Potential Human Health Hazard of Post-Hurricane Harvey Sediments in Galveston Bay and Houston Ship Channel: A Case Study of Using In Vitro Bioactivity Data to Inform Risk Management Decisions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sediment Deposition Model
2.2. Chemicals and Biologicals
2.3. Sediment Sample Collection, Extraction, and Bioactivity Screening
2.4. Concentration-Response Modeling and Data Integration
2.5. Associations of Sediment Spatial Locations and Biological/PAH Measurements
2.6. Mapping of PAH Concentrations and In Vitro Bioactivity Data
2.7. Prediction between Chemical and Biological Measurements
2.8. Hazard Index and Cancer Risk Calculation for PAHs
2.9. Comparing Alternative Remediation Goals
3. Results and Discussion
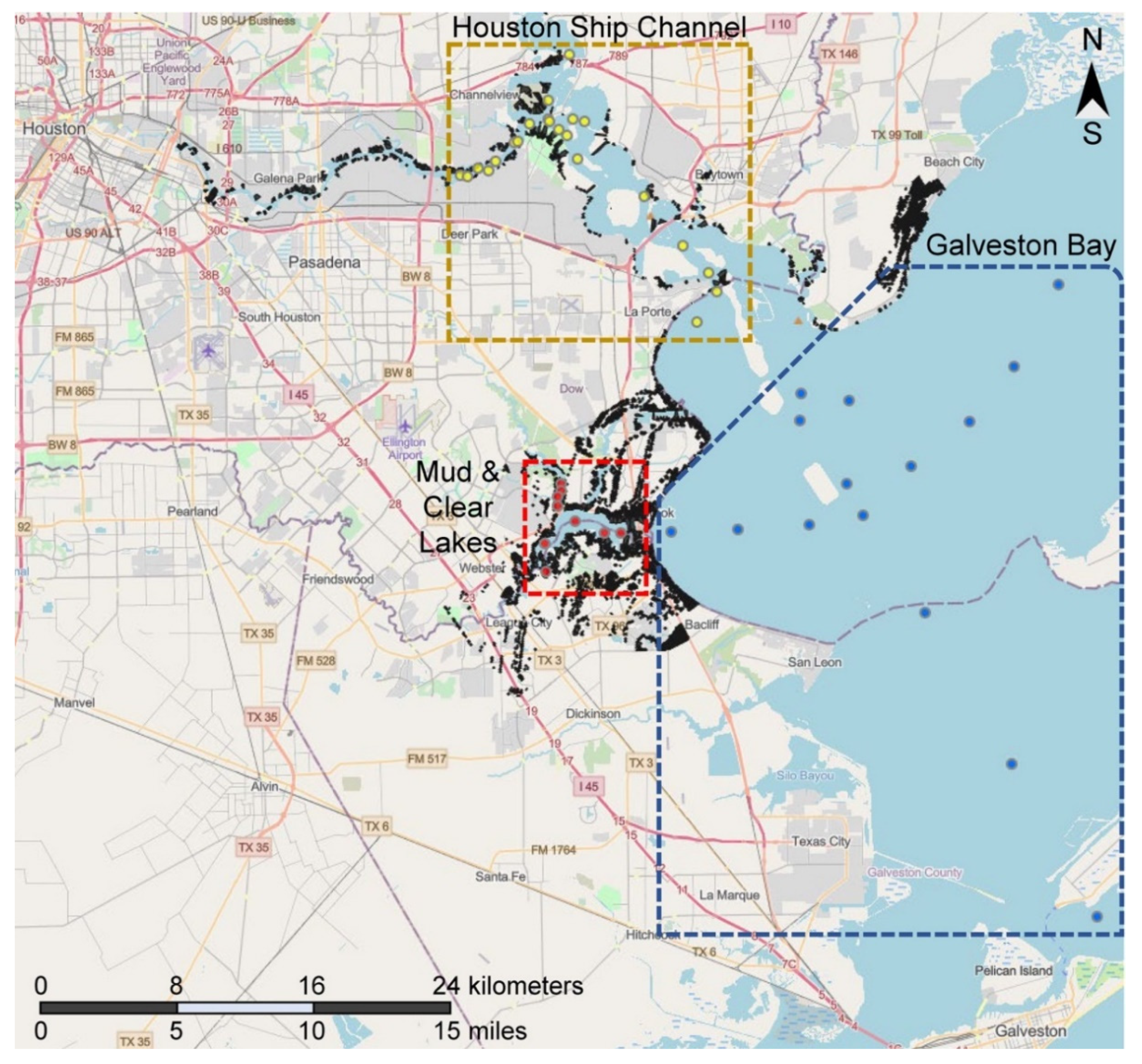
3.1. Study Area and Sample Selection Rationale
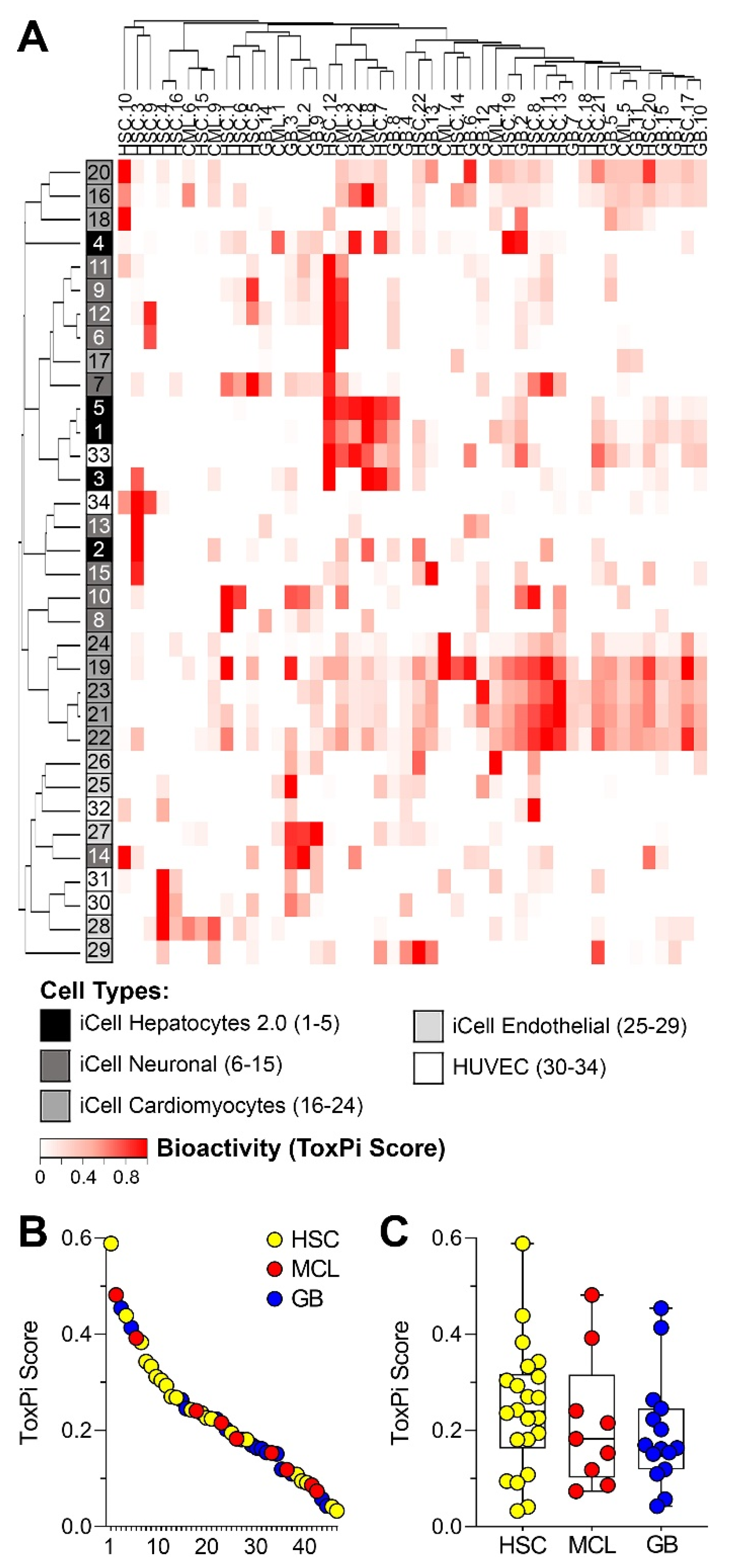
3.2. Bioactivity of Sediment Samples
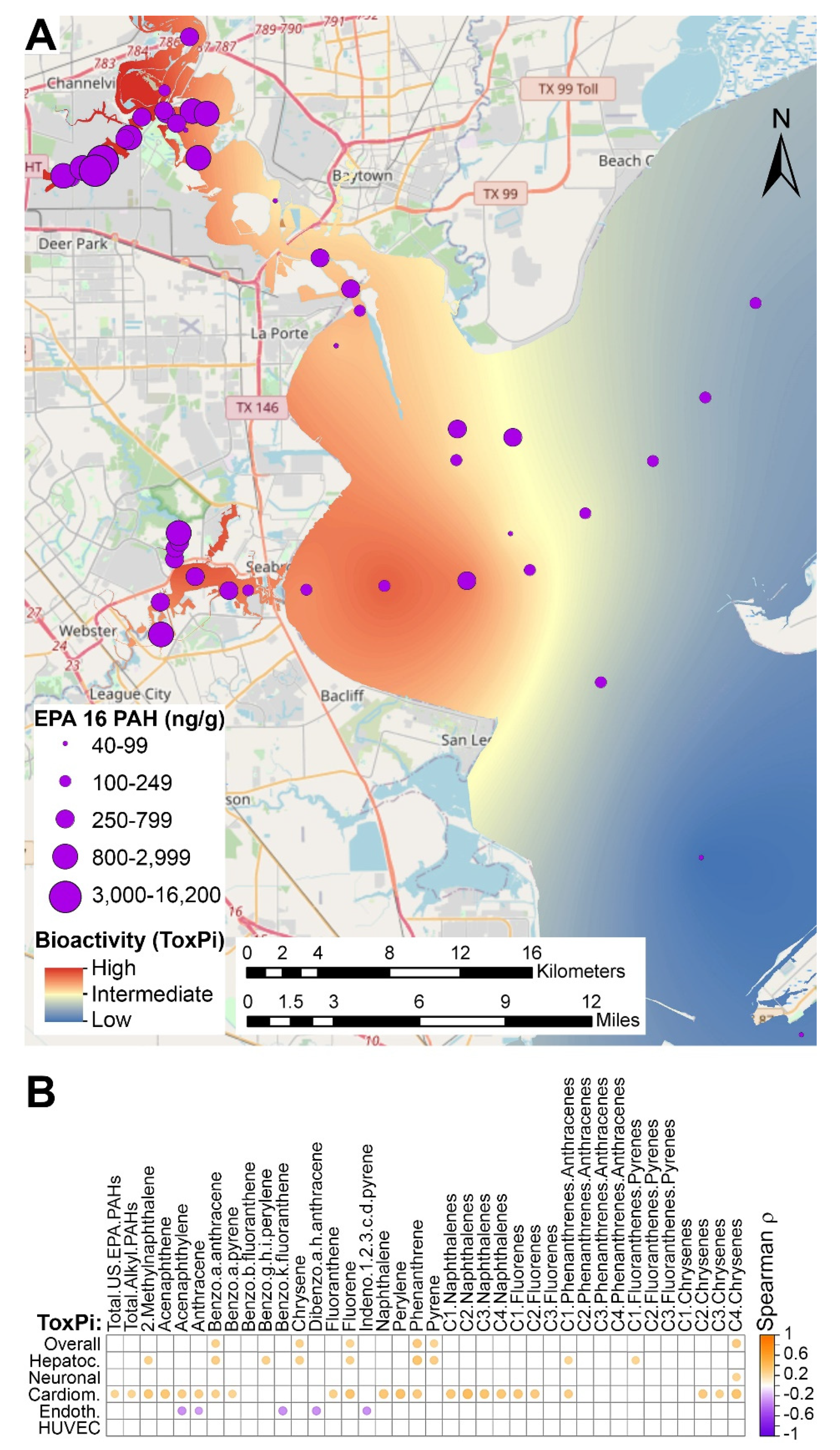
3.3. Spatial Associations of Bioactivity and PAH Concentration Data in Sediment Samples
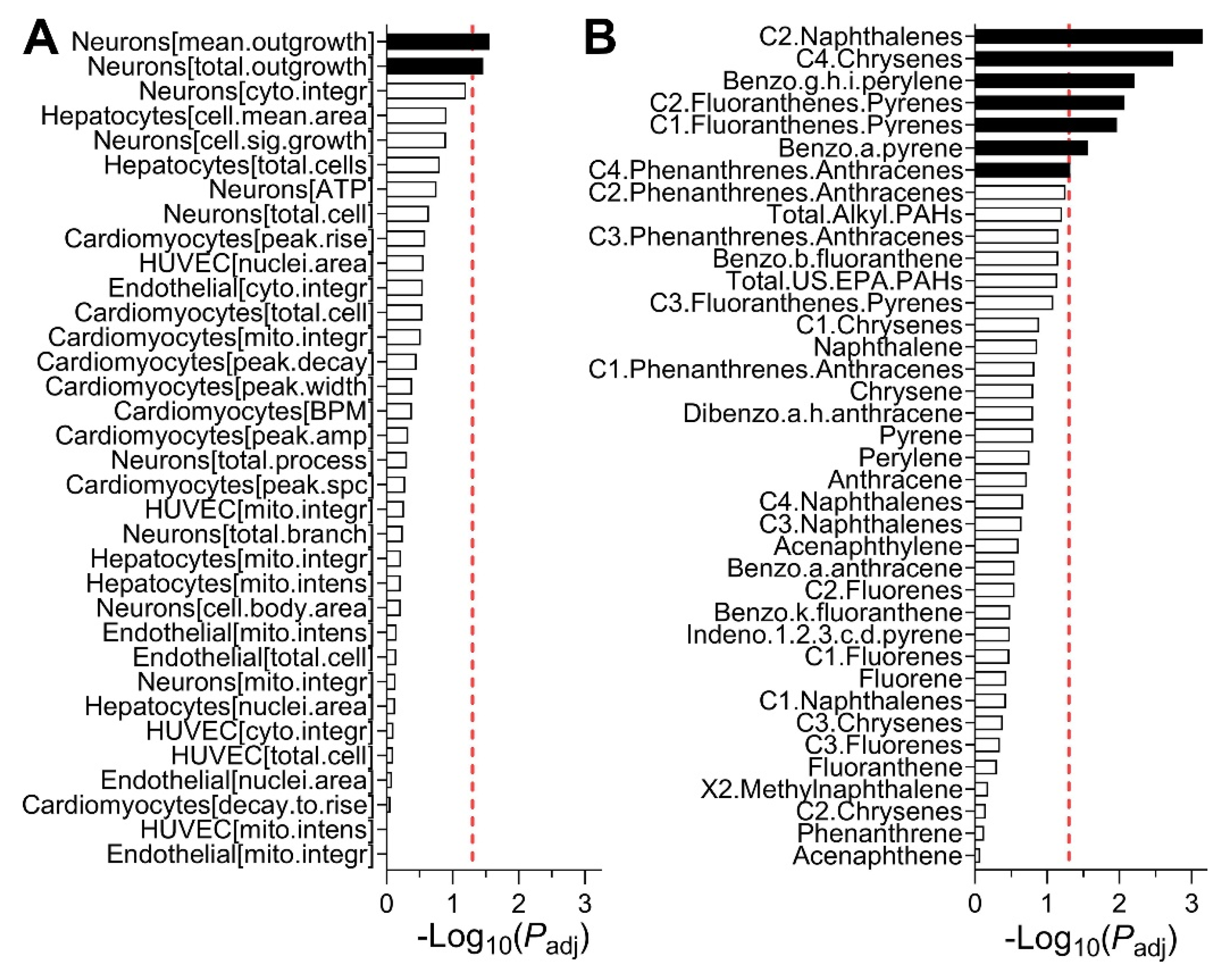
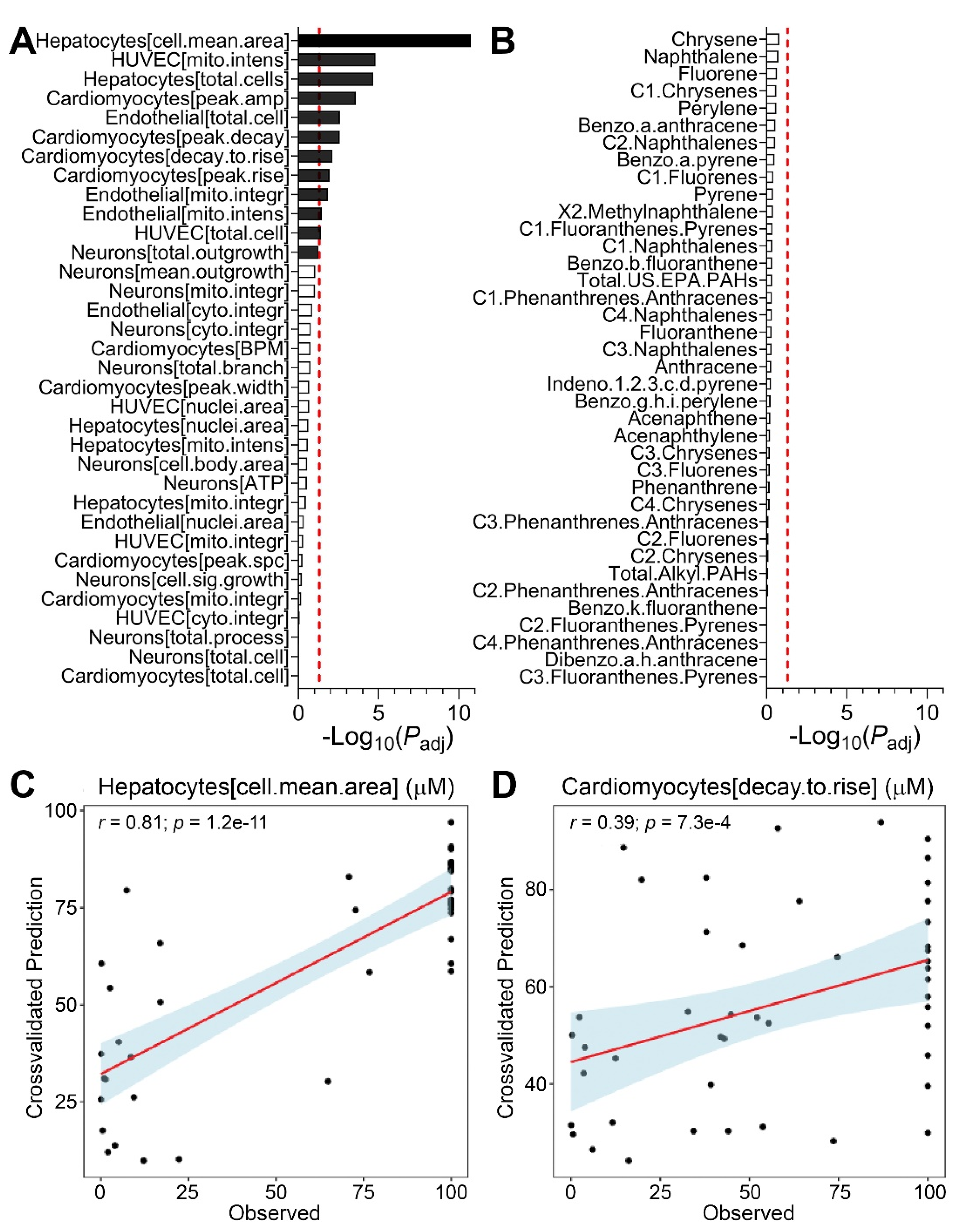
3.4. Predictions of Bioactivity and PAH Concentrations in Sediment Samples
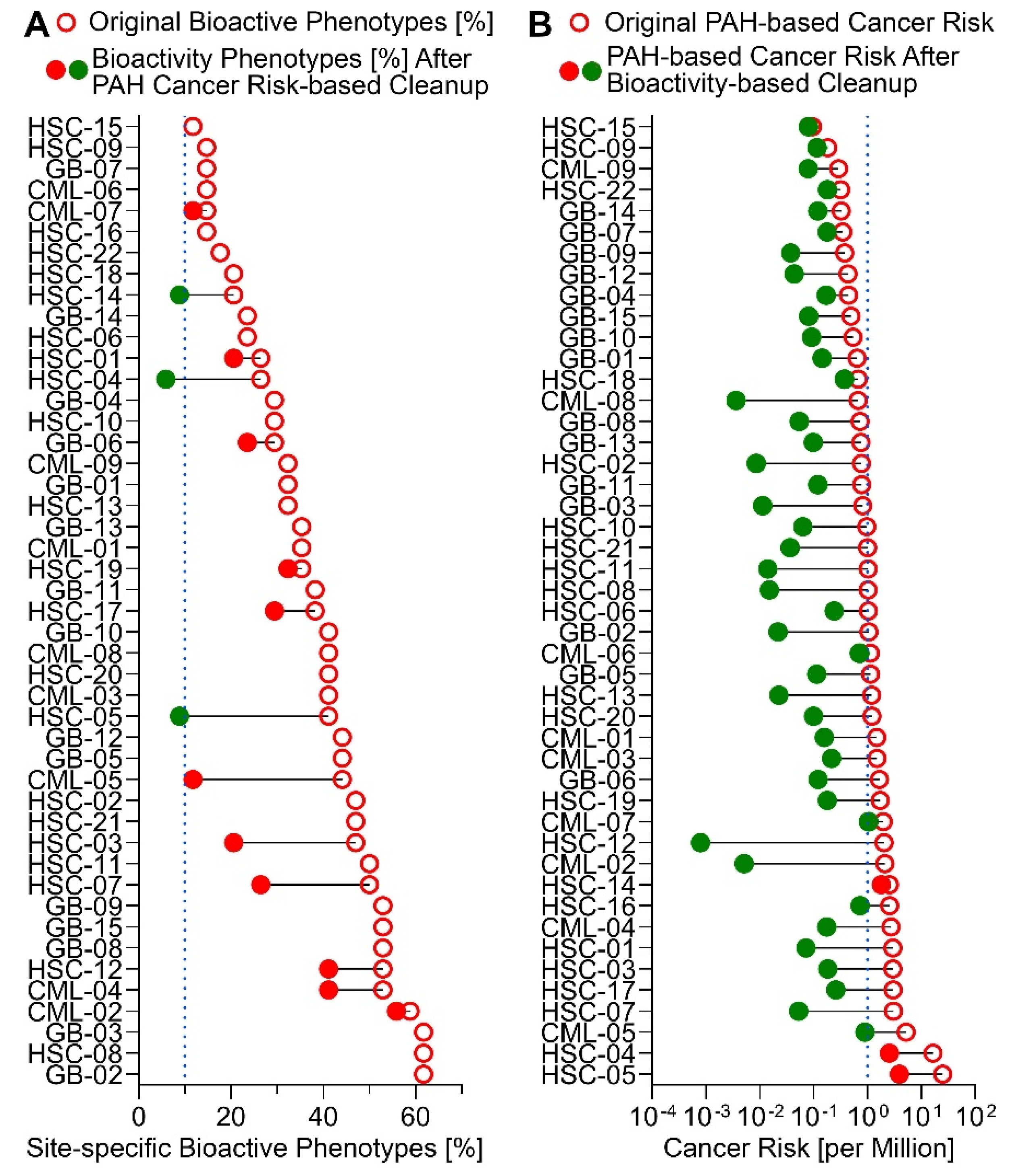
3.5. Risk Characterization and Site Remediation Scenarios from In Vitro Bioactivity or PAH data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CR | cancer risk |
| DMSO | dimethyl sulfoxide |
| GB | Galveston Bay |
| HI | hazard index |
| HSC | Houston Ship Channel |
| HUVEC | human umbilical vein endothelial cell |
| iPSC | induced pluripotent stem cell |
| NAM | new approach method |
| PAH | Polycyclic Aromatic Hydrocarbon |
| PCB | polycyclic biphenyls |
| POD | point of departure |
| RSL | regional screening levels |
| ToxPi | toxicological priority index |
References
- Knap, A.H.; Rusyn, I. Environmental exposures due to natural disasters. Rev. Environ. Health 2016, 31, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Santschi, P.H.; Presley, B.J.; Wade, T.L.; Garcia-Romero, B.; Baskaran, M. Historical contamination of PAHs, PCBs, DDTs, and heavy metals in Mississippi River Delta, Galveston Bay and Tampa Bay sediment cores. Mar. Environ. Res. 2001, 52, 51–79. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Steichen, J.; Kamalanathan, M.; Windham, R.; Lubguban, A.; Labonte, J.M.; Kaiser, K.; Hala, D.; Santschi, P.H.; Quigg, A. Polycyclic aromatic hydrocarbons (PAHs) and putative PAH-degrading bacteria in Galveston Bay, TX (USA), following Hurricane Harvey (2017). Environ. Sci. Pollut. Res. Int. 2020, 27, 34987–34999. [Google Scholar] [CrossRef] [PubMed]
- Camargo, K.; Sericano, J.L.; Bhandari, S.; Hoelscher, C.; McDonald, T.J.; Chiu, W.A.; Wade, T.L.; Dellapenna, T.M.; Liu, Y.; Knap, A.H. Polycyclic aromatic hydrocarbon status in post-hurricane Harvey sediments: Considerations for environmental sampling in the Galveston Bay/Houston Ship Channel region. Mar. Pollut. Bull. 2021, 162, 111872. [Google Scholar] [CrossRef]
- Sansom, G.T.; Kirsch, K.R.; Casillas, G.A.; Camargo, K.; Wade, T.L.; Knap, A.H.; Baker, E.S.; Horney, J.A. Spatial Distribution of Polycyclic Aromatic Hydrocarbon Contaminants after Hurricane Harvey in a Houston Neighborhood. J. Health Pollut. 2021, 11, 210308. [Google Scholar] [CrossRef]
- Stone, K.W.; Casillas, G.A.; Karaye, I.; Camargo, K.; McDonald, T.J.; Horney, J.A. Using Spatial Analysis to Examine Potential Sources of Polycyclic Aromatic Hydrocarbons in an Environmental Justice Community After Hurricane Harvey. Environ. Justice 2019, 12, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Escher, B.I.; Stapleton, H.M.; Schymanski, E.L. Tracking complex mixtures of chemicals in our changing environment. Science 2020, 367, 388–392. [Google Scholar] [CrossRef]
- Hollender, J.; Schymanski, E.L.; Singer, H.P.; Ferguson, P.L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go? Environ. Sci. Technol. 2017, 51, 11505–11512. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.S.; Aly, N.A.; McCord, J.; Strynar, M.J.; Chiu, W.A.; Dodds, J.N.; Baker, E.S.; Rusyn, I. Rapid Characterization of Emerging Per- and Polyfluoroalkyl Substances in Aqueous Film-Forming Foams Using Ion Mobility Spectrometry-Mass Spectrometry. Environ. Sci. Technol. 2020, 54, 15024–15034. [Google Scholar] [CrossRef] [PubMed]
- Valdiviezo, A.; Aly, N.A.; Luo, Y.-S.; Cordova, A.; Casillas, G.; Foster, M.; Baker, E.S.; Rusyn, I. Analysis of per- and polyfluoroalkyl substances in Houston Ship Channel and Galveston Bay following a large-scale industrial fire using ion-mobility-spectrometry-mass spectrometry. J. Environ. Sci. 2022, 115, 350–362. [Google Scholar] [CrossRef]
- Chen, Z.; Lloyd, D.; Zhou, Y.H.; Chiu, W.A.; Wright, F.A.; Rusyn, I. Risk Characterization of Environmental Samples Using In Vitro Bioactivity and Polycyclic Aromatic Hydrocarbon Concentrations Data. Toxicol. Sci. 2021, 179, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Gomez, L.; Niegowska, M.; Navarro, A.; Amendola, L.; Arukwe, A.; Ait-Aissa, S.; Balzamo, S.; Barreca, S.; Belkin, S.; Bittner, M.; et al. Estrogenicity of chemical mixtures revealed by a panel of bioassays. Sci. Total Environ. 2021, 785, 147284. [Google Scholar] [CrossRef]
- Neale, P.A.; O'Brien, J.W.; Glauch, L.; Konig, M.; Krauss, M.; Mueller, J.F.; Tscharke, B.; Escher, B.I. Wastewater treatment efficacy evaluated with in vitro bioassays. Water Res. X 2020, 9, 100072. [Google Scholar] [CrossRef] [PubMed]
- Neale, P.A.; Braun, G.; Brack, W.; Carmona, E.; Gunold, R.; Konig, M.; Krauss, M.; Liebmann, L.; Liess, M.; Link, M.; et al. Assessing the Mixture Effects in In Vitro Bioassays of Chemicals Occurring in Small Agricultural Streams during Rain Events. Environ. Sci. Technol. 2020, 54, 8280–8290. [Google Scholar] [CrossRef]
- Neale, P.A.; Altenburger, R.; Ait-Aissa, S.; Brion, F.; Busch, W.; Umbuzeiro, G.D.; Denison, M.S.; Du Pasquier, D.; Hilscherova, K.; Hollert, H.; et al. Development of a bioanalytical test battery for water quality monitoring: Fingerprinting identified micropollutants and their Contribution to effects in surface water. Water Res. 2017, 123, 734–750. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, N.H.; Chen, Z.; Rusyn, I.; Chiu, W.A. Risk Characterization and Probabilistic Concentration-Response Modeling of Complex Environmental Mixtures Using New Approach Methodologies (NAMs) Data from Organotypic in Vitro Human Stem Cell Assays. Environ. Health Perspect. 2021, 129, 17004. [Google Scholar] [CrossRef]
- Lesser, G.R.; Roelvink, J.A.; Ven Kester, J.A.T.M.; Stelling, G.S. Development and validation of a three-dimensional morphological model. Coast. Eng. 2004, 51, 883–915. [Google Scholar] [CrossRef]
- Powell, M.D.; Houston, S.H.; Amat, L.R.; Morisseau-Leroy, N. The HRD real-time hurricane wind analysis system. J. Wind Eng. Ind. Aerod. 1998, 77–78, 53–64. [Google Scholar] [CrossRef]
- Ellison, R.J. The Use of the Dimethyl Sulphoxide (DMSO) Extract by the IP 346 Method as an Indicator of the Carcinogenicity of Lubricant Base Oils and Distillate Aromatic Extracts; CONCAWE Health Management Group: Brussels, Belgium, 1994. [Google Scholar]
- Chen, Z.; Liu, Y.; Wright, F.A.; Chiu, W.A.; Rusyn, I. Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. ALTEX 2020, 37, 623–638. [Google Scholar] [CrossRef]
- Sirenko, O.; Grimm, F.A.; Ryan, K.R.; Iwata, Y.; Chiu, W.A.; Parham, F.; Wignall, J.A.; Anson, B.; Cromwell, E.F.; Behl, M.; et al. In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol. Appl. Pharmacol. 2017, 322, 60–74. [Google Scholar] [CrossRef] [Green Version]
- Grimm, F.A.; Iwata, Y.; Sirenko, O.; Bittner, M.; Rusyn, I. High-Content Assay Multiplexing for Toxicity Screening in Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Hepatocytes. Assay Drug Dev. Technol. 2015, 13, 529–546. [Google Scholar] [CrossRef]
- Sirenko, O.; Hesley, J.; Rusyn, I.; Cromwell, E.F. High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 2014, 12, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Sirenko, O.; Cromwell, E.F.; Crittenden, C.; Wignall, J.A.; Wright, F.A.; Rusyn, I. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol. Appl. Pharmacol. 2013, 273, 500–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirenko, O.; Crittenden, C.; Callamaras, N.; Hesley, J.; Chen, Y.W.; Funes, C.; Rusyn, I.; Anson, B.; Cromwell, E.F. Multiparameter in vitro assessment of compound effects on cardiomyocyte physiology using iPSC cells. J. Biomole. Screen. 2013, 18, 39–53. [Google Scholar] [CrossRef] [Green Version]
- Klaren, W.D.; Rusyn, I. High-Content Assay Multiplexing for Muscle Toxicity Screening in Human-Induced Pluripotent Stem Cell-Derived Skeletal Myoblasts. Assay Drug Dev. Technol. 2018, 16, 333–342. [Google Scholar] [CrossRef]
- Iwata, Y.; Klaren, W.D.; Lebakken, C.S.; Grimm, F.A.; Rusyn, I. High-Content Assay Multiplexing for Vascular Toxicity Screening in Induced Pluripotent Stem Cell-Derived Endothelial Cells and Human Umbilical Vein Endothelial Cells. Assay Drug Dev. Technol. 2017, 15, 267–279. [Google Scholar] [CrossRef]
- Grimm, F.A.; Klaren, W.D.; Li, X.; Lehmler, H.J.; Karmakar, M.; Robertson, L.W.; Chiu, W.A.; Rusyn, I. Cardiovascular Effects of Polychlorinated Biphenyls and Their Major Metabolites. Environ. Health Perspect. 2020, 128, 77008. [Google Scholar] [CrossRef] [PubMed]
- Blanchette, A.D.; Burnett, S.D.; Grimm, F.A.; Rusyn, I.; Chiu, W.A. A Bayesian Method for Population-wide Cardiotoxicity Hazard and Risk Characterization Using an In Vitro Human Model. Toxicol. Sci. 2020, 178, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Blanchette, A.D.; Grimm, F.A.; Dalaijamts, C.; Hsieh, N.H.; Ferguson, K.; Luo, Y.S.; Rusyn, I.; Chiu, W.A. Thorough QT/QTc in a Dish: An In Vitro Human Model That Accurately Predicts Clinical Concentration-QTc Relationships. Clin. Pharmacol. Ther. 2019, 105, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Burnett, S.D.; Blanchette, A.D.; Grimm, F.A.; House, J.S.; Reif, D.M.; Wright, F.A.; Chiu, W.A.; Rusyn, I. Population-based toxicity screening in human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Appl. Pharmacol. 2019, 381, 114711. [Google Scholar] [CrossRef]
- Grimm, F.A.; House, J.S.; Wilson, M.R.; Sirenko, O.; Iwata, Y.; Wright, F.A.; Ball, N.; Rusyn, I. Multi-dimensional in vitro bioactivity profiling for grouping of glycol ethers. Reg. Toxicol. Pharmac. 2019, 101, 91–102. [Google Scholar] [CrossRef] [PubMed]
- House, J.S.; Grimm, F.A.; Klaren, W.D.; Dalzell, A.; Kuchi, S.; Zhang, S.D.; Lenz, K.; Boogaard, P.J.; Ketelslegers, H.B.; Gant, T.W.; et al. Grouping of UVCB substances with new approach methodologies (NAMs) data. ALTEX 2021, 38, 123–137. [Google Scholar] [CrossRef]
- Grimm, F.A.; Iwata, Y.; Sirenko, O.; Chappell, G.A.; Wright, F.A.; Reif, D.M.; Braisted, J.; Gerhold, D.L.; Yeakley, J.M.; Shepard, P.; et al. A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives. Green Chem. 2016, 18, 4407–4419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Sances, S.; Workman, M.J.; Svendsen, C.N. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 2020, 26, 309–329. [Google Scholar] [CrossRef] [PubMed]
- Grimm, F.A.; Blanchette, A.; House, J.S.; Ferguson, K.; Hsieh, N.H.; Dalaijamts, C.; Wright, A.A.; Anson, B.; Wright, F.A.; Chiu, W.A.; et al. A human population-based organotypic in vitro model for cardiotoxicity screening. ALTEX 2018, 35, 441–452. [Google Scholar] [CrossRef]
- Burnett, S.D.; Blanchette, A.D.; Chiu, W.A.; Rusyn, I. Human induced pluripotent stem cell (iPSC)-derived cardiomyocytes as an in vitro model in toxicology: Strengths and weaknesses for hazard identification and risk characterization. Expert Opin. Drug Metab. Toxicol. 2021, 17, 887–902. [Google Scholar] [CrossRef]
- Anson, B.D.; Kolaja, K.L.; Kamp, T.J. Opportunities for use of human iPS cells in predictive toxicology. Clin. Pharmacol. Ther. 2011, 89, 754–758. [Google Scholar] [CrossRef] [Green Version]
- Paul Friedman, K.; Gagne, M.; Loo, L.H.; Karamertzanis, P.; Netzeva, T.; Sobanski, T.; Franzosa, J.A.; Richard, A.M.; Lougee, R.R.; Gissi, A.; et al. Utility of In Vitro Bioactivity as a Lower Bound Estimate of In Vivo Adverse Effect Levels and in Risk-Based Prioritization. Toxicol. Sci. 2020, 173, 202–225. [Google Scholar] [CrossRef]
- Marvel, S.W.; To, K.; Grimm, F.A.; Wright, F.A.; Rusyn, I.; Reif, D.M. ToxPi Graphical User Interface 2.0: Dynamic exploration, visualization, and sharing of integrated data models. BMC Bioinf. 2018, 19, 80. [Google Scholar] [CrossRef] [Green Version]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Zhou, Y.H.; Mayhew, G.; Sun, Z.; Xu, X.; Zou, F.; Wright, F.A. Space Time Clustering and the Permutation Moments of Quadratic Form. Stat 2013, 2, 292–302. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B Met. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Aly, N.A.; Luo, Y.S.; Liu, Y.; Casillas, G.; McDonald, T.J.; Kaihatu, J.M.; Jun, M.; Ellis, N.; Gossett, S.; Dodds, J.N.; et al. Temporal and spatial analysis of per and polyfluoroalkyl substances in surface waters of Houston ship channel following a large-scale industrial fire incident. Environ. Pollut. 2020, 265 (Pt B), 115009. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; McDonald, T.J.; Bhandari, S.; Rusyn, I.; Chiu, W.A. Spatial and temporal distribution of surface water contaminants in the Houston Ship Channel after the Intercontinental Terminal Company Fire. J. Expo. Sci. Environ. Epidemiol. 2021, 31, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, D.; Howell, N.L.; Rifai, H.S.; Koenig, L. Spatial and temporal variation of polychlorinated biphenyls in the Houston Ship Channel. Chemosphere 2010, 80, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Willett, K.L.; McDonald, S.J.; Steinberg, M.A.; Beatty, K.B.; Kennicutt, M.C.; Safe, S.H. Biomarker sensitivity for polynuclear aromatic hydrocarbon contamination in two marine fish species collected in Galveston Bay, Texas. Environ. Toxicol. Chem. 1997, 16, 1472–1479. [Google Scholar]
- Bera, G.; Camargo, K.; Sericano, J.L.; Liu, Y.; Sweet, S.T.; Horney, J.; Jun, M.; Chiu, W.; Rusyn, I.; Wade, T.L.; et al. Baseline data for distribution of contaminants by natural disasters: Results from a residential Houston neighborhood during Hurricane Harvey flooding. Heliyon 2019, 5, e02860. [Google Scholar] [CrossRef]
- Horney, J.A.; Casillas, G.A.; Baker, E.; Stone, K.W.; Kirsch, K.R.; Camargo, K.; Wade, T.L.; McDonald, T.J. Comparing residential contamination in a Houston environmental justice neighborhood before and after Hurricane Harvey. PLoS ONE 2018, 13, e0192660. [Google Scholar] [CrossRef] [Green Version]
- US EPA. Guidelines for the Health Risk Assessment of Chemical Mixtures. In Risk Assessment Forum; U.S. Environmental Protection Agency: Washington, DC, USA, 1986. [Google Scholar]
- Escher, B.I.; Aїt-Aїssa, S.; Behnisch, P.A.; Brack, W.; Brion, F.; Brouwer, A.; Buchinger, S.; Crawford, S.E.; Du Pasquier, D.; Hamers, T.; et al. Effect-based trigger values for in vitro and in vivo bioassays performed on surface water extracts supporting the environmental quality standards (EQS) of the European Water Framework Directive. Sci. Total Environ. 2018, 628-629, 748–765. [Google Scholar] [CrossRef] [PubMed]
- Harmon, M.; Pait, A.S.; Hameedi, M.J. Sediment Contamination, Toxicity, and Macroinvertebrate Infaunal Community in Galveston Bay; NOAA: Silver Spring, MD, USA, 2003.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Jang, S.; Kaihatu, J.M.; Zhou, Y.-H.; Wright, F.A.; Chiu, W.A.; Rusyn, I. Potential Human Health Hazard of Post-Hurricane Harvey Sediments in Galveston Bay and Houston Ship Channel: A Case Study of Using In Vitro Bioactivity Data to Inform Risk Management Decisions. Int. J. Environ. Res. Public Health 2021, 18, 13378. https://doi.org/10.3390/ijerph182413378
Chen Z, Jang S, Kaihatu JM, Zhou Y-H, Wright FA, Chiu WA, Rusyn I. Potential Human Health Hazard of Post-Hurricane Harvey Sediments in Galveston Bay and Houston Ship Channel: A Case Study of Using In Vitro Bioactivity Data to Inform Risk Management Decisions. International Journal of Environmental Research and Public Health. 2021; 18(24):13378. https://doi.org/10.3390/ijerph182413378
Chicago/Turabian StyleChen, Zunwei, Suji Jang, James M. Kaihatu, Yi-Hui Zhou, Fred A. Wright, Weihsueh A. Chiu, and Ivan Rusyn. 2021. "Potential Human Health Hazard of Post-Hurricane Harvey Sediments in Galveston Bay and Houston Ship Channel: A Case Study of Using In Vitro Bioactivity Data to Inform Risk Management Decisions" International Journal of Environmental Research and Public Health 18, no. 24: 13378. https://doi.org/10.3390/ijerph182413378






