Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Materials and Methods
2.1. Fluoroquinolones and Metalloantibiotics Preparation
2.2. Antimicrobial Activity Assays
2.2.1. Bacterial Strains and Culture Conditions
2.2.2. Determination of Minimum Inhibitory Concentrations (MICs)
2.2.3. Combination Synergy Study
Disk Diffusion Method
Checkerboard Method
2.3. Enzymatic Inhibitory Activity Assays
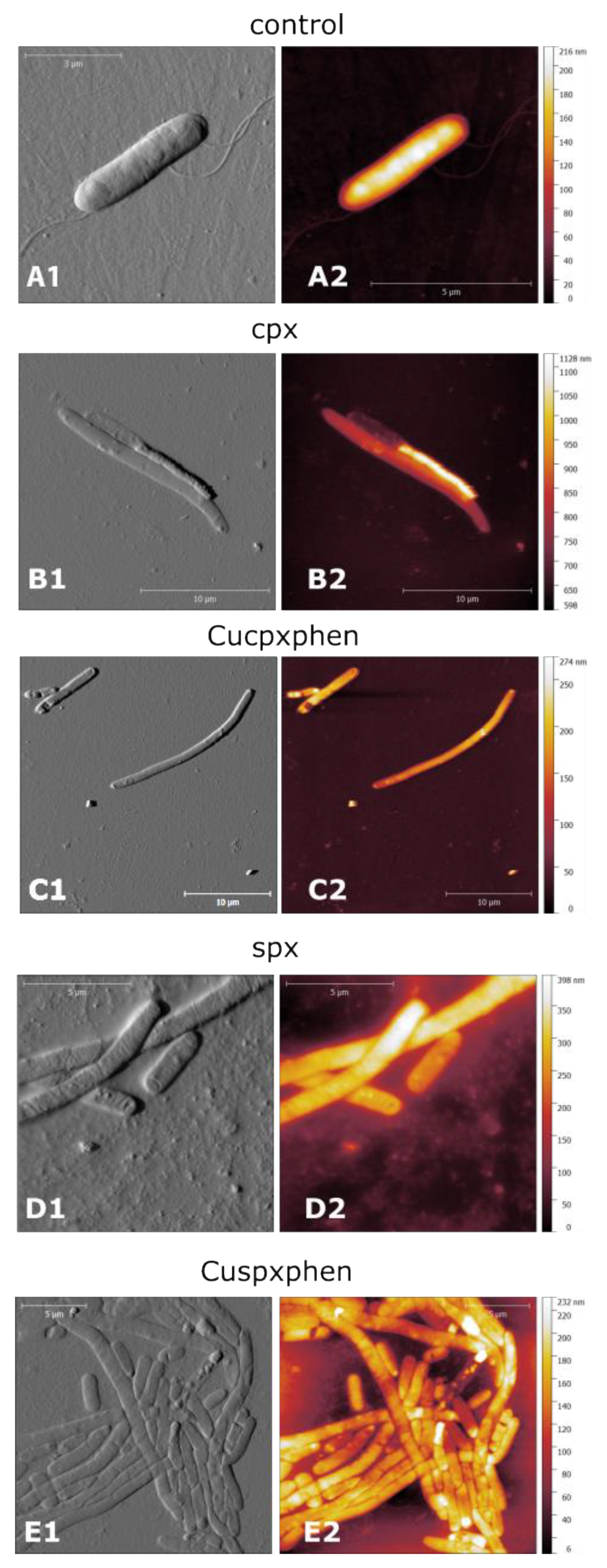
2.4. Evaluation of the Effect of Compounds on Bacterial Membranes by Atomic Force Microscopy
3. Results and Discussion
3.1. Antimicrobial Activity against Multidrug-Resistant Clinical Isolates
3.2. Combinatorial Effect of Metalloantibiotics with Antibiotics
3.3. Studies of the Mechanism of Action of the Metalloantibiotics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernández, L.; Hancock, R.E.W. Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Graef, F.; Vukosavljevic, B.; Michel, J.-P.; Wirth, M.; Ries, O.; De Rossi, C.; Windbergs, M.; Rosilio, V.; Ducho, C.; Gordon, S.; et al. The bacterial cell envelope as delimiter of anti-infective bioavailability – An in vitro permeation model of the Gram-negative bacterial inner membrane. J. Control. Release 2016, 243, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Strahl, H.; Errington, J. Bacterial Membranes: Structure, Domains, and Function. Annu. Rev. Microbiol. 2017, 71, 519–538. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Epand, R.F. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta Biomembr. 2009, 1788, 289–294. [Google Scholar] [CrossRef]
- Willey, J.M.; Sherwood, L.M.; Woolverton, C.J. Prescott, Harley, and Klein’s Microbiology, 7th ed.; McGraw-Hill Higher Education: New York, NY, USA, 2008; pp. 44–61. [Google Scholar]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef]
- Masi, M.; Pagès, J.-M. Structure, Function and Regulation of Outer Membrane Proteins Involved in Drug Transport in Enterobactericeae: The OmpF/C–TolC Case. Open Microbiol. J. 2013, 7, 22–33. [Google Scholar] [CrossRef]
- Rollauer, S.E.; Sooreshjani, M.A.; Noinaj, N.; Buchanan, S.K. Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.I.; MacGowan, A.P. Development of the quinolones. J. Antimicrob. Chemother. 2003, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, C.M.; Green, G.M. Quinolones: A comprehensive review. Am. Fam. Physician 2002, 65, 455–464. [Google Scholar] [PubMed]
- Appelbaum, P.C.; Hunter, P.A. The fluoroquinolone antibacterials: Past, present and future perspectives. Int. J. Antimicrob. Agents 2000, 16, 5–15. [Google Scholar] [CrossRef]
- Masi, M.; Réfregiers, M.; Pos, K.M.; Pagès, J.-M. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2017, 2, 17001. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of Quinolone Action and Resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef]
- Feio, M.J.; Sousa, I.; Ferreira, M.; Cunha-Silva, L.; Saraiva, R.G.; Queirós, C.; Alexandre, J.G.; Claro, V.; Mendes, A.; Ortiz, R.; et al. Fluoroquinolone–metal complexes: A route to counteract bacterial resistance? J. Inorg. Biochem. 2014, 138, 129–143. [Google Scholar] [CrossRef]
- Saraiva, R.; Lopes, S.; Ferreira, M.; Novais, F.; Pereira, E.; Feio, M.J.; Gameiro, P. Solution and biological behaviour of enrofloxacin metalloantibiotics: A route to counteract bacterial resistance? J. Inorg. Biochem. 2010, 104, 843–850. [Google Scholar] [CrossRef]
- Sousa, I.; Claro, V.; Pereira, J.L.; Amaral, A.L.; Cunha-Silva, L.; de Castro, B.; Feio, M.J.; Pereira, E.; Gameiro, P. Synthesis, characterization and antibacterial studies of a copper(II) levofloxacin ternary complex. J. Inorg. Biochem. 2012, 110, 64–71. [Google Scholar] [CrossRef]
- Gameiro, P.; Rodrigues, C.; Baptista, T.; Sousa, I.; de Castro, B. Solution studies on binary and ternary complexes of copper(II) with some fluoroquinolones and 1,10-phenanthroline: Antimicrobial activity of ternary metalloantibiotics. Int. J. Pharm. 2007, 334, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gil, J.; Perelló, L.; Ortiz, R.; Alzuet, G.; González-Álvarez, M.; Liu-González, M. Synthesis, structure and biological properties of several binary and ternary complexes of copper(II) with ciprofloxacin and 1,10 phenanthroline. Polyhedron 2009, 28, 138–144. [Google Scholar] [CrossRef]
- Serafin, A.; Stańczak, A. The complexes of metal ions with fluoroquinolones. Russ. J. Coord. Chem. 2009, 35, 81–95. [Google Scholar] [CrossRef]
- Ruíz, P.; Ortiz, R.; Perelló, L.; Alzuet, G.; González-Álvarez, M.; Liu-González, M.; Sanz-Ruíz, F. Synthesis, structure, and nuclease properties of several binary and ternary complexes of copper(II) with norfloxacin and 1,10 phenantroline. J. Inorg. Biochem. 2007, 101, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Efthimiadou, E.K.; Sanakis, Y.; Katsarou, M.; Raptopoulou, C.P.; Karaliota, A.; Katsaros, N.; Psomas, G. Neutral and cationic mononuclear copper(II) complexes with enrofloxacin: Structure and biological activity. J. Inorg. Biochem. 2006, 100, 1378–1388. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Available online: https://www.who.int/ (accessed on 11 December 2019).
- Ferreira, M.; Gameiro, P. Ciprofloxacin Metalloantibiotic: An Effective Antibiotic with an Influx Route Strongly Dependent on Lipid Interaction? J. Membr. Biol. 2015, 248, 125–136. [Google Scholar] [CrossRef]
- Lopes, S.C.; Ribeiro, C.; Gameiro, P. A New Approach to Counteract Bacteria Resistance: A Comparative Study Between Moxifloxacin and a New Moxifloxacin Derivative in Different Model Systems of Bacterial Membrane. Chem. Biol. Drug Des. 2013, 81, 265–274. [Google Scholar] [CrossRef]
- Ribeiro, C.; Lopes, S.C.; Gameiro, P. New insights into the translocation route of enrofloxacin and its metalloantibiotics. J. Membr. Biol. 2011, 241, 117–125. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. CLSI document M07-A10, Approved Standard-Tenth ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Sopirala, M.M.; Mangino, J.E.; Gebreyes, W.A.; Biller, B.; Bannerman, T.; Balada-Llasat, J.-M.; Pancholi, P. Synergy Testing by Etest, Microdilution Checkerboard, and Time-Kill Methods for Pan-Drug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4678–4683. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100S, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Chalkley, L.J.; Koornhof, H.J. Antimicrobial activity of ciprofloxacin against Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus determined by the killing curve method: Antibiotic comparisons and synergistic interactions. Antimicrob. Agents Chemother. 1985, 28, 331. [Google Scholar] [CrossRef] [PubMed]
- Rey-Mellano, M.E.; Senoro, D.B.; Tayo, L.L.; Wan, M.-W. Adsorption of Cu (II) and Ni(II) in Aqueous Solution using Biofilm Supported with Kaolinite Clay. In Proceedings of the 6th International Conference on Biological, Chemical & Environmental Sciences (BCES-2016), Pattaya, Thailand, 8–9 August 2016. [Google Scholar]
- Gugala, N.; Lemire, J.; Turner, R. The efficacy of different anti-microbial metals at preventing the formation of, and eradicating bacterial biofilms of pathogenic indicator strains. J. Antibiot. 2017, 70, 775–780. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.1. 2018. Available online: http://www.eucast.org (accessed on 16 December 2019).
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Tamil Selvi, P.; Palaniandavar, M. Copper(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Sousa, C.F.; Ferreira, M.; Abreu, B.; Medforth, C.J.; Gameiro, P. Interactions of a non-fluorescent fluoroquinolone with biological membrane models: A multi-technique approach. Int. J. Pharm. 2015, 495, 761–770. [Google Scholar] [CrossRef]
- Eaton, P.; West, P. Atomic Force Microscopy, 1st ed.; Oxford University Press: New York, NY, USA, 2010; pp. 164–167. [Google Scholar]
- Diver, J.M.; Wise, R. Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin. J. Antimicrob. Chemother. 1986, 18, 31–41. [Google Scholar] [CrossRef]
- Silva, F.; Lourenço, O.; Queiroz, J.; Domingues, F.C. Bacteriostatic versus bactericidal activity of ciprofloxacin in Escherichia coli assessed by flow cytometry using a novel far-red dye. J. Antibiot. 2011, 64, 321–325. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef]
- Monson, B.K.; Stringham, J.; Jones, B.B.; Abdel-Aziz, S.; Peck, C.M.C.; Olson, R.J. Scanning Electron Microscopy Visualization of Methicillin-Resistant Staphylococcus aureus After Contact With Gatifloxacin With and Without Preservative. J. Ocul. Pharmacol.Ther. 2010, 26, 133–136. [Google Scholar] [CrossRef]
- Greenwood, D.; O’Grady, F. Scanning Electron Microscopy of Staphylococcus aureus Exposed to Some Common Anti-staphylococcal Agents. J. Gen. Microbiol. 1972, 70, 263–270. [Google Scholar] [CrossRef] [PubMed]


| Compound | MIC Value (μmol dm−3) | |||
|---|---|---|---|---|
| E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | S. aureus ATCC 25923 | S. aureus ATCC 29213 | |
| cpx | 0.012 | 0.18 | 0.36–0.75 | 0.75–1.51 |
| Cucpxphen | 0.011 | 0.17–0.35 | 0.35–0.71 | 1.41 |
| erx | 0.022 | 2.78 | 0.33–0.70 | 0.33 |
| Cuerxphen | 0.022 | 2.97 | 0.37–0.74 | 0.18–0.37 |
| lvx | 0.022 | 1.38 | 0.33–0.69 | 0.33 |
| Culvxphen | 0.021–0.042 | 1.39 | 0.35–0.69 | 0.35 |
| mxfx | 0.018 | 1.14–2.28 | 0.07–0.14 | 0.14 |
| Cumxfxphen | 0.019–0.038 | 2.54 | 0.08–0.15 | 0.08–0.15 |
| spx | 0.010 | 0.64–1.27 | 0.08–0.15 | 0.15 |
| Cuspxphen | 0.005–0.010 | 0.65 | 0.32 | 0.08–0.16 |
| phen | 40.4 | 645.7 | 80.7–161.4 | 161.4 |
| Cu(II)/phen (1:1) | 72.8 | ≥1164.1 | 72.8 | 145.5 |
| Cu(NO3)2.3H2O | ≥4238.2 | ≥4238.2 | ≥4238.2 | ≥3642.2 |
| Compound | MIC Value (μmol dm−3) | |
|---|---|---|
| HSJ Ec002 | HSJ Ec003 | |
| cpx | 386.3 | 193.2 |
| Cucpxphen | 93.0 | 45.1–90.3 |
| erx | 178.1 | 178.1 |
| Cuerxphen | 95.0 | 95.0 |
| lvx | 22.1 | 88.6 |
| Culvxphen | 44.4 | 44.4–88.7 |
| mxfx | 18.3 | 18.3 |
| Cumxfxphen | 20.3 | 40.6 |
| spx | 40.8 | 81.6 |
| Cuspxphen | 41.6 | 83.1 |
| phen | 80.7 | 40.4 |
| Cu(II)/phen (1:1) | 145.5 | 72.8 |
| Cu(NO3)2.3H2O | >4238.2 | ≥4238.2 |
| Compound | MIC Value (μmol dm−3) | |||
|---|---|---|---|---|
| Sa1-SA3 | Sa3-SA3 | 19/35 | 26/01 | |
| cpx | 386.3 | 386.3–772.6 | ≥3090.5 | 1545.2 |
| Cucpxphen | 90.3 | 90.3 | 180.5 | 180.5 |
| erx | 22.3 | 44.5–89.0 | 178.1 | 712.3 |
| Cuerxphen | 95.0 | 95.0 | 95.0 | 95.0 |
| lvx | 708.4 | 44.3 | 177.1 | 1416.8 |
| Culvxphen | 88.7 | 88.7–177.5 | 88.7 | 88.7 |
| mxfx | 18.3 | 292.3 | 18.3 | 73.1 |
| Cumxfxphen | 10.2 | 10.2 | 20.3 | 40.6 |
| spx | 652.4 | 20.4 | 163.1 | 652.4 |
| Cuspxphen | 83.1–166.2 | 41.6 | 41.6 | 83.1 |
| phen | 2582.9 | 645.7 | 322.9 | 322.9–645.7 |
| Cu(II)/phen (1:1) | 72.8 | 72.8 | 145.5 | 145.5 |
| Cu(NO3)2.3H2O | ≥4238.2 | ≥4238.2 | ≥3642.2 | ≥3642.2 |
| Compound | Compound Alone | Compound + cpx | Compound + amp |
|---|---|---|---|
| MRSA Sa1-SA3 | |||
| Cucpxphen | 10.5–11 | 10 | 12 |
| Cuspxphen | 12 | 11 | 12 |
| phen | 0 | 0 | 12 |
| Cu(II)/phen (1:1) | 9–11 | 9 | 12 |
| Cu(NO3)2.3H2O | 0 | 0 | 10 |
| cpx (5 µg/disk) | 0 | - | - |
| amp (10 µg/disk) | 0 | - | - |
| MRSA Sa3-SA3 | |||
| Cucpxphen | 9–12 | 12 | 12 |
| Cuspxphen | 13–14 | 14 | 14 |
| phen | 0 | 0 | 10 |
| Cu(II)/phen (1:1) | 0 | 0 | 10 |
| Cu(NO3)2.3H2O | 8–9 | 9 | 11 |
| cpx (5 µg/disk) | 0 | - | - |
| amp (10 µg/disk) | 10 | - | - |
| MIC Value (μg mL−1) | ∑ FIC | ||||
|---|---|---|---|---|---|
| Alone | In Combination | ||||
| Clinical isolate | Cuspxphen | cpx | Cuspxphen | cpx | |
| HSJ Ec002 | 32 | 128 | 32 | 128 | 2 (I) |
| Clinical isolate | Cucpxphen | amp | Cucpxphen | amp | |
| Sa1-SA3 | 64 | 64 | 8 | 32 | 0.625 (A) |
| Clinical isolate | Cucpxphen | cpx | Cucpxphen | cpx | |
| Sa3-SA3 | 64 | 256 | 16 | 64 | 0.5 (S) |
| Clinical isolate | Cucpxphen | amp | Cucpxphen | amp | |
| Sa3-SA3 | 64 | 256 | 32 | 128 | 1 (A) |
| Assay | Bacterial Enzyme | Concentration of Compound Able to Inhibit the Enzyme/µmol dm−3 | ||
|---|---|---|---|---|
| cpx | Cucpxphen | Cuspxphen | ||
| DNA gyrase supercoiling inhibition assay | E. coli | 5 | 5 | 5 |
| S. aureus | 50 | 50 | 50 | |
| Topoisomerase IV relaxation inhibition assay | E. coli | 10 | 5–10 | 5–10 |
| S. aureus | 10 | 5 | 5 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, M.; Bessa, L.J.; Sousa, C.F.; Eaton, P.; Bongiorno, D.; Stefani, S.; Campanile, F.; Gameiro, P. Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 2020, 17, 3127. https://doi.org/10.3390/ijerph17093127
Ferreira M, Bessa LJ, Sousa CF, Eaton P, Bongiorno D, Stefani S, Campanile F, Gameiro P. Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus. International Journal of Environmental Research and Public Health. 2020; 17(9):3127. https://doi.org/10.3390/ijerph17093127
Chicago/Turabian StyleFerreira, Mariana, Lucinda J. Bessa, Carla F. Sousa, Peter Eaton, Dafne Bongiorno, Stefania Stefani, Floriana Campanile, and Paula Gameiro. 2020. "Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus" International Journal of Environmental Research and Public Health 17, no. 9: 3127. https://doi.org/10.3390/ijerph17093127
APA StyleFerreira, M., Bessa, L. J., Sousa, C. F., Eaton, P., Bongiorno, D., Stefani, S., Campanile, F., & Gameiro, P. (2020). Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus. International Journal of Environmental Research and Public Health, 17(9), 3127. https://doi.org/10.3390/ijerph17093127










