Batch-Mode Analysis of Thermophilic Methanogenic Microbial Community Changes in the Overacidification Stage in Beverage Waste Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance of Anaerobic Sludge
2.2. Methane Production at High Substrate Loading in Batch Culture Experiments
2.3. Analytical Methods
2.4. Amplicon Sequence Analysis of 16S rRNA Genes Using a Nanopore Sequencer
2.5. Data Analysis
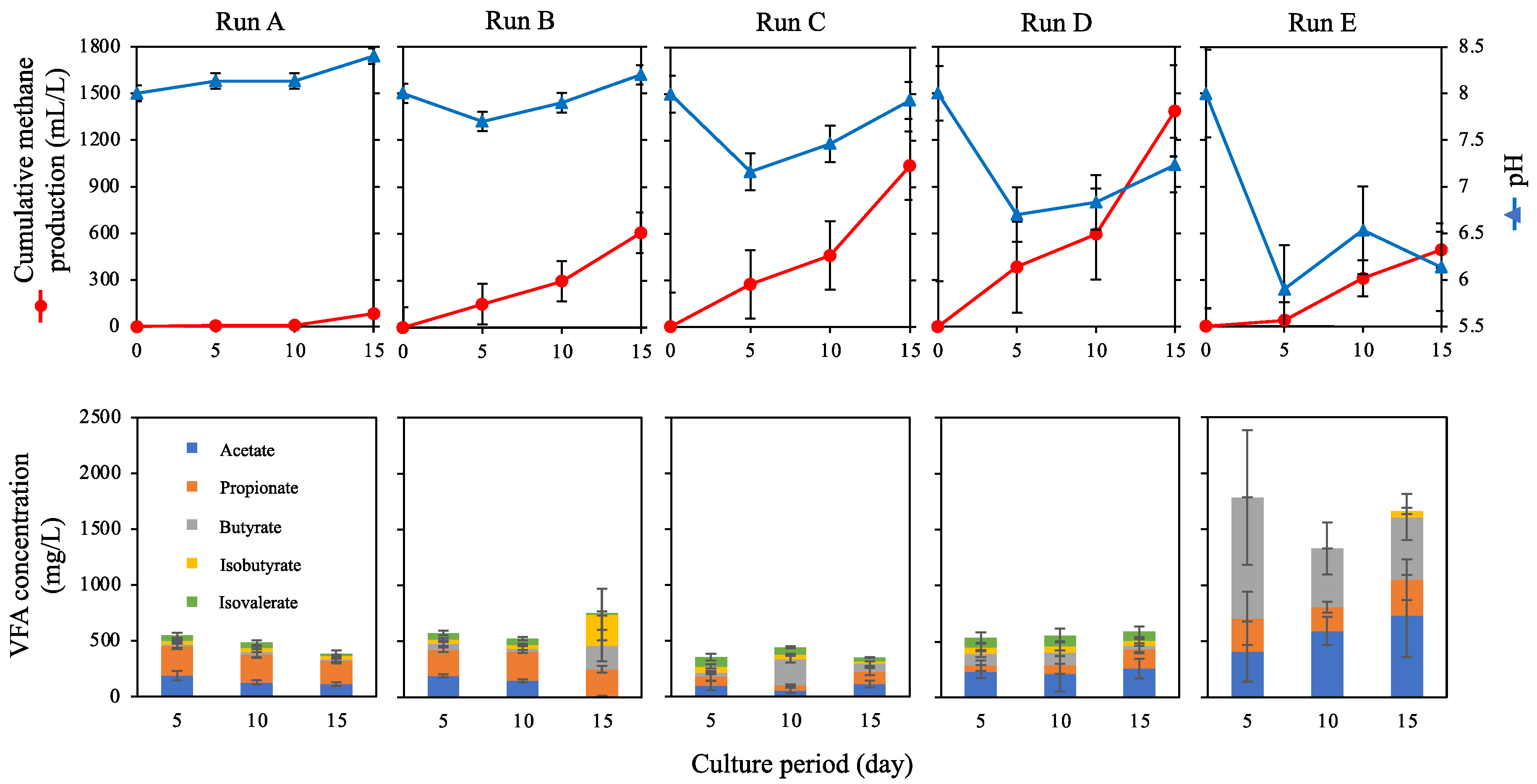
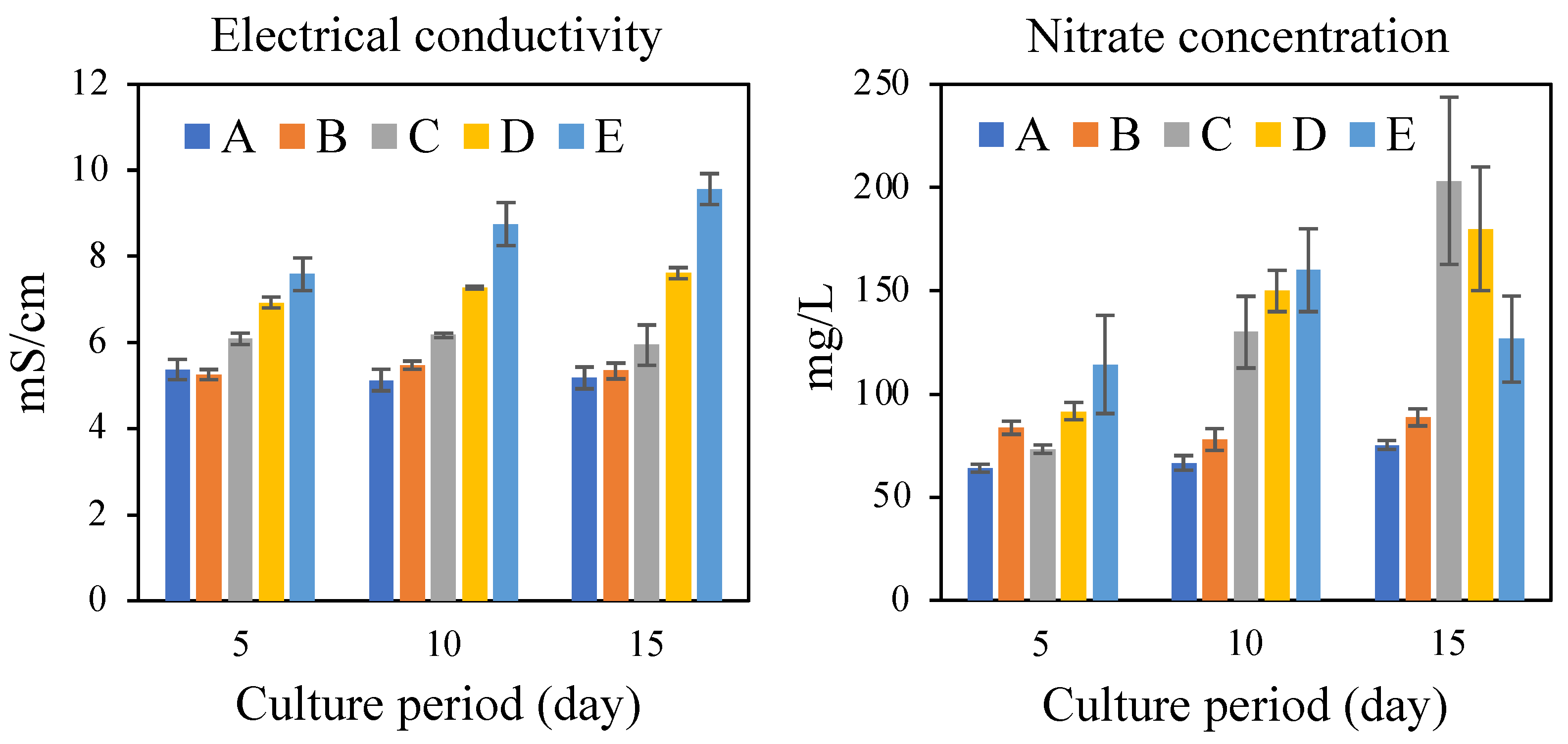
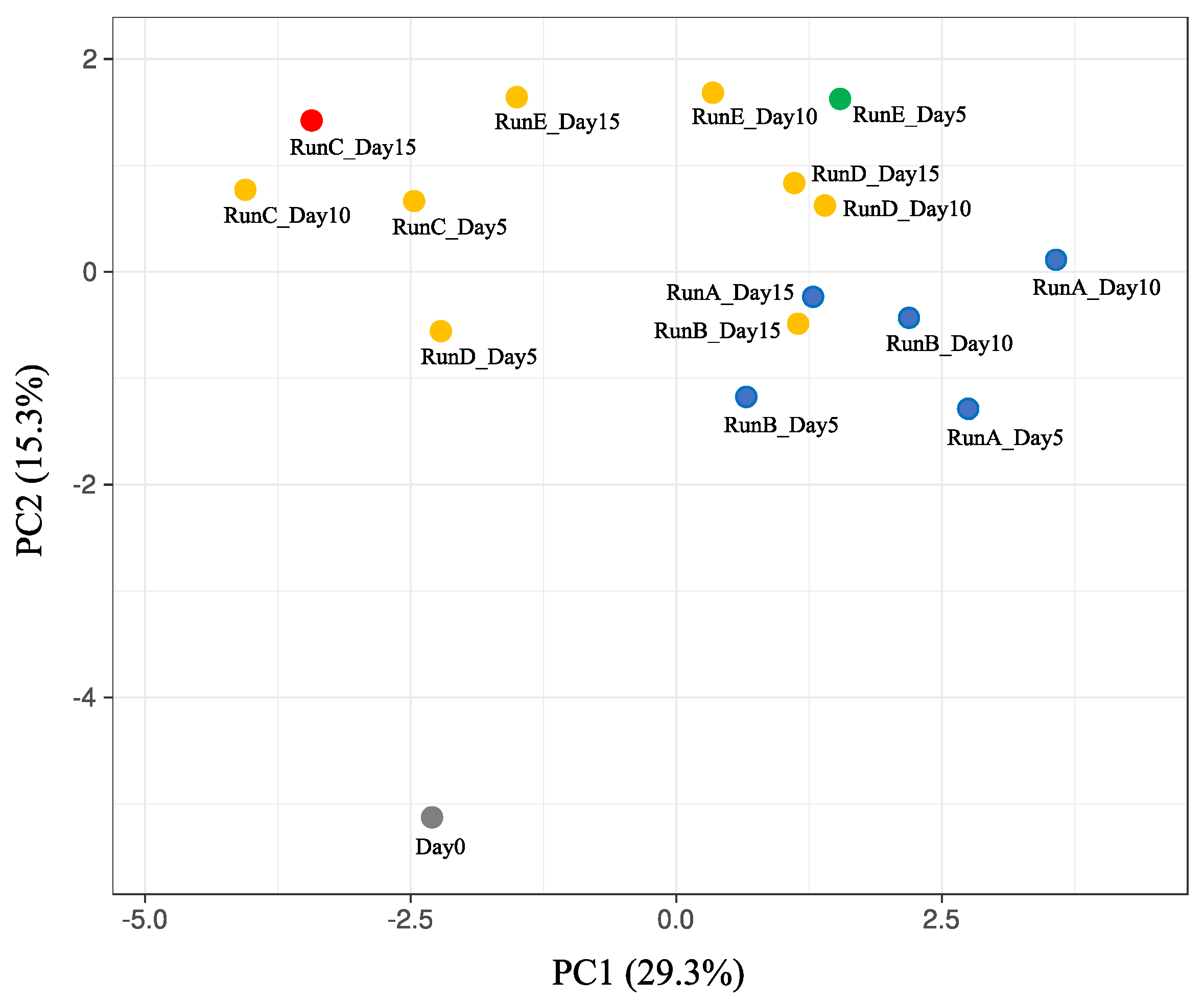
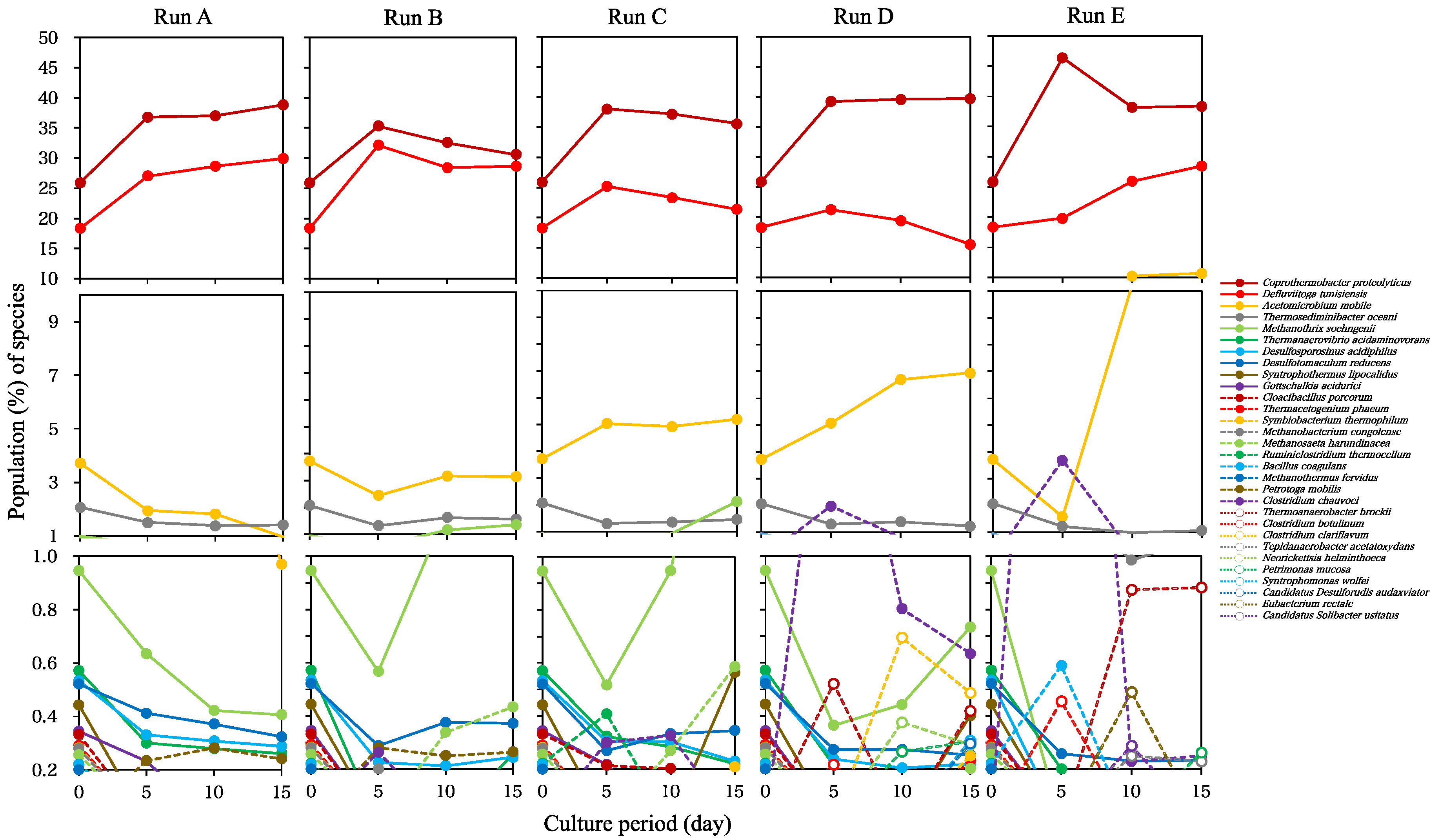
3. Results and Discussion
3.1. Methane Production in Batch Cultures Using SBWM
3.2. Changes in the Microbial Population in Batch Cultures with SBWM
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Meegoda, J.N.; Li, B.; Patel, K.; Wang, L.B. A review of the processes, parameters, and optimization of anaerobic digestion. Int. J. Environ. Res. Public Health 2018, 15, 2224. [Google Scholar] [CrossRef] [PubMed]
- Abouelenien, F.; Nakashimada, Y.; Nishio, N. Dry mesophilic fermentation of chicken manure for production of methane by repeated batch culture. J. Biosci. Bioeng. 2008, 107, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-H.; Hashimoto, A.G. Effects of pH and substrate: Inoculum ratio on batch methane fermentation. Bioresour. Technol. 1996, 56, 179–186. [Google Scholar] [CrossRef]
- Tekippe, J.A.; Hristov, A.N.; Heyler, K.S.; Zheljazkov, V.D.; Ferreira, J.F.S.; Cantrell, C.L.; Varga, G.A. Effects of plants and essential oils on ruminal in vitro batch culture methane production and fermentation. Can. J. Anim. Sci. 2012, 92, 395–408. [Google Scholar] [CrossRef]
- Yáñez-Ruiz, D.R.; Bannik, A.; Dijkstra, J.; Kebreab, E.; Morgavi, D.P.; O’Kiely, P.; Reynolds, C.K.; Schwarm, A.; Shingfield, K.J.; Yu, Z.; et al. Design, implementation and interpretation of in vitro batch culture experiments to assess enteric methane mitigation in ruminants—A review. Anim. Feed Sci. Technol. 2016, 216, 1–18. [Google Scholar] [CrossRef]
- Quested, T.; Ingle, R.; Parry, A. Household Food and Drink Waste in the United Kingdom 2012; Waste & Resources Action Programme: Banbury, UK, 2013; p. 40. [Google Scholar]
- Report of Survey on Recycling Status in Food Industries FY2017. Available online: https://www.maff.go.jp/j/shokusan/recycle/syoku_loss/attach/pdf/161227_8-56.pdf (accessed on 9 October 2020). (In Japanese).
- Tyagi, V.K.; Liu, J.; Poh, L.S.; Ng, W.J. Anaerobic-aerobic system for beverage effluent treatment: Performance evaluation and microbial community dynamics. Bioresour. Technol. Rep. 2019, 7, 100309. [Google Scholar] [CrossRef]
- Wickham, R.; Xie, S.; Galway, B.; Bustamante, H.; Nghiem, L.D. Anaerobic digestion of soft drink beverage waste and sewage sludge. Bioresour. Technol. 2018, 262, 141–147. [Google Scholar] [CrossRef]
- Tang, Y.-Q.; Shigematsu, T.; Morimura, S.; Kida, K. Dynamics of the microbial community during continuous methane fermentation in continuously stirred tank reactors. J. Biosci. Bioeng. 2015, 119, 375–383. [Google Scholar] [CrossRef]
- Leng, L.; Yang, P.; Singh, S.; Zhuang, H.; Xu, L.; Chen, W.-H.; Dolfing, J.; Li, D.; Zhang, Y.; Zeng, H.; et al. A review on the bioenergetics of anaerobic microbial metabolism close to the thermodynamic limits and its implications for digestion applications. Bioresour. Technol. 2018, 247, 1095–1106. [Google Scholar] [CrossRef]
- Wang, L.; Hossen, E.H.; Aziz, T.N.; Ducoste, J.J.; de los Reyes, F.L., III. Increased loading stress leads to convergence of microbial communities and high methane yields in adapted anaerobic co-digesters. Water Res. 2020, 169, 115155. [Google Scholar] [CrossRef]
- Blume, F.; Bergman, I.; Nettmann, E.; Schelle, H.; Rehde, G.; Mundt, K.; Klocke, M. Methanogenic population dynamics during semi-continuous biogas fermentation and acidification by overloading. J. Appl. Microbiol. 2010, 109, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Kleyböcker, A.; Liebrich, M.; Verstraete, W.; Kraume, M.; Würdemann, H. Early warning indicators for process failure due to organic loading by rapeseed oil in one-stage continuously stirred tank reactor, sewage sludge and waste digesters. Bioresour. Technol. 2012, 123, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Q.; Ma, Y.; Wang, X.; Peng, X. Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: Relationship between community structure and process stability. Bioresour. Technol. 2015, 189, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Tale, V.P.; Maki, J.S.; Struble, C.A.; Zitomer, D.H. Methanogen community structure-activity relationship and bioaugmentation of overloaded anaerobic digesters. Water Res. 2011, 45, 5249–5256. [Google Scholar] [CrossRef]
- Wu, S.; Xu, S.; Chen, X.; Sun, H.; Hu, M.; Bai, Z.; Zhuang, G.; Zhuang, X. Bacterial communities changes during food waste spoilage. Sci. Rep. 2018, 8, 8220. [Google Scholar] [CrossRef]
- Marchaim, U.; Krause, C. Propionic to acetic acid ratios in overloaded anaerobic digestion. Bioresour. Technol. 1993, 43, 195–203. [Google Scholar] [CrossRef]
- Zhu, H.; Qu, F.; Zhu, L.-H. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 1993, 21, 5279–5280. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data (BETA). Available online: https://biit.cs.ut.ee/clustvis/ (accessed on 18 September 2020).
- Kin, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar]
- Steinbusch, K.J.J.; Arvaniti, E.; Hamelers, H.V.M.; Buisman, C.J.N. Selective inhibition of methanogenesis to enhance ethanol and n-butyrate production through acetate reduction in mixed culture fermentation. Bioresour. Technol. 2009, 100, 3261–3267. [Google Scholar] [CrossRef]
- Kaur, G.; Johnravindar, D.; Wong, J.W.C. Enhanced volatile fatty acid degradation and methane production efficiency by biochar addition in food waste-sludge co-digestion: A step towards increased organic loading efficiency in co-digestion. Bioresour. Technol. 2020, 308, 123250. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Han, R.; Zhang, Y.; He, C.; Liu, H. Differentiated stimulating effects of activated carbon on methanogenic degradation of acetate, propionate and butyrate. Waste Manag. 2018, 76, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Redzwan, G.; Banks, C.J. An Evaluation of Soft-drink Wastewater Treatment by Anaerobic Digestion Process. Malays. J. Sci. 2007, 26, 35–42. [Google Scholar]
- Allison, C.; Macfarlane, G.T. Effect of nitrate on methane production and fermentation by slurries of human faecal bacteria. J. Gen. Microbiol. 1988, 134, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, P.C.; Kamra, N.; Agarwal, N.; Chaudhary, L.C. Effect of sodium nitrate and nitrate reducing bacteria on in vitro methane production and fermentation with buffalo rumen liquor. Asian Aust. J. Anim. Sci. 2012, 25, 812–817. [Google Scholar] [CrossRef]
- Zhou, Z.; Yu, Z.; Meng, Q. Effects on nitrate on methane production, fermentation, and microbial populations in in vitro ruminal cultures. Bioresour. Technol. 2012, 103, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Conrad, R. Effect of methanogenic precursors (acetate, hydrogen, propionate) on the suppression of methane production by nitrate in anoxic rice field soil. FEMS Microbiol. Ecol. 1999, 28, 49–61. [Google Scholar] [CrossRef]
- Niu, Q.; Qiao, W.; Qiang, H.; Li, Y.-Y. Microbial community shifts and biogas conversion computation during steady, inhibited and recovered stages of thermophilic methan fermentation on chicken manure with a wide variation of ammonia. Bioresour. Technol. 2013, 146, 223–233. [Google Scholar] [CrossRef]
- Gagliano, M.C.; Braguglia, C.M.; Rossetti, S. In situ identification of the synthrophic protein fermentative Coprothermobacter spp. involved in the themophilic anaerobic digestion process. FEMS Microbiol. Lett. 2014, 358, 55–63. [Google Scholar] [CrossRef]
- Gagliano, M.C.; Braguglia, C.M.; Petruccioli, M.; Rossetti, S. Ecology and biotechnological potential of the thermophilic fermentative Coprothermobacter spp. FEMS Microbiol. Ecol. 2015, 91, fiv018. [Google Scholar] [CrossRef]
- Hania, W.B.; Godbane, R.; Postec, A.; Hamdi, M.; Ollivier, B.; Fardeau, M.-L. Defluviitoga tunisiensis gen. nov., sp. nov., a thermophilic bacterium isolated from a mesothermic and anaerobic whey digester. Int. J. Syst. Evol. Microbiol. 2012, 62, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Wagner, I.D.; Brice, M.E.; Kevbrin, V.V.; Mills, G.L.; Romanck, C.S.; Wiegel, J. Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter lithoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru Margin. Extremophiles 2005, 9, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Fröschle, B.; Messelhäusser, U.; Höller, C.; Lebuhn, M. Fate of Clostridium botulinum and incidence of pathogenic clostridia in biogas processes. J. Appl. Microbiol. 2015, 119, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Wainaina, S.; Awasthi, L.M.K.; Taherzadeh, M.J. Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: A critical review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef]
- Huser, B.A.; Wuhrmann, K.; Zehnder, A.J.B. Methanothrix soehngenii gen. nov. sp. nov., a new acetotrophic non-hydrogen-oxidizing methane bacterium. Arch. Microbiol. 1982, 132, 1–9. [Google Scholar] [CrossRef]
- Ennouri, H.; Miladi, B.; Diaz, S.Z.; Güelfo, L.A.F.; Solera, R.; Hamdi, M.; Bouallagui, H. Effect of thermal pretreatment on the biogas production and microbial communities balance during anaerobic digestion of urban and industrial waste activated sludge. Bioresour. Technol. 2016, 214, 184–191. [Google Scholar] [CrossRef]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical background and biotechnological applications. AMB Expr. 2018, 8, 1. [Google Scholar] [CrossRef]
- Scully, S.M.; Orlygsson, J. Brabched-chain alcohol formation from branched-chain amino acids by Thermoanaerobacter brockii and Themoanaerobacter yonseiensis. Anaerobe 2014, 30, 82–84. [Google Scholar] [CrossRef]
- Ma, K.; Liu, X.; Dong, X. Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int. J. Syst. Evol. Microbiol. 2006, 56, 127–131. [Google Scholar] [CrossRef]
- Huang, Y.L.; Mann, K.; Novak, J.M.; Yang, S.-T. Acetic acid production from fructose by Clostridium formicoaceticum immobilized in a fibrous-bed bioreactor. Biotechnol. Prog. 1998, 14, 800–806. [Google Scholar] [CrossRef]
- Hahnke, S.; Langer, T.; Koeck, D.E.; Klocke, M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 2016, 66, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.; Gößner, A.S.; Drake, H.L. Anaerobic trophic interactions of contrasting methane-emitting mire soils: Processes versus taxa. FEMS Microbiol. Ecol. 2015, 91, fiv045. [Google Scholar] [CrossRef]
- Ueki, A.; Akasaka, H.; Suzuki, D.; Ueki, K. Paludibacter propionicigenes gen. nov., sp. nov., a novel strictly anaerobic, Gram-negative, propionate-producing bacterium isolated from plant residue in irrigated rice-field soil in Japan. Int. J. Syst. Evol. Microbiol. 2006, 56, 39–44. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; Yao, Q. Contrasting P acquisition strategies of the bacterial communities associated with legume and grass in subtropical orchard soil. Environ. Microbiol. Rep. 2018, 10, 310–319. [Google Scholar] [CrossRef]
- Mitani, Y.; Lezhava, A.; Kawai, Y.; Kikuchi, T.; Oguchi-Katayama, A.; Kogo, Y.; Itoh, M.; Miyagi, T.; Takakura, H.; Hoshi, K.; et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat. Method. 2007, 4, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Lezhava, A.; Ishidao, T.; Ishizu, Y.; Naito, K.; Hanami, T.; Katayama, A.; Kogo, Y.; Soma, T.; Ikeda, S.; Murakami, K.; et al. Exciton primer-mediated SNP detection in SmartAmp2 reactions. Hum. Mutat. 2010, 31, 208–217. [Google Scholar] [CrossRef]
| Run | SBWM (mL) | Sterilizaed Water (mL) | Inoculum (mL) |
|---|---|---|---|
| A | 0 | 10.0 | 10.0 |
| B | 0.5 | 9.5 | 10.0 |
| C | 2.0 | 8.0 | 10.0 |
| D | 5.0 | 5.0 | 10.0 |
| E | 10.0 | 0 | 10.0 |
| Run | Simpson index | Shannon index | ||||
|---|---|---|---|---|---|---|
| Day 5 | Day 10 | Day 15 | Day 5 | Day 10 | Day 15 | |
| A | 0.630 | 0.629 | 0.556 | 1.900 | 1.872 | 1.517 |
| B | 0.587 | 0.640 | 0.663 | 1.833 | 1.988 | 1.983 |
| C | 0.627 | 0.636 | 0.713 | 1.929 | 1.925 | 2.181 |
| D | 0.721 | 0.708 | 0.725 | 2.293 | 2.248 | 2.369 |
| E | 0.670 | 0.667 | 0.652 | 2.023 | 1.959 | 1.834 |
| Culture Period (Day) | Run A | Run B | Run C | Run D | Run E | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 0 | 5 | 10 | 15 | 0 | 5 | 10 | 15 | 0 | 5 | 10 | 15 | 0 | 5 | 10 | 15 | |
| Species | ||||||||||||||||||||
| Candidatus Solibacter usitatus | 0.007 | 0.004 | 0.002 | 0.003 | 0.007 | 0.005 | 0.004 | 0.004 | 0.007 | 0.019 | 0.005 | 0.004 | 0.007 | 0.066 | 0.008 | 0.005 | 0.007 | 0.007 | 0.287 | 0.017 |
| Clostridium chauvoei | VL | VL | VL | 0.002 | VL | 0.263 | 0.185 | 0.073 | VL | 0.300 | 0.327 | 0.076 | VL | 1.982 | 0.802 | 0.634 | VL | 3.696 | 0.874 | 0.882 |
| Clostridium formicaceticum | 0.004 | 0.002 | 0.001 | VL | 0.004 | VL | VL | 0.001 | 0.004 | 0.004 | 0.001 | 0.001 | 0.004 | 0.017 | 0.003 | 0.006 | 0.004 | VL | 0.101 | 0.005 |
| Methanosaeta harundinacea | 0.256 | 0.186 | 0.168 | 0.122 | 0.256 | 0.183 | 0.338 | 0.434 | 0.256 | 0.153 | 0.269 | 0.586 | 0.256 | 0.119 | 0.145 | 0.202 | 0.256 | 0.050 | 0.037 | 0.020 |
| Methanothrix soehngenii | 0.945 | 0.635 | 0.422 | 0.001 | 0.945 | 0.566 | 1.164 | 1.358 | 0.945 | 0.517 | 0.947 | 2.126 | 0.945 | 0.364 | 0.442 | 0.734 | 0.945 | 0.147 | 0.110 | 0.072 |
| Neorickettsia helminthoeca | 0.027 | 0.031 | 0.019 | 0.028 | 0.027 | 0.061 | 0.052 | 0.080 | 0.027 | 0.137 | 0.157 | 0.143 | 0.027 | 0.002 | 0.376 | 0.291 | 0.027 | 0.002 | 0.001 | VL |
| Paludibacter propionicigenes | ND | 0.001 | ND | 0.002 | ND | 0.005 | 0.006 | 0.008 | ND | 0.027 | 0.032 | 0.038 | ND | 0.018 | 0.110 | 0.108 | ND | ND | ND | 0.109 |
| Petrimonas mucosa | 0.004 | 0.002 | 0.002 | 0.241 | 0.004 | 0.012 | 0.021 | 0.015 | 0.004 | 0.071 | 0.100 | 0.099 | 0.004 | 0.051 | 0.265 | 0.304 | 0.004 | ND | VL | 0.262 |
 < 0.001;
< 0.001;  0.001–0.01;
0.001–0.01;  0.01–0.1;
0.01–0.1;  0.1–1.0;
0.1–1.0;  >1.0. The blank (white) panels indicate ‘not changed by more than 10-fold’ in the runs. ND and VL indicate ‘not detectable’ and ‘very low level less than 0.001%’, respectively.
>1.0. The blank (white) panels indicate ‘not changed by more than 10-fold’ in the runs. ND and VL indicate ‘not detectable’ and ‘very low level less than 0.001%’, respectively.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuda, S.; Yamato, T.; Mochizuki, Y.; Sekiguchi, Y.; Ohtsuki, T. Batch-Mode Analysis of Thermophilic Methanogenic Microbial Community Changes in the Overacidification Stage in Beverage Waste Treatment. Int. J. Environ. Res. Public Health 2020, 17, 7514. https://doi.org/10.3390/ijerph17207514
Matsuda S, Yamato T, Mochizuki Y, Sekiguchi Y, Ohtsuki T. Batch-Mode Analysis of Thermophilic Methanogenic Microbial Community Changes in the Overacidification Stage in Beverage Waste Treatment. International Journal of Environmental Research and Public Health. 2020; 17(20):7514. https://doi.org/10.3390/ijerph17207514
Chicago/Turabian StyleMatsuda, Shuhei, Takahiro Yamato, Yoshiyuki Mochizuki, Yoshinori Sekiguchi, and Takashi Ohtsuki. 2020. "Batch-Mode Analysis of Thermophilic Methanogenic Microbial Community Changes in the Overacidification Stage in Beverage Waste Treatment" International Journal of Environmental Research and Public Health 17, no. 20: 7514. https://doi.org/10.3390/ijerph17207514
APA StyleMatsuda, S., Yamato, T., Mochizuki, Y., Sekiguchi, Y., & Ohtsuki, T. (2020). Batch-Mode Analysis of Thermophilic Methanogenic Microbial Community Changes in the Overacidification Stage in Beverage Waste Treatment. International Journal of Environmental Research and Public Health, 17(20), 7514. https://doi.org/10.3390/ijerph17207514






