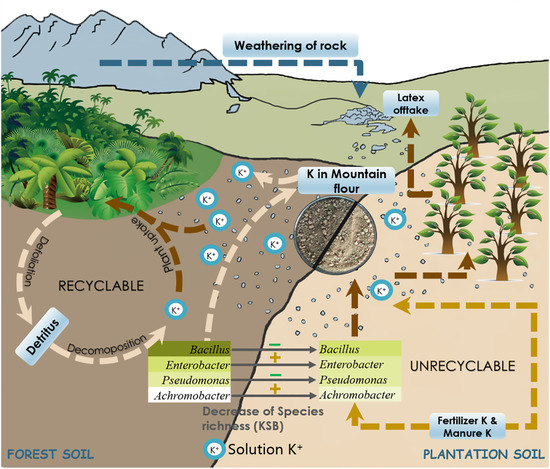
Differences in Distribution of Potassium-Solubilizing Bacteria in Forest and Plantation Soils in Myanmar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Elemental Analysis
2.2. DNA Isolation and Illumina MiSeq Sequencing
2.3. Isolation of KSB from Soil Samples in the Laboratory
2.4. Amplified Ribosomal DNA Restriction Analysis (ARDRA)
2.5. Sequencing and Analysis of Data
2.6. Determination of Potassium-Solubilizing Capability
3. Results
3.1. Characteristics of Soil Samples
3.2. Enumeration of Total Bacteria
3.3. Bacterial Abundance and Diversity
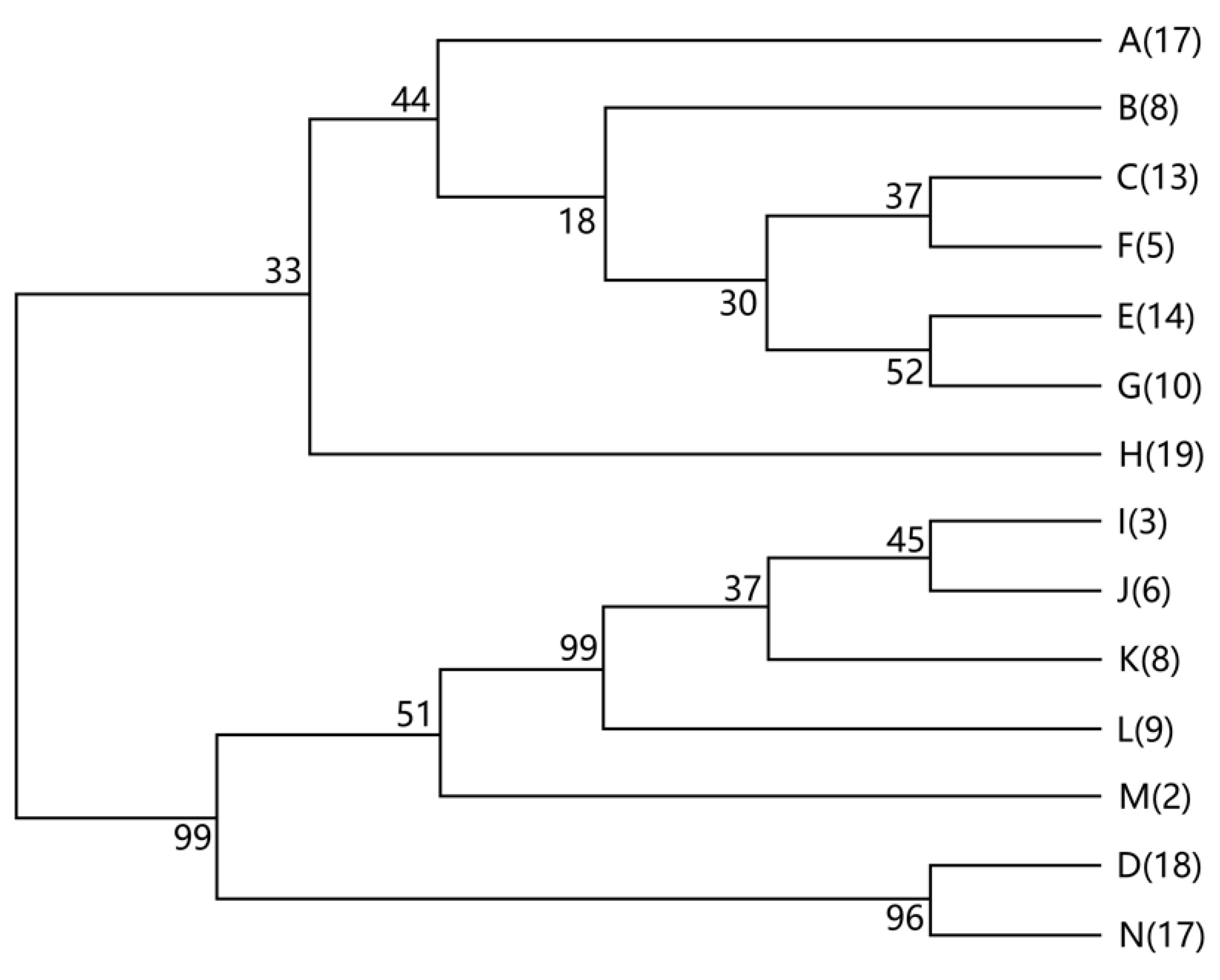
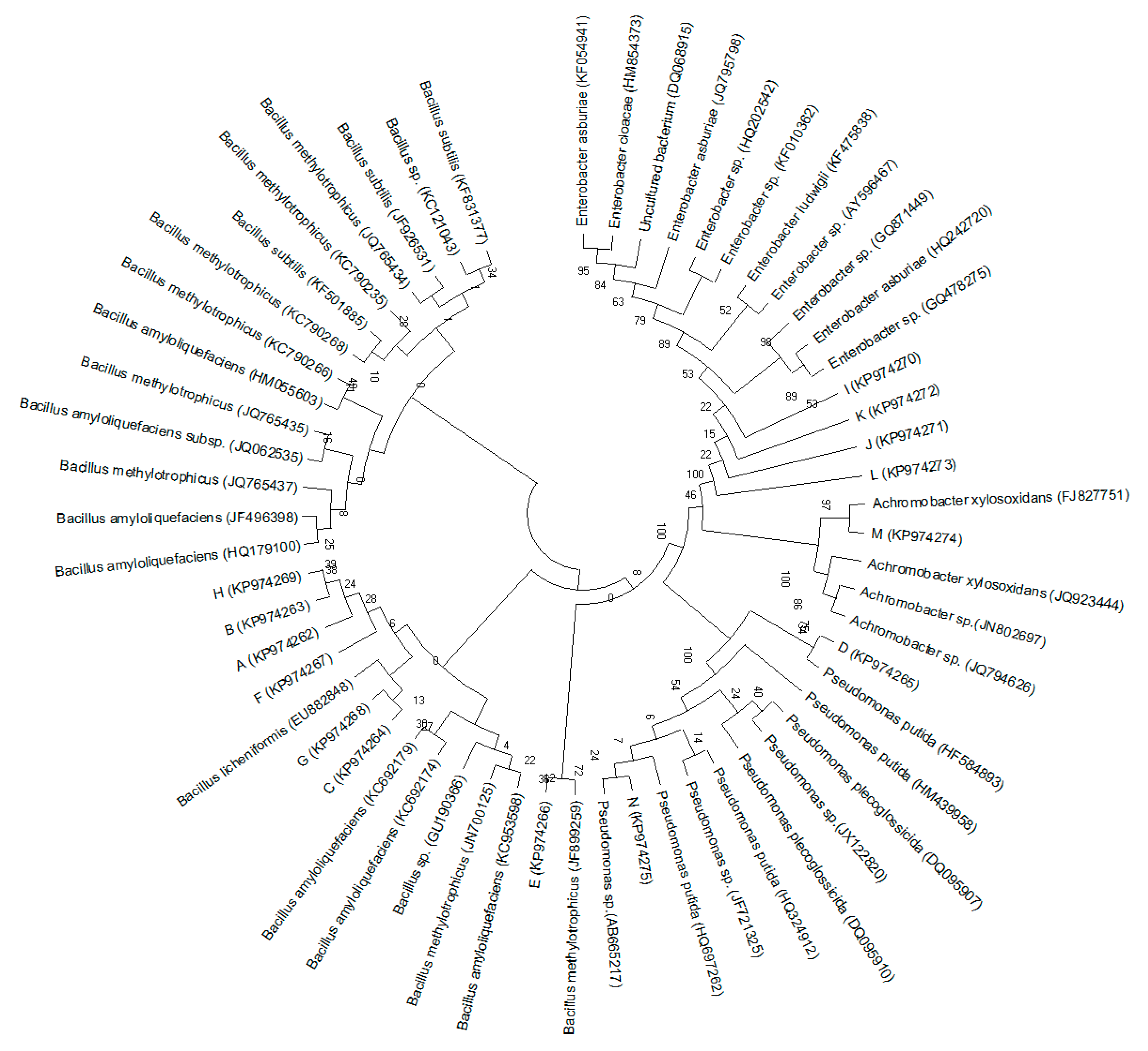
3.4. Amplified Ribosomal DNA Restriction Analysis (ARDRA) and 16S rDNA Sequencing Analysis
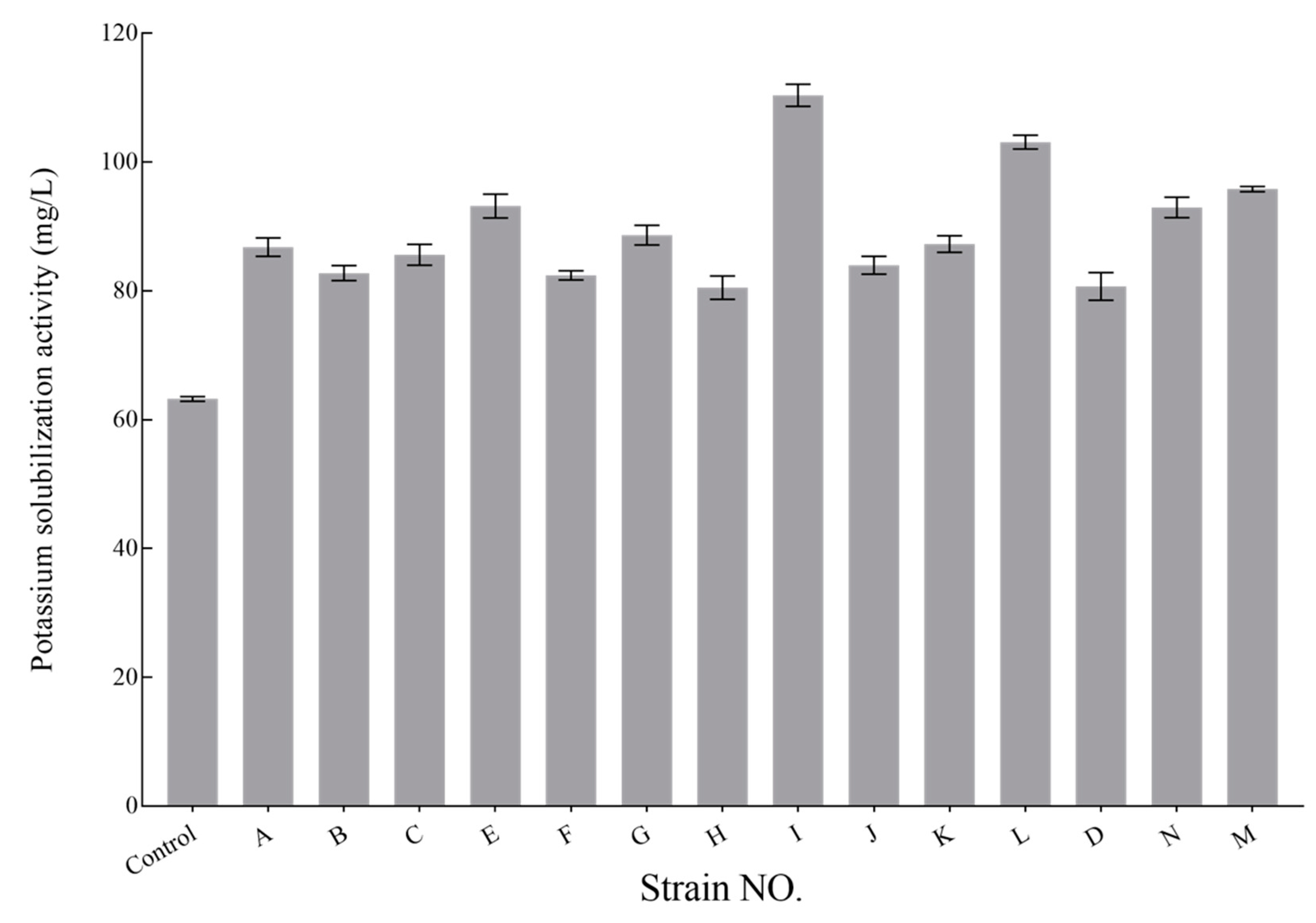
3.5. K-Solubilizing Capacity Analysis of KSB Isolates
3.6. Impacts of the KSB–Soil Interactions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raj, S.; Das, G.; Pothen, J.; Dey, S.K. Relationship between latex yield of Hevea brasiliensis and antecedent environmental parameters. Int. J. Biometeorol. 2005, 49, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Michels, T.; Eschbach, J.-M.; Lacote, R.; Benneveau, A.; Papy, F. Tapping panel diagnosis, an innovative on-farm decision support system for rubber tree tapping. Agron. Sustain. Dev. 2012, 32, 791–801. [Google Scholar] [CrossRef]
- Zörb, C.; Senbayram, M.; Peiter, E. Potassium in agriculture—Status and perspectives. J. Plant. Physiol. 2014, 171, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Goulding, K.; Loveland, P. The classification and mapping of potassium reserves in soils of England and Wales. J. Soil Sci. 1986, 37, 555–565. [Google Scholar] [CrossRef]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, W.T. Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol. Plant. 2008, 133, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Ovreas, L. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Garza, D.R.; Dutilh, B.E. From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 2015, 72, 4287–4308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartram, A.K.; Lynch, M.D.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef] [PubMed]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparks, D.L. Potassium Dynamics in Soils. In Advances in Soil Science; Springer: Berlin, Germany, 1987; pp. 1–63. [Google Scholar]
- Sugumaran, P.; Janarthanam, B. Solubilization of potassium containing minerals by bacteria and their effect on plant growth. World J. Agric. Sci. 2007, 3, 350–355. [Google Scholar]
- Parmar, P.; Sindhu, S. Potassium solubilization by rhizosphere bacteria: influence of nutritional and environmental conditions. J. Microbiol. Res. 2013, 3, 25–31. [Google Scholar]
- Masood, S.; Bano, A. Mechanism of Potassium Solubilization in the Agricultural Soils by the Help of Soil Microorganisms. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 137–147. [Google Scholar]
- Sheng, X.F. Growth promotion and increased potassium uptake of cotton and rape by a potassium releasing strain of Bacillus edaphicus. Soil Biol. Biochem. 2005, 37, 1918–1922. [Google Scholar] [CrossRef]
- Basak, B.B.; Biswas, D.R. Co-inoculation of potassium solubilizing and nitrogen fixing bacteria on solubilization of waste mica and their effect on growth promotion and nutrient acquisition by a forage crop. Biol. Fert. Soils 2010, 46, 641–648. [Google Scholar] [CrossRef]
- Zarjani, J.K.; Aliasgharzad, N.; Oustan, S.; Emadi, M.; Ahmadi, A. Isolation and characterization of potassium solubilizing bacteria in some Iranian soils. Arch. Agron. Soil Sci. 2013, 59, 1713–1723. [Google Scholar] [CrossRef]
- Zhang, C.S.; Kong, F.Y. Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil Ecol. 2014, 82, 18. [Google Scholar] [CrossRef]
- Sheng, X.; He, L.; Huang, W. The conditions of releasing potassium by a silicate dissolving bacterial strain NBT. Agric. Sci. China 2002, 1, 662–666. [Google Scholar]
- Nutaratat, P.; Monprasit, A.; Srisuk, N. High-yield production of indole-3-acetic acid by Enterobacter sp. DMKU-RP206, a rice phyllosphere bacterium that possesses plant growth-promoting traits. 3 Biotech 2017, 7, 305. [Google Scholar] [CrossRef] [PubMed]
- Basak, B.; Biswas, D. Influence of potassium solubilizing microorganism (Bacillus mucilaginosus) and waste mica on potassium uptake dynamics by sudan grass (Sorghum vulgare Pers.) grown under two Alfisols. Plant. Soil 2009, 317, 235–255. [Google Scholar] [CrossRef]
- Han, H.; Lee, K. Phosphate and potassium solubilizing bacteria effect on mineral uptake, soil availability and growth of eggplant. Res. J. Agric. Biol. Sci. 2005, 1, 176–180. [Google Scholar]
- Cheng, X.; Yu, M.; Wu, T. Effect of Forest Structural Change on Carbon Storage in a Coastal Metasequoia glyptostroboides Stand. Sci. World J. 2013. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Z.; Yang, L.; Guo, T.; Lv, L.; Yin, C.H.; Xu, Q.; Zhou, F.; Yan, H. Classification of potassium-releasing bacteria isolated from four agricultural soil samples. J. Pure Appl. Microbiol. 2013, 7, 3001–3008. [Google Scholar]
- Coico, R. Gram staining. Curr. Protoc. Microbiol. 2005. [Google Scholar] [CrossRef]
- Liu, X.; Yang, L.; Liu, Y.; Shanqi, M.; Lv, L.; Zhang, C.; Xu, Q.; Yan, H. Isolation and characterization of cellulose-degrading bacteria from primeval forest soil. Fresen. Environ. Bull. 2014, 23, 1802–1811. [Google Scholar]
- Ulrich, A.; Wirth, S. Phylogenetic diversity and population densities of culturable cellulolytic soil bacteria across an agricultural encatchment. Microb. Ecol. 1999, 37, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Dominique, C.; Philippot, L.; Hartmann, A.; Catherine, H.; Jean-laude, G. 16s rdna analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol. Ecol. 2001, 34, 121–128. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstruction of phylogenetic trees. J. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Hernesmaa, A.; Björklöf, K.; Kiikkilä, O.; Fritze, H.; Haahtela, K.; Romantschuk, M. Structure and function of microbial communities in the rhizosphere of Scots pine after tree-felling. Soil Biol. Biochem. 2005, 37, 777–785. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Q.; Wang, B.; Wang, X.; Franks, A.E.; Teng, Y.; Li, Z.; Luo, Y. Changes in the abundance and structure of bacterial communities under long-term fertilization treatments in a peanut monocropping system. Plant. Soil 2015, 395, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Sarathchandra, S.; Ghani, A.; Yeates, G.; Burch, G.; Cox, N. Effect of nitrogen and phosphate fertilisers on microbial and nematode diversity in pasture soils. Soil Biol. Biochem. 2001, 33, 953–964. [Google Scholar] [CrossRef]
- Esperschütz, J.; Gattinger, A.; Mäder, P.; Schloter, M.; Fließbach, A. Response of soil microbial biomass and community structures to conventional and organic farming systems under identical crop rotations. FEMS Microbiol. Ecol. 2007, 61, 26–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melero, S.; Madejón, E.; Herencia, J.F.; Ruiz, J.C. Effect of implementing organic farming on chemical and biochemical properties of an irrigated loam soil. Agron. J. 2008, 100, 136. [Google Scholar] [CrossRef]
- Chen, D.M.; Xing, W.; Lan, Z.C.; Saleem, M.; Wu, Y.; Hu, S.J.; Bai, Y.F. Direct and indirect effects of nitrogen enrichment on soil organisms and carbon and nitrogen mineralization in a semi-arid grassland. Funct. Ecol. 2018, 33, 175–187. [Google Scholar] [CrossRef]
- Girvan, M.S.; Bullimore, J.; Pretty, J.N.; Osborn, A.M.; Ball, A.S. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 2003, 69, 1800–1809. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Prakash, V.; Kundu, S.; Kumar, N.; Mina, B.L. Soil enzymatic activity as affected by long term application of farm yard manure and mineral fertilizer under a rainfed soybean–wheat system in NW Himalaya. Eur. J. Soil Biol. 2008, 44, 309–315. [Google Scholar] [CrossRef]
- Bottomley, P.; Yarwood, R.; Kageyama, S.; Waterstripe, K.; Williams, M.; Cromack, K.; Myrold, D. Responses of soil bacterial and fungal communities to reciprocal transfers of soil between adjacent coniferous forest and meadow vegetation in the Cascade Mountains of Oregon. Plant. Soil 2006, 289, 35–45. [Google Scholar] [CrossRef]
- Sun, H.; Wang, Q.-X.; Liu, N.; Li, L.; Zhang, C.-G.; Liu, Z.-B.; Zhang, Y.-Y. Effects of different leaf litters on the physicochemical properties and bacterial communities in Panax ginseng-growing soil. Appl. Soil Ecol. 2017, 111, 17–24. [Google Scholar] [CrossRef]
- Smit, E.; Leeflang, P.; Gommans, S.; van den Broek, J.; van Mil, S.; Wernars, K. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef] [PubMed]
- Hartman, W.H.; Richardson, C.J.; Vilgalys, R.; Bruland, G.L. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. USA 2008, 105, 17842–17847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, B.C.; Ostle, N.; McNamara, N.; Bailey, M.J.; Whiteley, A.S.; Griffiths, R.I. Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. 2010, 59, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Cederlund, H.; Wessén, E.; Enwall, K.; Jones, C.M.; Juhanson, J.; Pell, M.; Philippot, L.; Hallin, S. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl. Soil Ecol. 2014, 84, 62–68. [Google Scholar] [CrossRef]
- Yuan, Z.S.; Liu, F.; Zhang, G.F. Characteristics and biodiversity of endophytic phosphorus- and potassium-solubilizing bacteria in Moso Bamboo (Phyllostachys edulis). Acta Biol. Hung. 2015, 66, 449–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayer, E.J. Using experimental manipulation to assess the roles of leaf litter in the functioning of forest ecosystems. Biol. Rev. 2006, 81, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms—A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Stephan, A.; Meyer, A.H.; Schmid, B. Plant diversity affects culturable soil bacteria in experimental grassland communities. J. Ecol. 2000, 88, 988–998. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, F.; Cao, Y.; Zhang, X. Effect of plant types on release of mineral potassium from gneiss. Nutr. Cycl. Agroecosyst. 2000, 56, 37–43. [Google Scholar] [CrossRef]
- Moritsuka, N.; Yanai, J.; Kosaki, T. Possible processes releasing nonexchangeable potassium from the rhizosphere of maize. Plant. Soil 2004, 258, 261–268. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Shen, Q.-H.; Zhou, J.-M.; Wang, J.; Du, C.-W.; Chen, X.-Q. Plants use alternative strategies to utilize nonexchangeable potassium in minerals. Plant. Soil 2011, 343, 209–220. [Google Scholar] [CrossRef]

| Soil Sample | K+ (mg/g) | Mg2+ (mg/g) | P (mg/g) | N (mg/g) | Si (mg/g) |
|---|---|---|---|---|---|
| S1 | 0.029 ± 0.007 | 0.006 ± 0.001 | 0.022 ± 0.002 | 5.366 ± 0.223 | 0.099 ± 0.007 |
| S2 | 0.009 ± 0.002 | 0.044 ± 0.005 | 0.016 ± 0.004 | 3.805 ± 0.346 | 0.04 ± 0.012 |
| S3 | 0.004 ± 0.002 | 0.056 ± 0.012 | 0.012 ± 0.006 | 5.476 ± 0.478 | 0.042 ± 0.005 |
| Soil Sample | S1 | S2 | S3 |
|---|---|---|---|
| TB | 653 | 531 | 494 |
| KSB | 80 | 29 | 40 |
| KSB/TB (%) | 12.25% | 5.46% | 8.10% |
| Sample | Sequencing Results | Diversity Estimates | ||||
|---|---|---|---|---|---|---|
| Reads | OTUs | ACE | Chao | Shannon | Simpson | |
| S1 | 16,013 | 738 | 764 | 764 | 5.82 | 0.0054 |
| S2 | 16,224 | 702 | 781 | 782 | 5.43 | 0.0097 |
| S3 | 26,151 | 803 | 848 | 858 | 5.32 | 0.0151 |
| Gram | Genus | ARDRA | S1 | S2 | S3 | Total |
|---|---|---|---|---|---|---|
| + | Bacillus | Group (I) | ||||
| A | 17 | 0 | 0 | 17 | ||
| B | 8 | 0 | 0 | 8 | ||
| C | 12 | 0 | 1 | 13 | ||
| E | 14 | 0 | 0 | 14 | ||
| F | 0 | 4 | 1 | 5 | ||
| G | 2 | 8 | 0 | 10 | ||
| H | 10 | 7 | 2 | 19 | ||
| − | Enterobacter | Group (II) | ||||
| I | 0 | 3 | 0 | 3 | ||
| J | 0 | 5 | 1 | 6 | ||
| K | 0 | 1 | 7 | 8 | ||
| L | 1 | 0 | 8 | 9 | ||
| − | Pseudomonas | Group (III) | ||||
| D | 16 | 1 | 1 | 18 | ||
| N | 0 | 0 | 17 | 17 | ||
| − | Achromobacter | Group (IV) | ||||
| M | 0 | 0 | 2 | 2 | ||
| Total | 80 | 29 | 40 | 149 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Lv, L.; Wang, W.; Liu, Y.; Yin, C.; Xu, Q.; Yan, H.; Fu, J.; Liu, X. Differences in Distribution of Potassium-Solubilizing Bacteria in Forest and Plantation Soils in Myanmar. Int. J. Environ. Res. Public Health 2019, 16, 700. https://doi.org/10.3390/ijerph16050700
Dong X, Lv L, Wang W, Liu Y, Yin C, Xu Q, Yan H, Fu J, Liu X. Differences in Distribution of Potassium-Solubilizing Bacteria in Forest and Plantation Soils in Myanmar. International Journal of Environmental Research and Public Health. 2019; 16(5):700. https://doi.org/10.3390/ijerph16050700
Chicago/Turabian StyleDong, Xin, Le Lv, Weijun Wang, Yongzhi Liu, Chunhua Yin, Qianqian Xu, Hai Yan, Jinxia Fu, and Xiaolu Liu. 2019. "Differences in Distribution of Potassium-Solubilizing Bacteria in Forest and Plantation Soils in Myanmar" International Journal of Environmental Research and Public Health 16, no. 5: 700. https://doi.org/10.3390/ijerph16050700