf. 2019, 171, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Wang, Y.; Tong, T.; Wang, S. The fate of antibiotic resistance genes and their potential hosts during bio-electrochemical treatment of high-salinity pharmaceutical wastewater. Water Res. 2018, 133, 79–86. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, M.; Qi, F.; Sun, P.; Van Ginkel, S. Effect of trace tetracycline concentrations on the structure of a microbial community and the development of tetracycline resistance genes in sequencing batch reactors. Bioresour. Technol. 2013, 150, 9–14. [Google Scholar] [CrossRef]
- Leski, T.; Bangura, U.; Jimmy, D.; Ansumana, R.; Lizewski, S.; Stenger, D.; Taitt, C.; Vora, G. Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int. J. Antimicrob. Agents 2013, 42, 83–86. [Google Scholar] [CrossRef]
- Huang, L.; Xu, Y.; Xu, J.; Ling, J.; Chen, J.; Zhou, J.; Zheng, L.; Du, Q. Antibiotic resistance genes (ARGs) in duck and fish production ponds with integrated or non-integrated mode. Chemosphere 2017, 168, 1107–1114. [Google Scholar] [CrossRef]
- Wu, L.; Ning, D.; Zhang, B.; Li, Y.; Zhang, P.; Shan, X.; Zhang, Q.; Brown, M.; Li, Z.; Van Nostrand, J.; et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019, 4, 1183–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Italiano, F.; Agostiano, A.; Belviso, B.D.; Caliandro, R.; Carrozzini, B.; Comparelli, R.; Melillo, M.T.; Mesto, E.; Tempesta, G. Interaction between the photosynthetic anoxygenic microorganism Rhodobacter sphaeroides and soluble gold compounds. From toxicity to gold nanoparticle synthesis. Colloid Surf. B 2018, 172, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Feng, K.; Li, S.; Zhang, Y.; Chen, H.; Yin, H.; Xu, M.; Deng, Y. Exploring abundance, diversity and variation of a widespread antibiotic resistance gene in wastewater treatment plants. Environ. Int. 2018, 117, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Asvapathanagul, P.; Olson, B.; Gedalanga, P.; Hashemi, A.; Huang, Z.; La, J. Identification and quantification of Thiothrix eikelboomii using qPCR for early detection of bulking incidents in a full-scale water reclamation plant. Appl. Microbiol. Biotechnol. 2015, 99, 4045–4057. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, H.; Du, B. Bacteria and bacteriophage communities in bulking and non-bulking activated sludge in full-scale municipal wastewater treatment systems. Biochem. Eng. J. 2017, 119, 101–111. [Google Scholar] [CrossRef]
- Han, X.; Zhou, Z.; Mei, X.; Ma, Y.; Xie, Z. Influence of fermentation liquid from waste activated sludge on anoxic/oxic- membrane bioreactor performance: Nitrogen removal, membrane fouling and microbial community. Bioresour. Technol. 2018, 250, 699–707. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, X.; Xia, X.; Zhong, H.; Zhao, L.J. Versatile aromatic compound-degrading capacity and microdiversity of Thauera strains isolated from a coking wastewater treatment bioreactor. J. Ind. Microbiol. Biotechnol. 2010, 37, 927–934. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-ocurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [Green Version]


| Target Gene | Primer Name | Primer Sequence (5′-3′) | Amplicon Size (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|---|
| 16Sr RNA | 338-F | ACTCCTACGGGAGGCAGCAG | 181 | 55 | [13] |
| 518-R | ATTACCGCGGCTGCTGG | ||||
| tetA | tetA-F | GCTACATCCTGCTTGCCTTC | 210 | 57 | [9] |
| tetA-R | CATAGATCGCCGTGAAGAGG | ||||
| tetC | tetC-F | CTTGAGAGCCTTCAACCCAG | 418 | 60 | [13] |
| tetC-R | ATGGTCGTCATCTACCTGCC | ||||
| tetL | tetM-F | AGTGGAGAAATCCCTGCTCGGT | 149 | 60 | [9] |
| tetM-R | TGACTATTTGGACGACGGGGCT | ||||
| tetO | tetO-F | ACGGARAGTTTATTGTATACC | 171 | 55 | [11] |
| tetO-R | TGGCGTATCTATAATGTTGAC | ||||
| tetW | tetW-F | GAGAGCCTGCTATATGCCAGC | 168 | 60 | [11] |
| tetW-R | GGGCGTATCCACAATGTTAAC | ||||
| tetX | tetX-F | AGCCTTACCAATGGGTGTAAA | 278 | 60 | [13] |
| tetX-R | TTCTTACCTTGGACATCCCG | ||||
| sul1 | sul1-F | CGCACCGGAAACATCGCTGCAC | 163 | 60 | [40] |
| sul1-R | TGAAGTTCCGCCGCAAGGCTCG | ||||
| sul2 | sul2-F | CTCCGATGGAGGCCGGTAT | 190 | 60 | [2] |
| sul2-R | GGGAATGCCATCTGCCTTGA | ||||
| sul3 | sul3-F | TCCGTTCAGCGAATTGGTGCAG | 128 | 60 | [40] |
| sul3-R | TTCGTTCACGCCTTACACCAGC | ||||
| intI1 | intI1-F | CCTCCCGCACGATGATC | 280 | 60 | [35] |
| intI1-R | TCCACGCATCGTCAGGC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, B.; Yang, Q.; Wang, R.; Wang, R.; Wang, Q.; Xin, Y. Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole. Int. J. Environ. Res. Public Health 2019, 16, 4681. https://doi.org/10.3390/ijerph16234681
Du B, Yang Q, Wang R, Wang R, Wang Q, Xin Y. Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole. International Journal of Environmental Research and Public Health. 2019; 16(23):4681. https://doi.org/10.3390/ijerph16234681
Chicago/Turabian StyleDu, Bingbing, Qingxiang Yang, Ruifei Wang, Ruimin Wang, Qiang Wang, and Yuan Xin. 2019. "Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole" International Journal of Environmental Research and Public Health 16, no. 23: 4681. https://doi.org/10.3390/ijerph16234681
APA StyleDu, B., Yang, Q., Wang, R., Wang, R., Wang, Q., & Xin, Y. (2019). Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole. International Journal of Environmental Research and Public Health, 16(23), 4681. https://doi.org/10.3390/ijerph16234681





