Nitrofurantoin—Microbial Degradation and Interactions with Environmental Bacterial Strains
Abstract
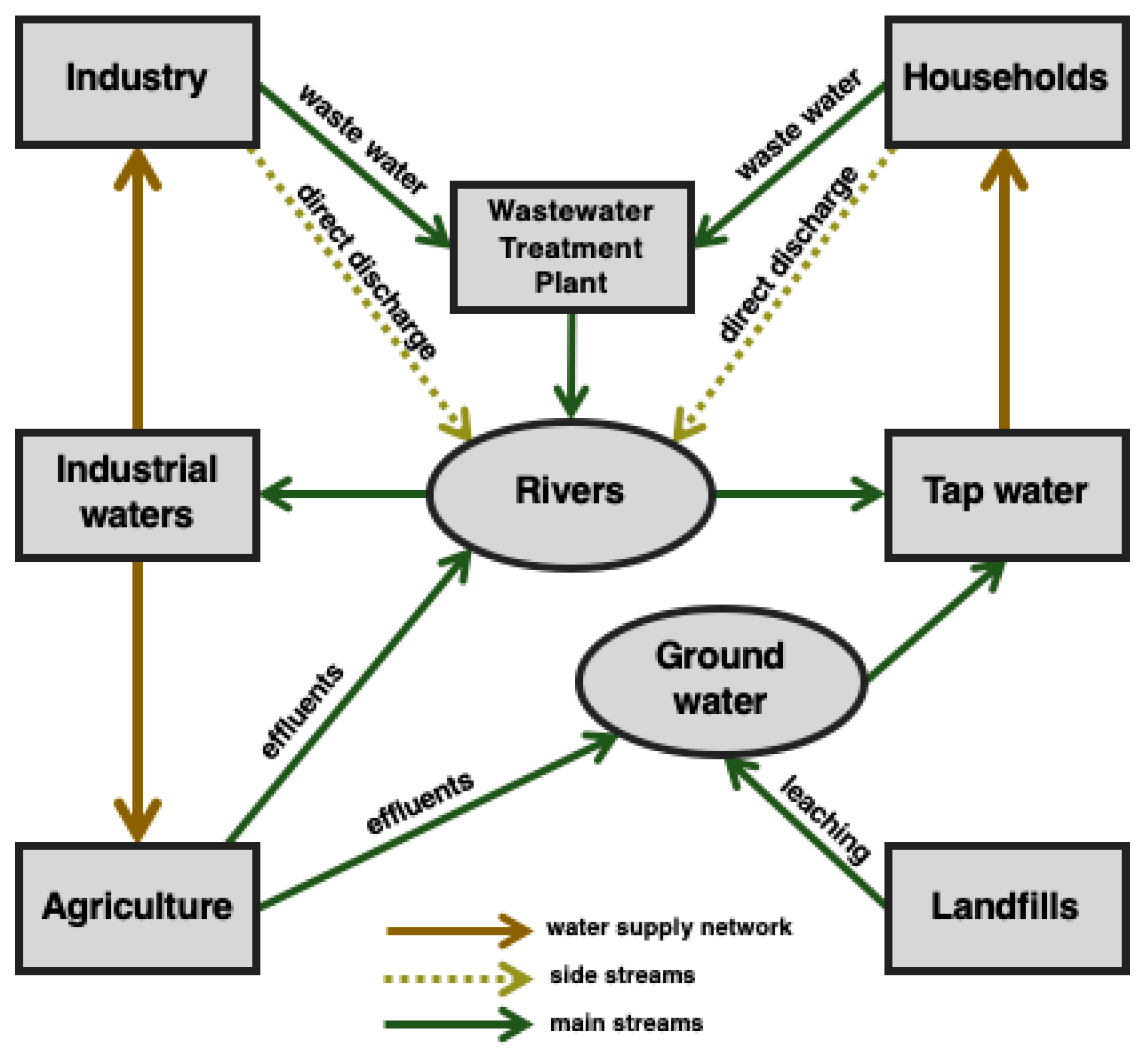
1. Introduction
2. Materials and Methods
2.1. Chemicals
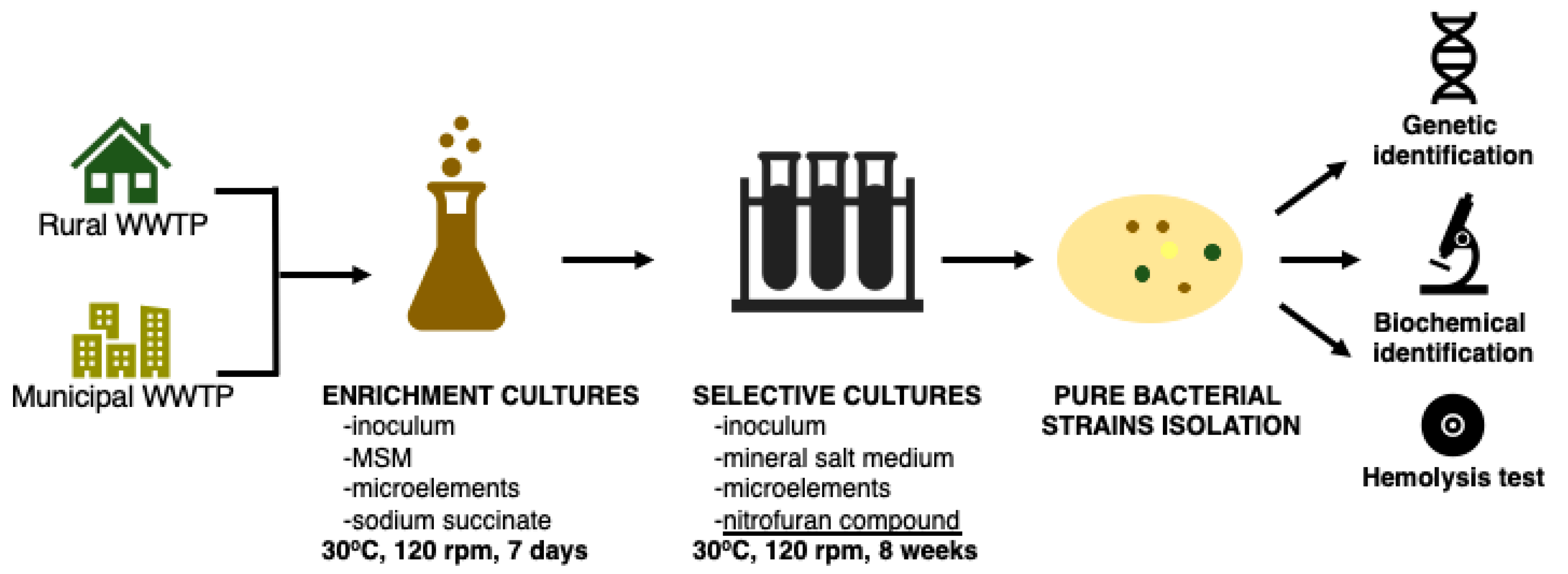
2.2. Isolation and Identification of Bacterial Strains
2.3. Biochemical Characterization of Bacterial Strains
2.4. Growing Conditions
2.5. Cell Membrane Permeability and Cell Surface Hydrophobicity
2.6. Microbes Viability after Contact with NFT
2.7. Nitrofurantoin Biodegradation with Kinetic Study
2.8. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Identification of Microbial Strains
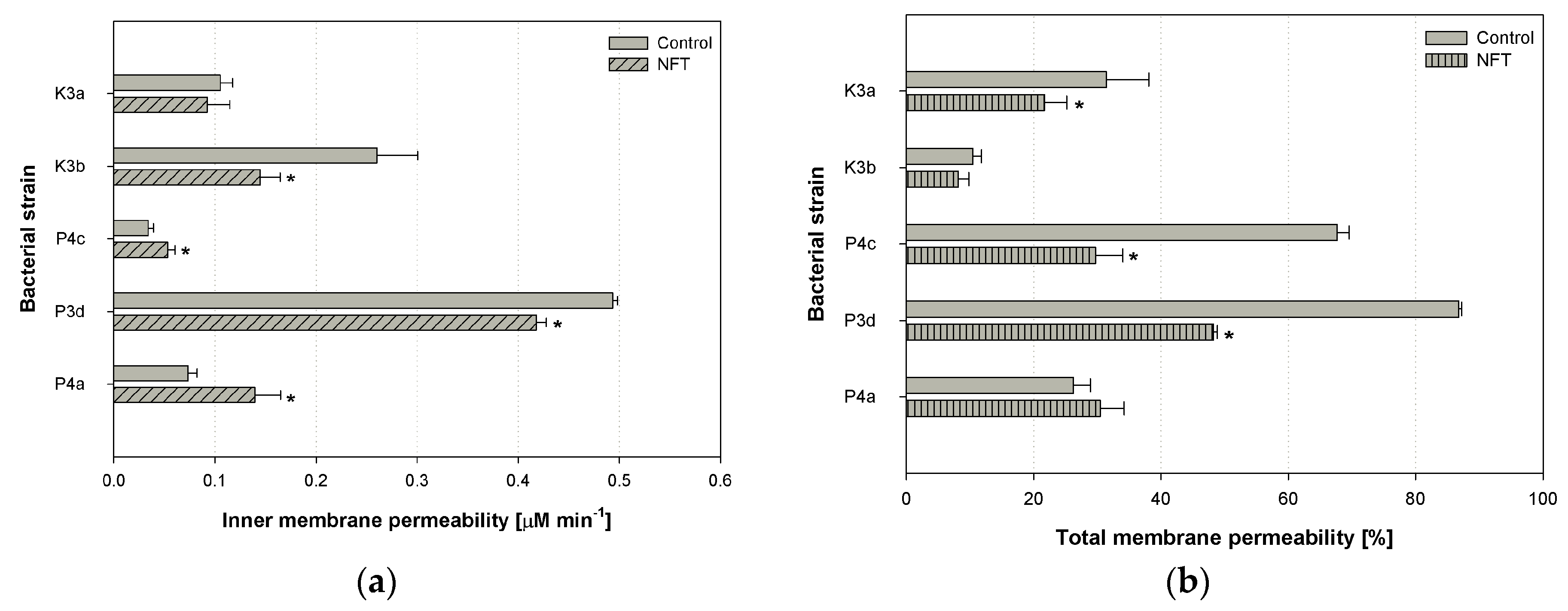
3.2. Inner and Total Membrane Permeability
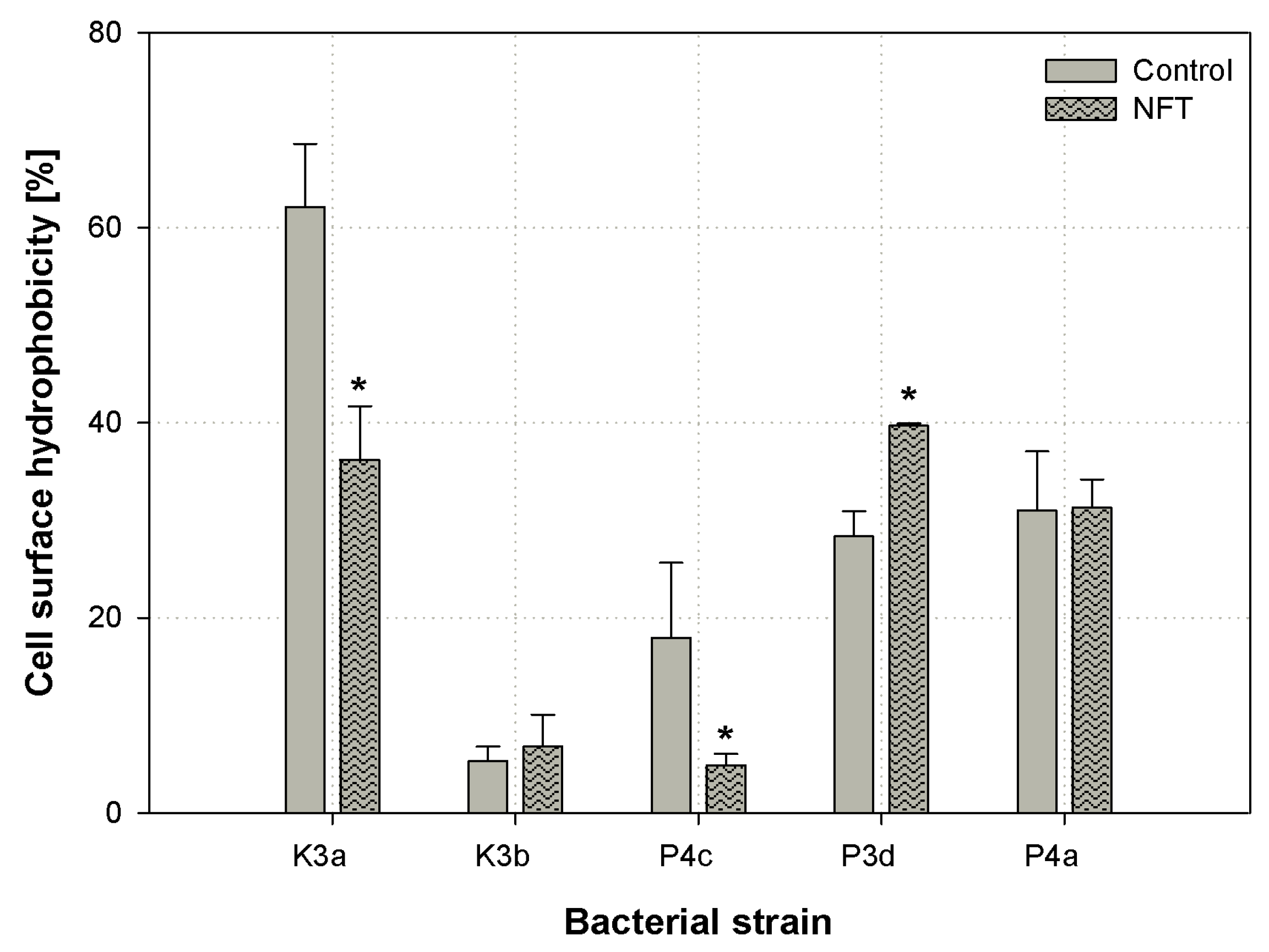
3.3. Cell Surface Hydrophobicity
3.4. Cytotoxity Analysis of NFT
3.5. Nitrofurantoin Removal
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Caracciolo, A.B.; Topp, E.; Grenni, P. Pharmaceuticals in the environment: Biodegradation and effects on natural microbial communities. A review. J. Pharm. Biomed. Anal. 2015, 106, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Bergheim, M.; Gminski, R.; Spangenberg, B.; Debiak, M.; Bürkle, A.; Mersch-Sundermann, V.; Kümmerer, K.; Gieré, R. Antibiotics and sweeteners in the aquatic environment: biodegradability, formation of phototransformation products, and in vitro toxicity. Environ. Sci. Pollut. 2015, 22, 18017–18030. [Google Scholar] [CrossRef] [PubMed]
- Edhlund, B.L.; Arnold, W.A.; McNeill, K. Aquatic photochemistry of nitrofuran antibiotics. Environ. Sci. Technol. 2006, 40, 5422–5427. [Google Scholar] [CrossRef]
- Li, Y.; Dang, Z.; Huang, Y.; Yang, C.; Guo, C. Effects of cytotoxicity of erythromycin on PAH-degrading strains and degrading efficiency. RSC Adv. 2016, 6, 114396–114404. [Google Scholar]
- Kümmerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef]
- Biošić, M.; Škorić, I.; Beganović, J.; Babić, S. Nitrofurantoin hydrolytic degradation in the environment. Chemosphere 2017, 186, 660–668. [Google Scholar] [CrossRef]
- Santos, F.; Mucha, A.P.; Alexandrino, D.A.; Almeida, C.M.R.; Carvalho, M.F. Biodegradation of enrofloxacin by microbial consortia obtained from rhizosediments of two estuarine plants. J. Environ. Manag. 2019, 231, 1145–1153. [Google Scholar] [CrossRef]
- Harrabi, M.; Alexandrino, D.A.; Aloulou, F.; Elleuch, B.; Liu, B.; Jia, Z.; Almeida, C.M.R.; Mucha, A.P.; Carvalho, M.F. Biodegradation of oxytetracycline and enrofloxacin by autochthonous microbial communities from estuarine sediments. Sci. Total. Environ. 2019, 648, 962–972. [Google Scholar] [CrossRef]
- Liu, N.; Hou, T.; Yin, H.; Han, L.; Huang, G. Effects of amoxicillin on nitrogen transformation and bacterial community succession during aerobic composting. J. Hazard. Mater. 2019, 362, 258–265. [Google Scholar] [CrossRef]
- Jepsen, R.; He, K.; Blaney, L.; Swan, C. Effects of antimicrobial exposure on detrital biofilm metabolism in urban and rural stream environments. Sci. Total. Environ. 2019, 666, 1151–1160. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Chen, J.; Qi, W.; Wang, F.; Zhou, Y. Impact of biofilm formation and detachment on the transmission of bacterial antibiotic resistance in drinking water distribution systems. Chemosphere 2018, 203, 368–380. [Google Scholar] [CrossRef]
- Lv, G.; Li, Z.; Elliott, L.; Schmidt, M.J.; MacWilliams, M.P.; Zhang, B. Impact of tetracycline-clay interactions on bacterial growth. J. Hazard. Mater. 2019, 370, 91–97. [Google Scholar] [CrossRef]
- Du, B.; Wang, R.; Yang, Q.; Hu, H.; Li, X.; Duan, X. Impact of tetracycline on the performance and abundance of functional bacteria of a lab-scale anaerobic-aerobic wastewater treatment system. Biochem. Eng. J. 2018, 138, 98–105. [Google Scholar] [CrossRef]
- Manzetti, S.; Ghisi, R. The environmental release and fate of antibiotics. Mar. Pollut. 2014, 79, 7–15. [Google Scholar] [CrossRef]
- Vumma, R.; Bang, C.S.; Kruse, R.; Johansson, K.; Persson, K. Antibacterial effects of nitric oxide on uropathogenic Escherichia coli during bladder epithelial cell colonization—A comparison with nitrofurantoin. J. Antibiot. 2016, 69, 183–186. [Google Scholar] [CrossRef]
- Purohit, V.; Basu, A.K. Mutagenicity of Nitroaromatic Compounds. Chem. Toxicol. 2000, 13, 673–692. [Google Scholar] [CrossRef]
- Kijima, A.; Ishii, Y.; Takasu, S.; Matsushita, K.; Kuroda, K.; Hibi, D.; Suzuki, Y.; Nohmi, T.; Umemura, T. Chemical structure-related mechanisms underlying in vivo genotoxicity induced by nitrofurantoin and its constituent moieties in gpt delta rats. Toxicology 2015, 331, 125–135. [Google Scholar] [CrossRef]
- Vass, M.; Hruska, K.; Fránek, M. Nitrofuran antibiotics: a review on the application, prohibition and residual analysis. Vet. Med. 2008, 53, 469–500. [Google Scholar] [CrossRef]
- Lewkowski, J.; Rogacz, D.; Rychter, P. Hazardous ecotoxicological impact of two commonly used nitrofuran-derived antibacterial drugs: Furazolidone and nitrofurantoin. Chemosphere 2019, 222, 381–390. [Google Scholar] [CrossRef]
- Wang, Y.; Chan, K.K.J.; Chan, W. Plant uptake and metabolism of nitrofuran antibiotics in spring onion grown in nitrofuran—Contaminated soil. J. Agric. Food Chem. 2017, 65, 4255–4261. [Google Scholar] [CrossRef]
- Kaczorek, E.; Sałek, K.; Guzik, U.; Dudzińska-Bajorek, B. Cell surface properties and fatty acids composition of Stenotrophomonas maltophilia under the influence of hydrophobic compounds and surfactants. New Biotechnol. 2013, 30, 173–182. [Google Scholar] [CrossRef]
- Youssef, N.H.; Duncan, K.E.; Nagle, D.P.; Savage, K.N.; Knapp, R.M.; McInerney, M.J. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 2004, 56, 339–347. [Google Scholar] [CrossRef]
- Hassanshahian, M. Isolation and characterization of biosurfactant producing bacteria from Persian Gulf (Bushehr provenance). Mar. Pollut. 2014, 86, 361–366. [Google Scholar] [CrossRef]
- Pacholak, A.; Simlat, J.; Zgoła-Grześkowiak, A.; Kaczorek, E. Biodegradation of clotrimazole and modification of cell properties after metabolic stress and upon addition of saponins. Ecotoxicol. Environ. Saf. 2018, 161, 676–682. [Google Scholar] [CrossRef]
- Devi, K.P.; Sakthivel, R.; Nisha, S.A.; Suganthy, N.; Pandian, S.K. Eugenol alters the integrity of cell membrane and acts against the nosocomial pathogen Proteus mirabilis. Arch. Pharmacal 2013, 36, 282–292. [Google Scholar] [CrossRef]
- Ambalam, P.; Kondepudi, K.K.; Nilsson, I.; Wadström, T.; Ljungh, Å. Bile stimulates cell surface hydrophobicity, Congo red binding and biofilm formation of Lactobacillus strains. FEMS Microbiol. Lett. 2012, 333, 10–19. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Wang, F.; Wei, D.; Wang, X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J. Microbiol. Methods 2010, 82, 330–333. [Google Scholar] [CrossRef]
- Bekins, B.A.; Warren, E.; Godsy, E.M. A Comparison of Zero-Order, First-Order, and Monod Biotransformation Models. Ground Water 1998, 36, 261–268. [Google Scholar] [CrossRef]
- Poszytek, K.; Karczewska-Golec, J.; Ciok, A.; Decewicz, P.; Dziurzynski, M.; Gorecki, A.; Jakusz, G.; Krucon, T.; Lomza, P.; Romaniuk, K.; et al. Genome-Guided Characterization of Ochrobactrum sp. POC9 enhancing sewage sludge utilization—biotechnological potential and biosafety considerations. Int. J. Environ. Res. Public. Health 2018, 15, 1501. [Google Scholar] [CrossRef]
- Wang, X.S.; Huang, L.P.; Li, Y.; Chen, J.; He, W.; Miao, H.H. Uptake of Cr (VI) by Sphingomonas paucimobilis Biomass from Aqueous Solutions. Sci. Technol. 2010, 45, 681–686. [Google Scholar]
- Thelusmond, J.-R.; Strathmann, T.J.; Cupples, A.M. Carbamazepine, triclocarban and triclosan biodegradation and the phylotypes and functional genes associated with xenobiotic degradation in four agricultural soils. Sci. Total. Environ. 2019, 657, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Murugesan, K.; Schmidt, S.; Bokare, V.; Jeon, J.-R.; Kim, E.-J.; Chang, Y.-S. Triclosan susceptibility and co-metabolism—A comparison for three aerobic pollutant-degrading bacteria. Bioresour. Technol. 2011, 102, 2206–2212. [Google Scholar] [CrossRef]
- Shao, S.; Hu, Y.; Cheng, J.; Chen, Y. Degradation of oxytetracycline (OTC) and nitrogen conversion characteristics using a novel strain. Chem. Eng. J. 2018, 354, 758–766. [Google Scholar] [CrossRef]
- Mulla, S.I.; Hu, A.; Sun, Q.; Li, J.; Suanon, F.; Ashfaq, M.; Yu, C.-P. Biodegradation of sulfamethoxazole in bacteria from three different origins. J. Environ. Manag. 2018, 206, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qiu, L.; Gong, A.; Yuan, X. Isolation and characterization of a high-efficiency erythromycin A-degrading Ochrobactrum sp. strain. Mar. Pollut. 2017, 114, 896–902. [Google Scholar] [CrossRef]
- Telke, A.; Kalyani, D.; Jadhav, J.; Govindwar, S. Kinetics and mechanism of reactive Red 141 degradation by a bacterial isolate Rhizobium radiobacter MTCC 816. Acta Chim. Slov. 2008, 55, 320–329. [Google Scholar]
- Parshetti, G.; Saratale, G.; Telke, A.; Govindwar, S. Biodegradation of hazardous triphenylmethane dye methyl violet by Rhizobium radiobacter (MTCC 8161). J. Basic Microbiol. 2009, 49, S36–S42. [Google Scholar] [CrossRef]
- Radó, J.; Kaszab, E.; Petrovics, T.; Pászti, J.; Kriszt, B.; Szoboszlay, S. Characterization of environmental Pseudomonas aeruginosa using multilocus sequence typing scheme. J. Med Microbiol. 2017, 66, 1457–1466. [Google Scholar] [CrossRef]
- Sindu, P.A.; Gautam, P. Studies on the biofilm produced by P. aeruginosa grown in different metal fatty acid salt media and its application in biodegradation of fatty acids and bioremediation of heavy metal ions. Can. J. Microbiol. 2017, 63, 61–73. [Google Scholar] [CrossRef]
- Rozitis, D.; Strade, E. COD reduction ability of microorganisms isolated from highly loaded pharmaceutical wastewater pre-treatment process. J. Mater. Environ. Sci. 2015, 6, 507–512. [Google Scholar]
- Guven, K.; Yolcu, M.; Gul-Guven, R.; Erdogan, S.; De Pomerai, D. The effects of organic pesticides on inner membrane permeability in Escherichia coli ML35. Cell Boil. Toxicol. 2005, 21, 73–81. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Dinesh Kumar, S.; Nam, J.; Jeon, D.; Kim, Y.; Lee, C.W.; Park, I.-S.; Shin, S.Y. Antimicrobial and anti-inflammatory activities of chemokine CXCL14-derived antimicrobial peptide and its analogs. Biochim. Biophys. Acta Biomembr. 2019, 1861, 256–267. [Google Scholar] [CrossRef]
- Sana, S.; Datta, S.; Biswas, D.; Sengupta, D. Assessment of synergistic antibacterial activity of combined biosurfactants revealed by bacterial cell envelop damage. Biochim. Biophys. Acta Biomembr 2018, 1860, 579–585. [Google Scholar] [CrossRef]
- Bharali, P.; Saikia, J.; Ray, A.; Konwar, B. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: A novel chemotaxis and antibacterial agent. Colloids Surf. B Biointerfaces 2013, 103, 502–509. [Google Scholar] [CrossRef]
- Kalia, V.; Miglani, R.; Purnapatre, K.P.; Mathur, T.; Singhal, S.; Khan, S.; Voleti, S.R.; Upadhyay, D.J.; Saini, K.S.; Rattan, A.; et al. Mode of Action of ranbezolid against Staphylococci and structural modeling studies of its interaction with ribosomes. Antimicrob. Agents Chemother. 2009, 53, 1427–1433. [Google Scholar] [CrossRef]
- Heipieper, H.J.; Neumann, G.; Cornelissen, S.; Meinhardt, F. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 2007, 74, 961–973. [Google Scholar] [CrossRef]
- Qiu, S.; Xu, H.; Xu, S.; Ma, F. The effect of tourmaline on cell membrane of nitrosomonaseuropaea and biodegradation of micropollutant: The effect of tourmaline on cell membrane. Surf. Interface Anal. 2014, 46, 564–569. [Google Scholar] [CrossRef]
- Branco, M.R.; Marinho, H.S.; Cyrne, L.; Antunes, F. Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 6501–6506. [Google Scholar] [CrossRef]
- Kaczorek, E.; Pacholak, A.; Zdarta, A.; Smułek, W. The impact of biosurfactants on microbial cell properties leading to hydrocarbon bioavailability increase. Colloids Interfaces 2018, 2, 35. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Microbiol. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Smułek, W.; Zdarta, A.; Guzik, U.; Dudzińska-Bajorek, B.; Kaczorek, E. Rahnella sp. strain EK12: Cell surface properties and diesel oil biodegradation after long-term contact with natural surfactants and diesel oil. Microbiol. Res. 2015, 176, 38–47. [Google Scholar] [CrossRef]
- Oliveira, I.M.; Borges, A.; Borges, F.; Simões, M. Repurposing ibuprofen to control Staphylococcus aureus biofilms. Eur. J. Med. Chem. 2019, 166, 197–205. [Google Scholar] [CrossRef]
- Zhang, W.; Niu, Z.; Yin, K.; Liu, F.; Chen, L. Degradation of furazolidone by bacteria Acinetobacter calcoaceticus T32, Pseudomonas putida SP1 and Proteus mirabilis V7. Int. Biodeterior. Biodegrad. 2013, 77, 45–50. [Google Scholar] [CrossRef]
- Samuelsen, O.B.; Solheim, E.; Lunestad, B.T. Fate and microbiological effects of furazolidone in a marine aquaculture sediment. Sci. Total. Environ. 1991, 108, 275–283. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y.; Li, B.; Yang, C.; Zhang, T. Aerobic Degradation of sulfadiazine by Arthrobacter spp.: kinetics, pathways, and genomic characterization. Environ. Sci. Technol. 2016, 50, 9566–9575. [Google Scholar] [CrossRef]
- Cheyns, K.; Mertens, J.; Diels, J.; Smolders, E.; Springael, D. Monod kinetics rather than a first-order degradation model explains atrazine fate in soil mini-columns: Implications for pesticide fate modelling. Environ. Pollut. 2010, 158, 1405–1411. [Google Scholar] [CrossRef]
- Sniegowski, K.; Mertens, J.; Diels, J.; Smolders, E.; Springael, D. Inverse modeling of pesticide degradation and pesticide-degrading population size dynamics in a bioremediation system: Parameterizing the Monod model. Chemosphere 2009, 75, 726–731. [Google Scholar] [CrossRef]

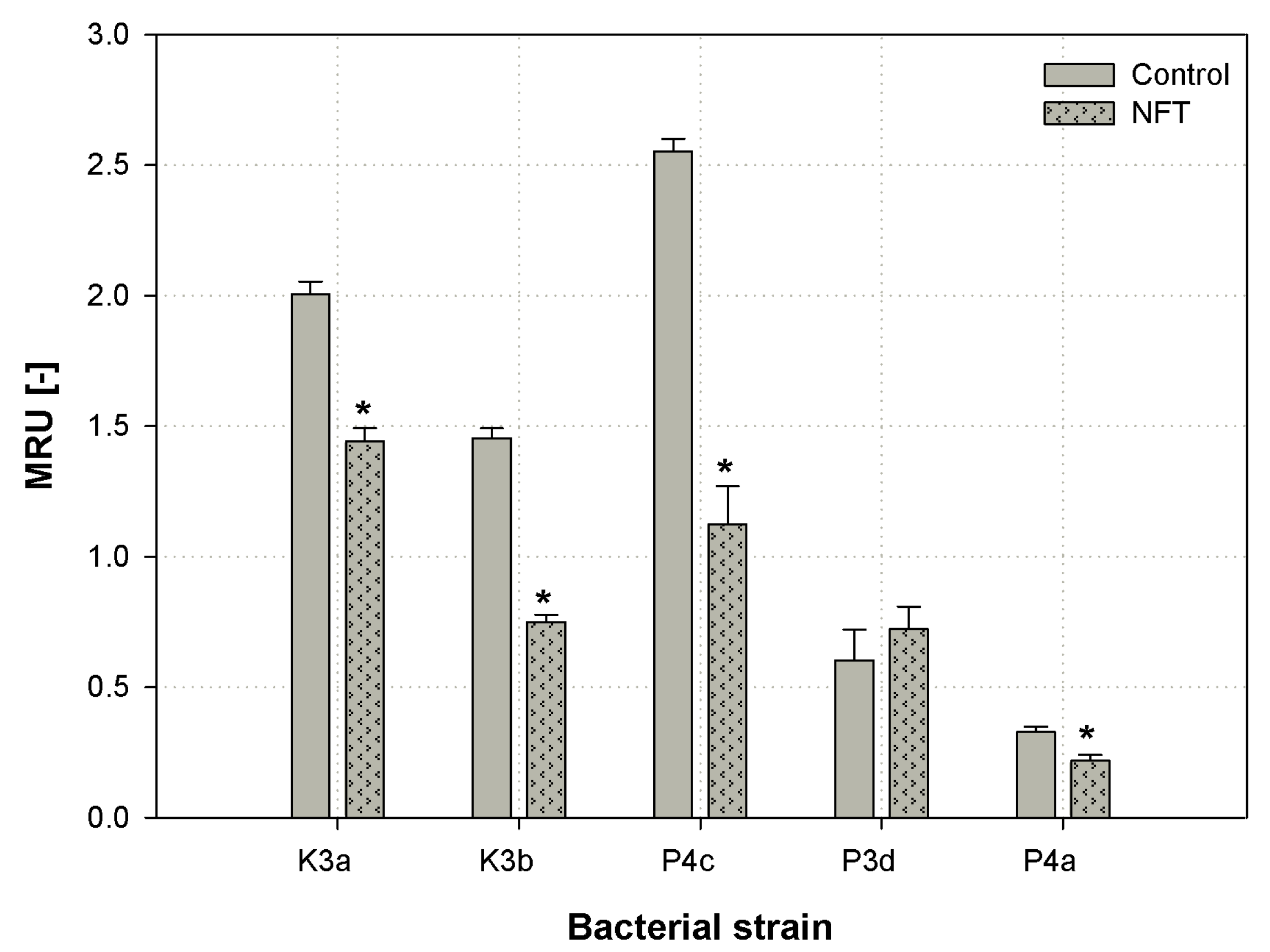

| Strain | APPA | ILATk | GlyA | O129R | ADO | dMAL | dTAG | AGLU | PyrA | AGLTp | dTRE | SUCT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K3a | + | + | − | − | − | + | − | + | + | − | − | − |
| K3b | − | + | + | − | + | − | + | − | + | − | − | + |
| P4c | − | − | + | − | − | − | − | − | + | − | − | − |
| P4a | − | + | − | + | − | − | − | − | − | + | (+) | − |
| P3d | − | − | − | − | − | − | − | + | − | − | − | − |
| IMLTa | IARL | dGLU | dMNE | CIT | ELLM | GGT | URE | MNT | CMT | BAlap | BGUR | |
| K3a | − | − | + | − | + | − | − | + | − | − | − | − |
| K3b | − | − | − | − | − | + | − | + | − | − | + | − |
| P4c | − | + | − | − | − | + | − | + | − | − | − | − |
| P4a | + | − | + | + | + | − | + | − | + | + | + | − |
| P3d | − | − | − | − | − | − | − | + | − | − | − | + |
| Bacterial Strain | Half-Saturation Constant Ks [mg L−1] | Substrate Utilization Rate vm [mg L−1 day−1] | R2 |
|---|---|---|---|
| K3a | 2.13 | 0.02 | 0.99 |
| K3b | 2.63 | 0.06 | 0.99 |
| P4c | 2.87 | 0.01 | 0.99 |
| P3d | 3.68 | 0.01 | 0.99 |
| P4a | 4.09 | 0.01 | 0.99 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacholak, A.; Smułek, W.; Zgoła-Grześkowiak, A.; Kaczorek, E. Nitrofurantoin—Microbial Degradation and Interactions with Environmental Bacterial Strains. Int. J. Environ. Res. Public Health 2019, 16, 1526. https://doi.org/10.3390/ijerph16091526
Pacholak A, Smułek W, Zgoła-Grześkowiak A, Kaczorek E. Nitrofurantoin—Microbial Degradation and Interactions with Environmental Bacterial Strains. International Journal of Environmental Research and Public Health. 2019; 16(9):1526. https://doi.org/10.3390/ijerph16091526
Chicago/Turabian StylePacholak, Amanda, Wojciech Smułek, Agnieszka Zgoła-Grześkowiak, and Ewa Kaczorek. 2019. "Nitrofurantoin—Microbial Degradation and Interactions with Environmental Bacterial Strains" International Journal of Environmental Research and Public Health 16, no. 9: 1526. https://doi.org/10.3390/ijerph16091526
APA StylePacholak, A., Smułek, W., Zgoła-Grześkowiak, A., & Kaczorek, E. (2019). Nitrofurantoin—Microbial Degradation and Interactions with Environmental Bacterial Strains. International Journal of Environmental Research and Public Health, 16(9), 1526. https://doi.org/10.3390/ijerph16091526








