Community Composition and Spatial Distribution of N-Removing Microorganisms Optimized by Fe-Modified Biochar in a Constructed Wetland
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Preparations and Characteristics of Biochar and Fe-Modified Biochar
2.2. Construction and Operation of HSCWs
2.3. Microorganism Sample Collection and DNA Extration
2.4. High-Throughput Sequencing of nirS and nirK Genes
2.5. Quantitative Analysis of Genes Involved in Denitrification and Anammox
2.6. Statistical Analysis
3. Results
3.1. The Richness and Diversity of Denitrifiers in the Three HSCWs
3.2. The Community Structures of Denitrifiers in the Three HSCWs
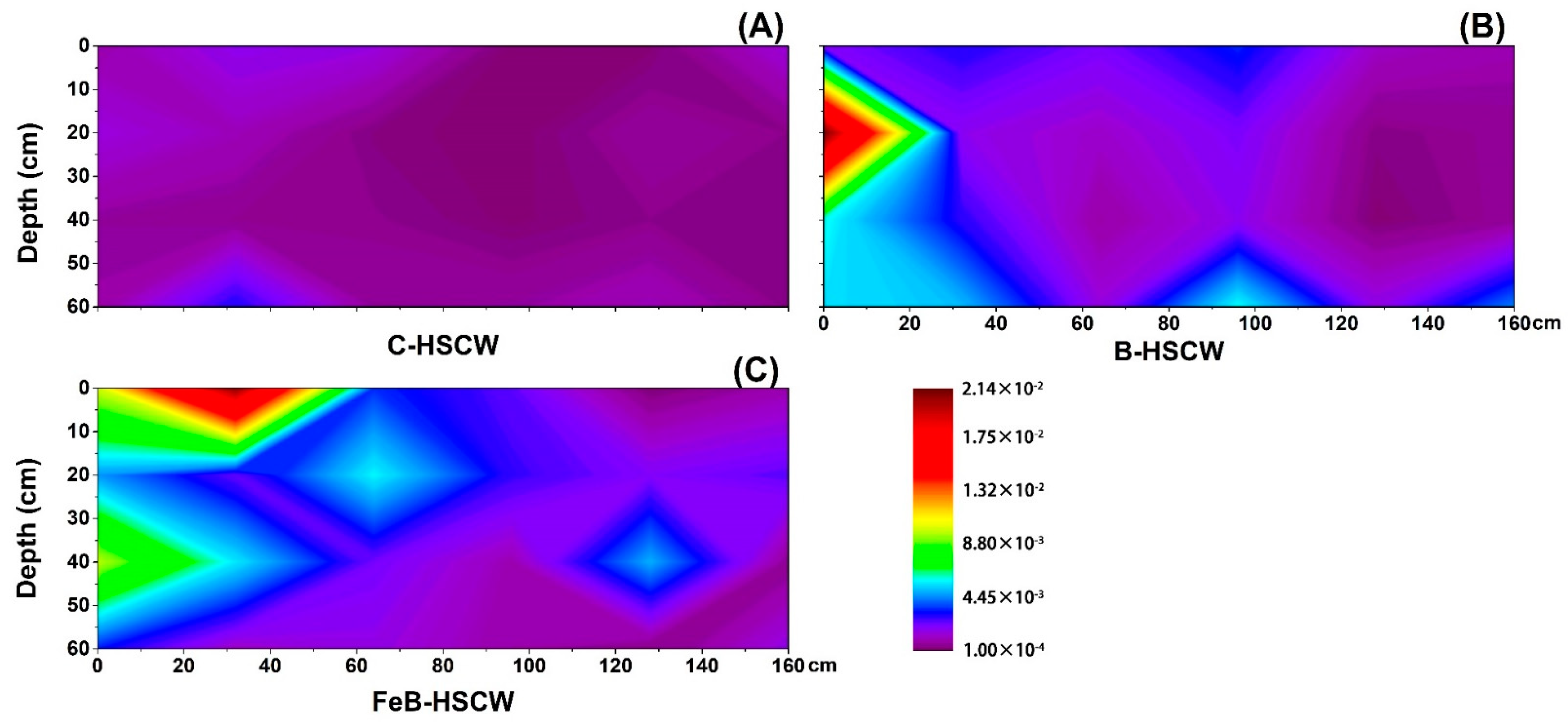
3.3. The Spatial Distribution of Denitrifying Functional Genes in the Three HSCWs
3.3.1. Spatial Distributions of narG and napA in HSCWs
3.3.2. Spatial Distributions of nirS and nirK in HSCWs
3.3.3. Spatial Distributions of qnorB and cnorB in HSCWs
3.3.4. Spatial Distributions of nosZ-I and nosZ-II in HSCWs
3.4. Spatial Distribution of Anammox Genes in HSCWs
4. Discussion
4.1. Effect of Fe-Modified Biochar on Denitrifier Richness and Diversity
4.2. Fe-Modified Biochar Optimizes Denitrifier Community Structures in FeB-SHCW
4.3. Fe-Modified Biochar Influences Spatial Distribution of Denitrifying Functional Genes in FeB-HSCW
4.4. FeB-Modified Biochar Improved the Spatial Distribution of Anammox Genes in FeB-HSCW
4.5. Strengthening Mechanism of Fe-Modified Biochar on Microbial N Removal in HSCW
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qu, J.; Fan, M. The current state of water quality and technology development for water pollution control in China. Crit. Rev. Environ. Sci. Technol. 2010, 40, 519–560. [Google Scholar] [CrossRef]
- Jahangir, M.M.R.; Fenton, O.; Müller, C.; Harrington, R.; Johnston, P.; Richards, K.G. In situ denitrification and dnra rates in groundwater beneath an integrated constructed wetland. Water Res. 2017, 111, 254–264. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, Y.; Zhou, Q.; Vymazal, J. Effects of plant biomass on nitrogen transformation in subsurface-batch constructed wetlands: A stable isotope and mass balance assessment. Water Res. 2014, 63, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Yan, B.X.; Xu, Y.Y.; Guan, J.N.; Liu, S.Y. Removal of nitrogen and COD in horizontal subsurface flow constructed wetlands under different influent C/N ratios. Ecol. Eng. 2014, 63, 58–63. [Google Scholar] [CrossRef]
- Long, Y.; Bing, Y.X.; Zhang, Z.K.; Cui, K.; Pan, X.K.; Yan, X.F.; Li, B.X.; Xie, S.G.; Guo, Q.W. Influence of plantation on microbial community in porous concrete treating polluted surface water. Int. Biodeterior. Biodegr. 2017, 117, 8–13. [Google Scholar] [CrossRef]
- Xiong, J.Q.; Ren, S.H.; He, Y.F.; Wang, X.C.C.; Bai, X.C.; Wang, J.X.; Dzakpasu, M. Bioretention cell incorporating Fe-biochar and saturated zones for enhanced stormwater runoff treatment. Chemosphere 2019, 237, 124424. [Google Scholar] [CrossRef]
- Feng, L.K.; Liu, Y.; Zhang, J.Y.; Li, C.; Wu, H.M. Dynamic variation in nitrogen removal of constructed wetlands modifed by biochar for treating secondary livestock effluent under varying oxygen supplying conditions. J. Environ. Manag. 2020, 260, 110152. [Google Scholar] [CrossRef]
- Liang, Y.K.; Wang, Q.H.; Huang, L.; Liu, M.L.; Wang, N.; Chen, Y.C. Insight into the mechanisms of biochar addition on pollutant removal enhancement and nitrous oxide emission reduction in subsurface flow constructed wetlands: Microbial community structure, functional genes and enzyme activity. Bioresour. Technol. 2020, 307, 123249. [Google Scholar] [CrossRef]
- Shang, X.; Yang, L.; Ouyang, D.; Zhang, B.; Zhang, W.Y.; Gu, M.Y.; Li, J.; Chen, M.F.; Huang, L.H.; Qian, L.B. Enhanced removal of 1,2,4-trichlorobenzene by modified biochar supported nanoscale zero-valent iron and palladium. Chemosphere 2020, 249, 126518. [Google Scholar] [CrossRef]
- Melton, E.D.; Swanner, E.D.; Behrens, S.; Schmidt, C.; Kappler, A. The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle. Nat. Rev. Microbiol. 2014, 12, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.F.; Xu, X.Q.; Sun, M.; Jiang, M.J.; Xue, G.; Li, X.; Liu, Z.H. How does iron facilitate the aerated biofilter for tertiary simultaneous nutrient and refractory organics removal from real dyeing wastewater? Water Res. 2019, 148, 344–358. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, T.H.; Zhang, X.; Qing, C.S.; Wang, J.; Yue, Z.B.; Liu, H.B.; Yang, Z. Simultaneous removal of nitrate and phosphate from wastewater by siderite based autotrophic denitrification. Chemosphere 2018, 199, 130–137. [Google Scholar] [CrossRef]
- Ma, Y.H.; Dai, W.Q.; Zheng, P.R.; Zheng, X.Y.; He, S.B.; Zhao, M. Iron scraps enhance simultaneous nitrogen and phosphorus removal in subsurface flow constructed wetlands. J. Hazard. Mater. 2020, 395, 122612. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.Y.; Seo, Y.D.; Kim, B.; Kim, I.Y.; Cha, D.K. Microbial reduction of nitrate in the presence of zero-valent iron and biochar. Bioresour. Technol. 2016, 200, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Li, P.J.; Lin, K.R.; Fang, Z.Q.; Wang, K.M. Enhanced nitrate removal by novel bimetallic Fe/Ni nanoparticles supported on biochar. J. Clean. Prod. 2017, 151, 21–33. [Google Scholar] [CrossRef]
- Wolthoorn, A.; Temminghoff, E.J.M.; Van Riemsdijk, W.H. Effect of synthetic iron colloids on the microbiological NH4+ removal process during groundwater purification. Water Res. 2004, 38, 1884–1892. [Google Scholar] [CrossRef]
- Li, J.H.; Lv, G.H.; Bai, W.B.; Liu, Q.; Zhang, Y.C.; Song, J.Q. Modification and use of biochar from wheat straw (Triticum aestivum L.) for nitrate and phosphate removal from water. Desalination Water Treat. 2016, 57, 4681–4693. [Google Scholar]
- Saeed, T.; Sun, G.Z. A review on nitrogen and organics removal mechanisms in subsurface flow constructed wetlands: Dependency on physic-chemical parameters, operating conditions and supporting media. J. Environ. Manag. 2012, 112, 429–448. [Google Scholar] [CrossRef]
- Sanchez, O. Constructed wetlands revisited: Microbial diversity in the -omics era. Microb. Ecol. 2017, 73, 722–733. [Google Scholar] [CrossRef]
- Wu, H.M.; Fan, J.L.; Zhang, J.; Ngo, H.H.; Guo, W.S.; Hu, Z.; Lv, J.L. Optimization of organics and nitrogen removal in intermittently aerated vertical flow constructed wetlands: Effects of aeration time and aeration rate. Int. Biodeterior. Biodegr. 2016, 113, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.L.; Huang, T.L.; Zhang, C.H.; Fang, K.K.; Xia, C.; Bai, S.Y.; Zeng, M.Z.; Qiu, X.P. Illumina MiSeq sequencing reveals the community composition of NirS-Type and NirK-Type denitrifiers in Zhoucun reservoir—A large shallow eutrophic reservoir in northern China. RSC Adv. 2016, 6, 91517–91528. [Google Scholar] [CrossRef]
- Ligi, T.; Truu, M.; Truu, J.; Nolvak, H.; Kaasik, A.; Mitsch, W.J.; Mander, U. Effects of soil chemical characteristics and water regime on denitrification genes (nirS, nirK, and nosZ) abundances in a created riverine wetland complex. Ecol. Eng. 2014, 72, 47–55. [Google Scholar] [CrossRef]
- Zhi, E.Q.; Song, Y.H.; Duan, L.; Yu, H.B.; Peng, J.F. Spatial distribution and diversity of microbial community in large-scale constructed wetland of the Liao River Conservation Area. Environ. Earth. Sci. 2015, 73, 5085–5094. [Google Scholar] [CrossRef]
- Fu, G.P.; Wu, J.F.; Han, J.Y.; Zhao, L.; Chan, G.; Leong, K.F. Effects of substrate type on denitrification efficiency and microbial community structure in constructed wetlands. Bioresour. Technol. 2020, 307, 123222. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, L.Y.; Wu, J.; Zhong, F.; Cheng, S.P. Step-feeding ratios affect nitrogen removal and related microbial communities in multi-stage vertical flow constructed wetlands. Sci. Total Environ. 2020, 721, 137689. [Google Scholar] [CrossRef]
- Huang, T.; Liu, W.; Zhang, Y.; Zhou, Q.H.; Wu, Z.B.; He, F. A stable simultaneous anammox, denitrifying anaerobic methane oxidation and denitrifcation process in integrated vertical constructed wetlands for slightly polluted wastewater. Environ. Pollut. 2020, 262, 114363. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, M.M.; Liu, F.; Li, Y.; He, Y.; Zhang, S.A.; Wu, J.S. Abundance and distribution of microorganisms involved in denitrification in sediments of a Myriophyllum elatinoides purification system for treating swine wastewater. Environ. Sci. Pollut. Res. 2015, 22, 17906–17916. [Google Scholar] [CrossRef]
- Gao, J.; Hou, L.J.; Zheng, Y.L.; Liu, M.; Yin, G.Y.; Li, X.F.; Lin, X.B.; Yu, C.D.; Wang, R.; Jiang, X.F.; et al. nirS-Encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl. Microbiol. Biotechnol. 2016, 100, 8573–8582. [Google Scholar] [CrossRef]
- Wan, Y.; Ruan, X.H.; Wang, J.; Shi, X.J. Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses. Int. J. Environ. Res. Public Health 2019, 16, 2298. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.S.T.; Irbis, C.; Wang, P.Y.; Inamura, T. Impact of plant harvest management on function and community structure of nitrifiers and denitrifiers in a constructed wetland. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Kalscheur, K.N.; Rojas, M.; Peterson, C.G.; Kelly, J.J.; Gray, K.A. Algal exudates and stream organic matter influence the structure and function of denitrifying bacterial communities. Microb. Ecol. 2012, 64, 881–892. [Google Scholar] [CrossRef]
- Palmer, K.; Biasi, C.; Horn, M.A. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 2012, 6, 1058–1077. [Google Scholar] [CrossRef] [Green Version]
- Mao, G.Z.; Chen, L.; Yang, Y.Y.; Wu, Z.; Tong, T.L.; Liu, Y.; Xie, S.G. Vertical profiles of water and sediment denitrifiers in two plateau freshwater lakes. Appl. Microbiol. Biotechnol. 2017, 101, 3361–3370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, Y.; Gao, G.; Jiang, J.H. Spatial-Temporal Variation of Bacterial Communities in Sediments in Lake Chaohu, a Large, Shallow Eutrophic Lake in China. Int. J. Environ. Res. Public Health 2019, 16, 3966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.L.; Wang, X.Z.; He, X.J.; Zhang, S.B.; Liang, R.B.; Shen, J. Effects of root exudates on denitrifier gene abundance, community structure and activity in a micro-polluted constructed wetland. Sci. Total Environ. 2017, 598, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Sun, X.; Gao, Y.; Yang, Y.C.; Yang, L.Y. Fe-modified biochar enhances microbial nitrogen removal capability of constructed wetland. Sci. Total. Environ. 2020, 740, 139534. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, A.; Sillanpää, M. A review of emerging adsorbents for nitrate removal from water. Chem. Eng. J. 2011, 168, 493–504. [Google Scholar] [CrossRef]
- Robertson, E.K.; Roberts, K.L.; Burdorf, L.D.W.; Cook, P.; Thamdrup, B. Dissimilatory nitrate reduction to ammonium coupled to Fe(II) oxidation in sediments of a periodically hypoxic estuary. Limnol. Oceanogr. 2016, 61, 365–381. [Google Scholar] [CrossRef] [Green Version]
- Li, B.X.; Chen, J.F.; Wu, Z.; Wu, S.F.; Xie, S.G.; Liu, Y. Seasonal and spatial dynamics of denitrification rate and denitrifier community in constructed wetland treating polluted river water. Int. Biodeterior. Biodegrad. 2018, 126, 143–151. [Google Scholar] [CrossRef]
- Pan, F.X.; Chapman, S.J.; Li, Y.Y.; Yao, H.Y. Straw amendment to paddy soil stimulates denitrification but biochar amendment promotes anaerobic ammonia oxidation. J. Soils Sediments 2017, 17, 2428–2437. [Google Scholar] [CrossRef]
- Zhang, P.F.; Peng, Y.K.; Lu, J.L.; Li, J.; Chen, H.P.; Xiao, L. Microbial communities and functional genes of nitrogen cycling in an electrolysis augmented constructed wetland treating wastewater treatment plant effluent. Chemosphere 2018, 211, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chandran, K.; Stensel, D. Microbial ecology of denitrification in biological wastewater treatment. Water Res. 2014, 64, 237–254. [Google Scholar] [CrossRef]
- Sanchez, C.; Minamisawa, K. Redundant roles of Bradyrhizobium oligotrophicum Cu-type (NirK) and cd (1)-type (NirS) nitrite reductase genes under denitrifying conditions. FEMS Microbiol. Lett. 2018, 365, fny015. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Ma, J.; Murinda, S.; Reddy, G.B. Bacterial community dynamics in surface flow constructed wetlands for the treatment of swine waste. Sci. Total Environ. 2016, 544, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Coates, J.D.; Chakraborty, R.; Lack, J.G.; O’Connor, S.M.; Cole, K.A.; Bender, K.S.; Achenbach, L.A. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 2001, 411, 1039–1043. [Google Scholar] [CrossRef]
- Bellini, M.I.; Gutierrez, L.; Tarlera, S.; Scavino, A.F. Isolation and functional analysis of denitrifiers in an aquifer with high potential for denitrification. Syst. Appl. Microbiol. 2013, 36, 505–516. [Google Scholar] [CrossRef]
- Shu, D.T.; He, Y.L.; Yue, H.; Wang, Q.Y. Microbial structures and community functions of anaerobic sludge in six full-scale wastewater treatment plants as revealed by 454 high-throughput pyrosequencing. Bioresour. Technol. 2015, 186, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, F.; Luo, P.; Chen, X.; Chen, J.; Huang, Z.; Peng, J.; Xiao, R.; Wu, J. Stimulation of optimized influent C:N ratios on nitrogen removal in surface flow constructed wetlands: Performance and microbial mechanisms. Sci. Total Environ. 2019, 694, 133575. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Han, J.; Yu, T.; Huangshen, L.; Zhao, L. The structure of denitrifying microbial communities in constructed mangrove wetlands in response to fluctuating salinities. J. Environ. Manag. 2019, 238, 1–9. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L. Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 2016, 34, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Osaka, T.; Shirotani, K.; Yoshie, S.; Tsuneda, S. Effects of carbon source on denitrification efficiency and microbial community structure in a saline wastewater treatment process. Water Res. 2008, 42, 3709–3718. [Google Scholar] [CrossRef]
- Zhao, H.M.; Zhao, J.Q.; Li, F.H.; Li, X.L. Performance of Denitrifying MicrobialFuel Cell with Biocathode over Nitrite. Front. Microbiol. 2016, 7, 344. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, X.H.; Cheng, Y.Y.; Jiang, M.J.; Xiang, X. Iron robustly stimulates simultaneous nitrification and denitrification under aerobic conditions. Environ. Sci. Technol. 2018, 52, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ho, K.L.; Liu, F.C.; Ho, M.; Wang, A.; Ren, N.; Lee, D.J. Autotrophic and heterotrophic denitrification by a newly isolated strain Pseudomonas sp. C27. Bioresour. Technol. 2013, 145, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Hosono, T.; Alvarez, K.; Lin, I.T.; Shimada, J. Nitrogen, carbon, and sulfur isotopic change during heterotrophic (Pseudomonas aureofaciens) and autotrophic (Thiobacillus denitrificans) denitrification reactions. J. Contam. Hydrol. 2015, 183, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Xiao, E.; Xu, P.; Lin, L.; Zhou, Q.; Xu, D.; Wu, Z. Bacterial community and nitrate removal by simultaneous heterotrophic and autotrophic denitrification in a bioelectrochemically-assisted constructed wetland. Bioresour. Technol. 2017, 245, 993–999. [Google Scholar] [CrossRef]
- He, S.; Ding, L.; Pan, Y.; Hu, H.; Ye, L.; Ren, H. Nitrogen loading efects on nitrifcation and denitrification with functional gene quantity/transcription analysis in biochar packed reactors at 5 °C. Sci. Rep. 2018, 8, 9844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa-Galeote, D.; Marco, D.E.; Tortosa1, G.; Bru, D.; Philippot, L.; Bedmar, E.J. Spatial distribution of N-cycling microbial communities showed complex patterns in constructed wetland sediments. FEMS Microbiol. Ecol. 2013, 83, 340–351. [Google Scholar] [CrossRef]
- Bru, D.; Sarr, A.; Philippot, L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Gutierrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 2004, 57, 399–407. [Google Scholar] [CrossRef]
- García-Lledo, A.; Vilar-Sanz, A.; Trias, R.; Hallin, S.; Bañeras, L. Genetic potential for N2O emissions from the sediment of a free water surface constructed wetland. Water Res. 2011, 45, 5621–5632. [Google Scholar] [CrossRef]
- Jones, C.M.; Stres, B.; Rosenquist, M.; Hallin, S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 2008, 25, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Braker, G.; Tiedje, J.M. Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 2003, 69, 3476–3483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, W.; Ji, G.D. Quantitative response relationships between nitrogen transformation rates and nitrogen functional genes in a tidal flow constructed wetland under C/N ratio constraints. Water Res. 2014, 64, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.M.; Graf, D.R.H.; Bru, D.; Philippot, L.; Hallin, S. The unaccounted yet abundant nitrous oxide-reducing microbial community: A potential nitrous oxide sink. ISME J. 2013, 7, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Keil, D.; Meyer, A.; Berner, D.; Poll, C. Influence of land-use intensity on the spatial distribution of N-cycling microorganisms in grassland soils. FEMS Microbiol. Ecol. 2011, 77, 95–106. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.K.; Bedardhaughn, A.; Siciliano, S.D.; Farrell, R.E. Relationship between nitrifier and denitrifier community composition and abundance in predicting nitrous oxide emissions from ephemeral wetland soils. Soil Biol. Biochem. 2008, 40, 1114–1123. [Google Scholar] [CrossRef]
- Fang, W.S.; Yan, D.D.; Wang, Q.X.; Huang, B.; Ren, Z.J.; Wang, X.L.; Wang, X.N.; Li, Y.; Ouyang, C.; Migheli, Q.; et al. Changes in the abundance and community composition of different nitrogen cycling groups in response to fumigation with 1,3-dichloropropene. Sci. Total Environ. 2019, 650, 44–55. [Google Scholar] [CrossRef]
- Fujiwara, T.; Fukumori, Y. Cytochrome cb-type nitric oxide reductase with cytochrome c oxidase activity from Paracoccus denitrificans ATCC 35512. J. Bacteriol. 1996, 178, 1866–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, A.; Vandegraaf, A.A.; Robertson, L.A.; Kuenen, J.G. Anaerobic ammonium oxidation discovered in a denitrifying fluidized-bed reactor. FEMS Microbiol. Ecol. 1995, 16, 177–183. [Google Scholar] [CrossRef]
- Harhangi, H.R.; Le Roy, M.; van Alen, T.; Hu, B.L.; Groen, J.; Kartal, B.; Tringe, S.G.; Quan, Z.X.; Jetten, M.S.M.; Op den Camp, H.J.M. Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl. Environ. Microbiol. 2012, 78, 752–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humbert, S.; Tarnawski, S.; Fromin, N.; Mallet, M.P.; Aragno, M.; Zopfi, J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2010, 4, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.C.; Jiang, Q.J.; Zhong, S.; Zhang, Y.Q.; Shi, X.J. Rice husk bioash impacts redox status and rice growth in a flooded soil from southwestern China. J. Residuals Sci. Technol. 2015, 12, S75–S78. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Yu, J.J.; Jia, X.Y.; Jin, R.C. Enhancement of anammox performance by Cu(II), Ni(II) and Fe(III) supplementation. Chemosphere 2014, 117, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.L.; Wang, Q.; Zhang, J.; Yang, W.H.; Zhou, X.W. Nutrients removal and nitrous oxide emission during simultaneous nitrification, denitrification, and phosphorus removal process: Effect of iron. Environ. Sci. Pollut. Res. 2016, 23, 15657–15664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Chen, Z.; Zhou, Y.; Ma, Y.P.; Ma, C.; Li, Y.; Liang, Y.H.; Jia, J.P. Impacts of the heavy metals Cu(II), Zn (II) and Fe (II) on an Anammox system treating synthetic wastewater in low ammonia nitrogen and low temperature: Fe(II) makes a difference. Sci. Total Environ. 2019, 648, 798–804. [Google Scholar] [CrossRef]
- Qiao, S.; Bi, Z.; Zhou, J.T.; Cheng, Y.J.; Zhang, J. Long term effects of divalent ferrous ion on the activity of anammox biomass. Bioresour. Technol. 2013, 142, 490–497. [Google Scholar] [CrossRef]
- Sun, Y.F.; Qi, S.Y.; Zheng, F.P.; Huang, L.L.; Pan, J.; Jiang, Y.Y.; Hou, W.Y.; Xiao, L. Organics removal, nitrogen removal and N2O emission in subsurface wastewater infiltration systems amended with/without biochar and sludge. Bioresour. Technol. 2018, 249, 57–61. [Google Scholar] [CrossRef]
- Qi, S.Y.; Zhao, Y.; Wang, S.Y.; Zheng, F.P.; Pan, J.; Fan, L.L.; Li, Z.Q.; Tan, C.Q.; Hou, W.Y. Nitrogen Removal and N2O Emission in Biochar-Sludge Subsurface Wastewater Infiltration Systems. Water Environ. Res. 2018, 90, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, X.Z.; Zhang, H.; Wu, H.M. Enhanced nitrogen removal of low C/N domestic wastewater using a biochar-amended aerated vertical flow constructed wetland. Bioresour. Technol. 2017, 241, 269–275. [Google Scholar] [CrossRef]
- Xiang, W.; Zhang, X.; Chen, J.; Zou, W.; He, F.; Hu, X.; Tsang, D.C.W.; Ok, Y.S.; Gao, B. Biochar technology in wastewater treatment: A critical review. Chemosphere 2020, 252, 126539. [Google Scholar] [CrossRef] [PubMed]
- Kizito, S.; Lv, T.; Wu, S.; Ajmal, Z.; Luo, H.; Dong, R. Treatment of Anaerobic Digested Effluent in Biochar-Packed Vertical Flow Constructed Wetland Columns: Role of Media and Tidal Operation. Sci. Total Environ. 2017, 592, 197–205. [Google Scholar] [CrossRef]
- Kappler, A.; Wuestner, M.L.; Ruecker, A.; Harter, J.; Halama, M.; Behrens, S. Biochar as an electron shuttle between bacteria and Fe(III) minerals. Environ. Sci. Technol Lett. 2014, 1, 339–344. [Google Scholar] [CrossRef]
- Wang, X.; Shu, D.T.; Yue, H. Taxonomical and functional microbial community dynamics in an Anammox-ASBR system under different Fe (III) supplementation. Appl. Microbiol. Biotechnol. 2016, 100, 10147–10163. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.E.J.; Bren, K.L. The chemistry and biochemistry of heme c: Functional bases for covalent attachment. Nat. Prod. Rep. 2008, 25, 1118–1130. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Zhang, H.M.; Yang, F.L.; Li, H.J.; Zhang, R. The effects of zero-valent iron (ZVI) and ferroferric oxide (Fe3O4) on anammox activity and granulation in anaerobic continuously stirred tank reactors (CSTR). Process Biochem. 2014, 49, 1970–1978. [Google Scholar] [CrossRef]
- Li, X.; Yuan, Y.; Huang, Y.; Liu, H.; Bi, Z.; Yuan, Y.; Yang, P. A novel method of simultaneous NH4+ and NO3− removal using Fe cycling as a catalyst: Feammox coupled with NAFO. Sci. Total Environ. 2018, 631–632, 153–157. [Google Scholar] [CrossRef] [PubMed]

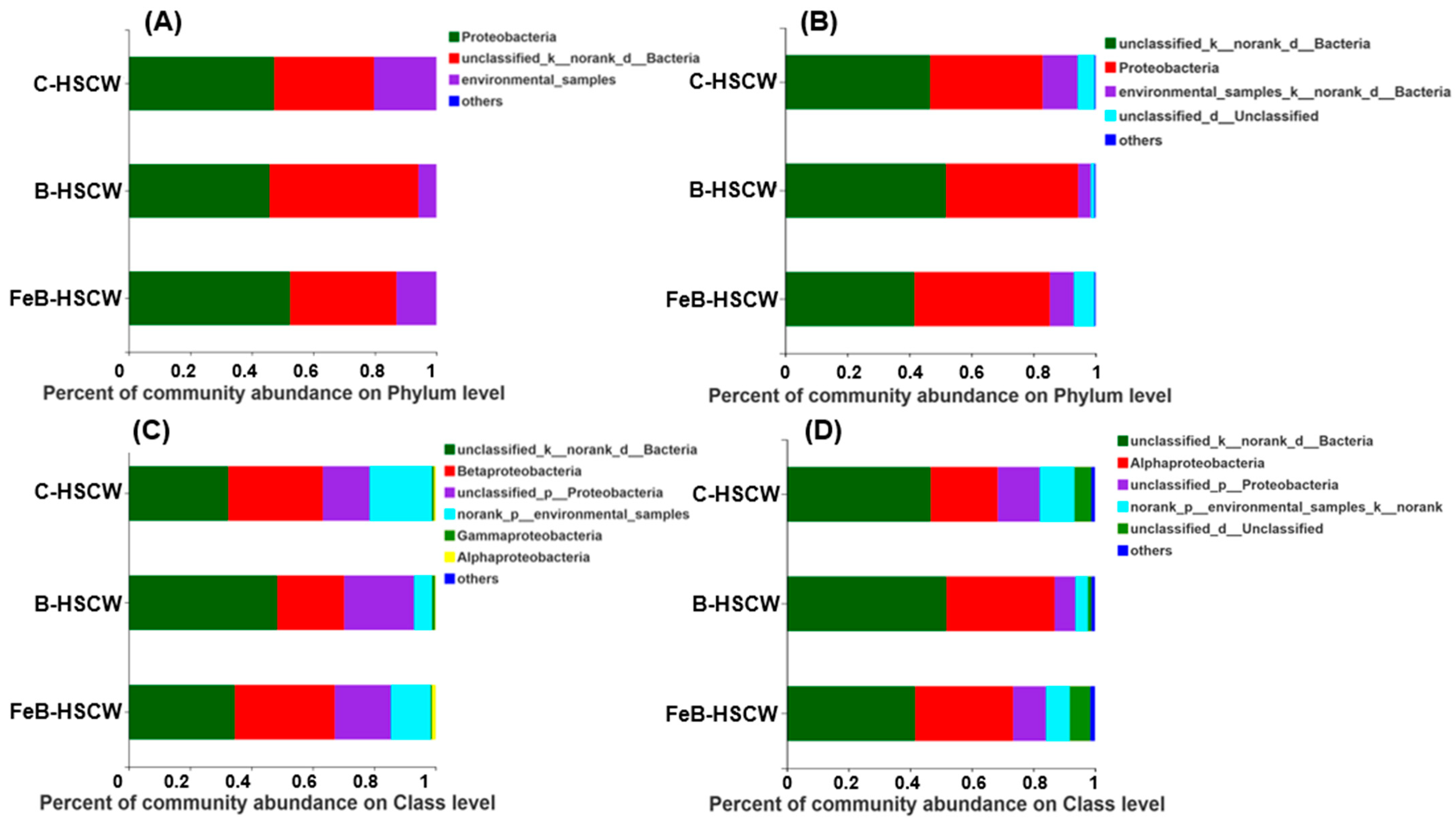
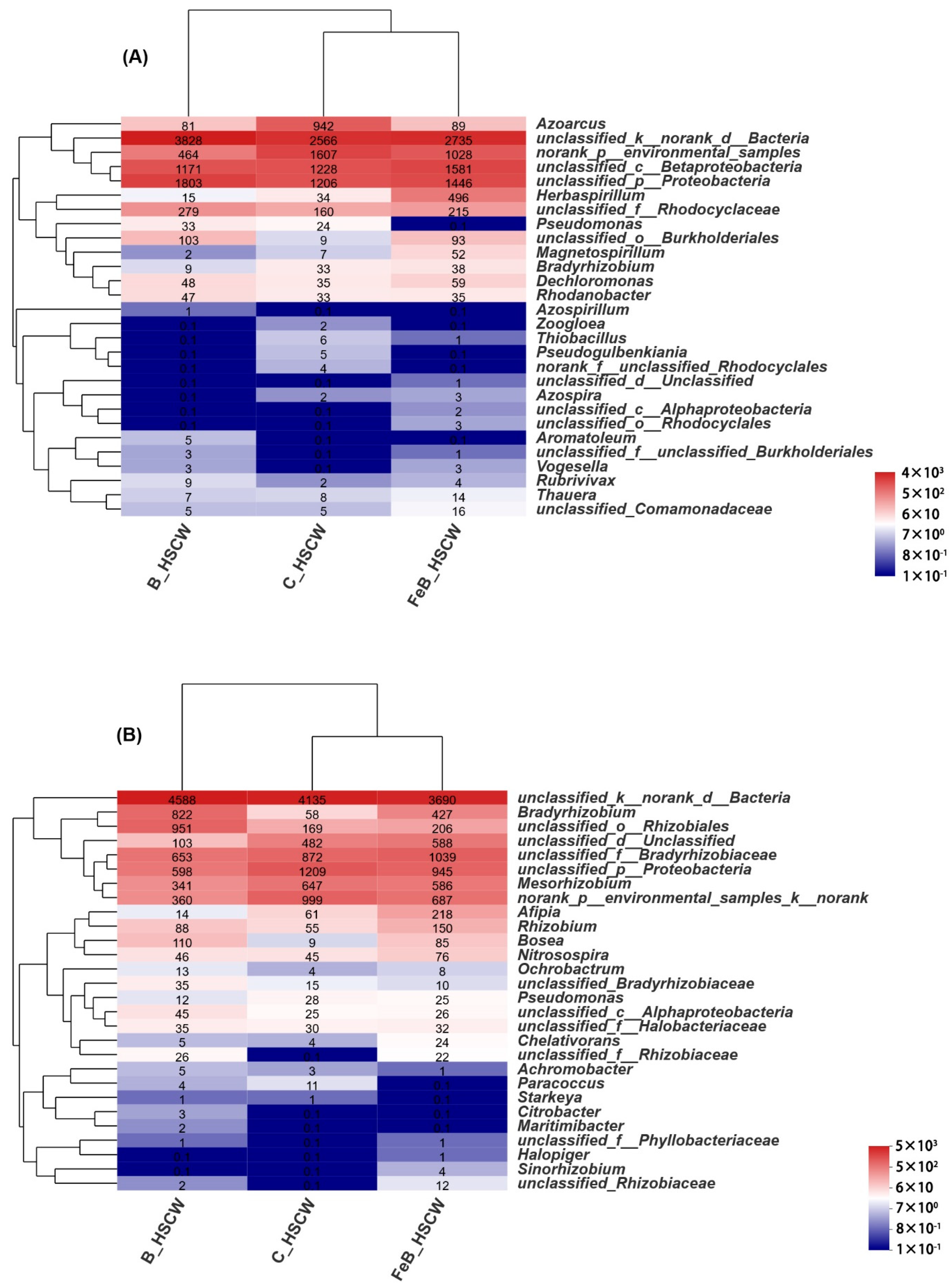
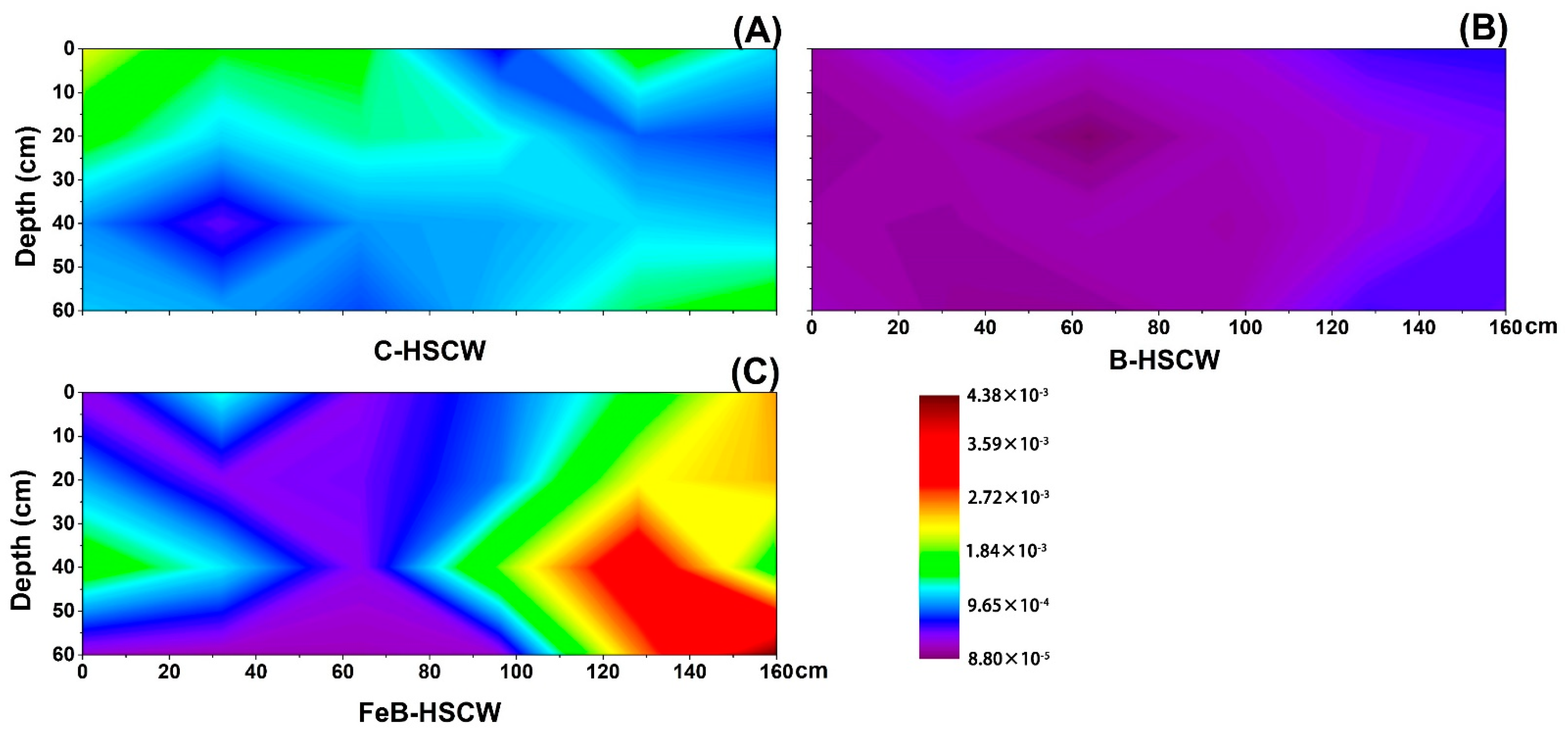
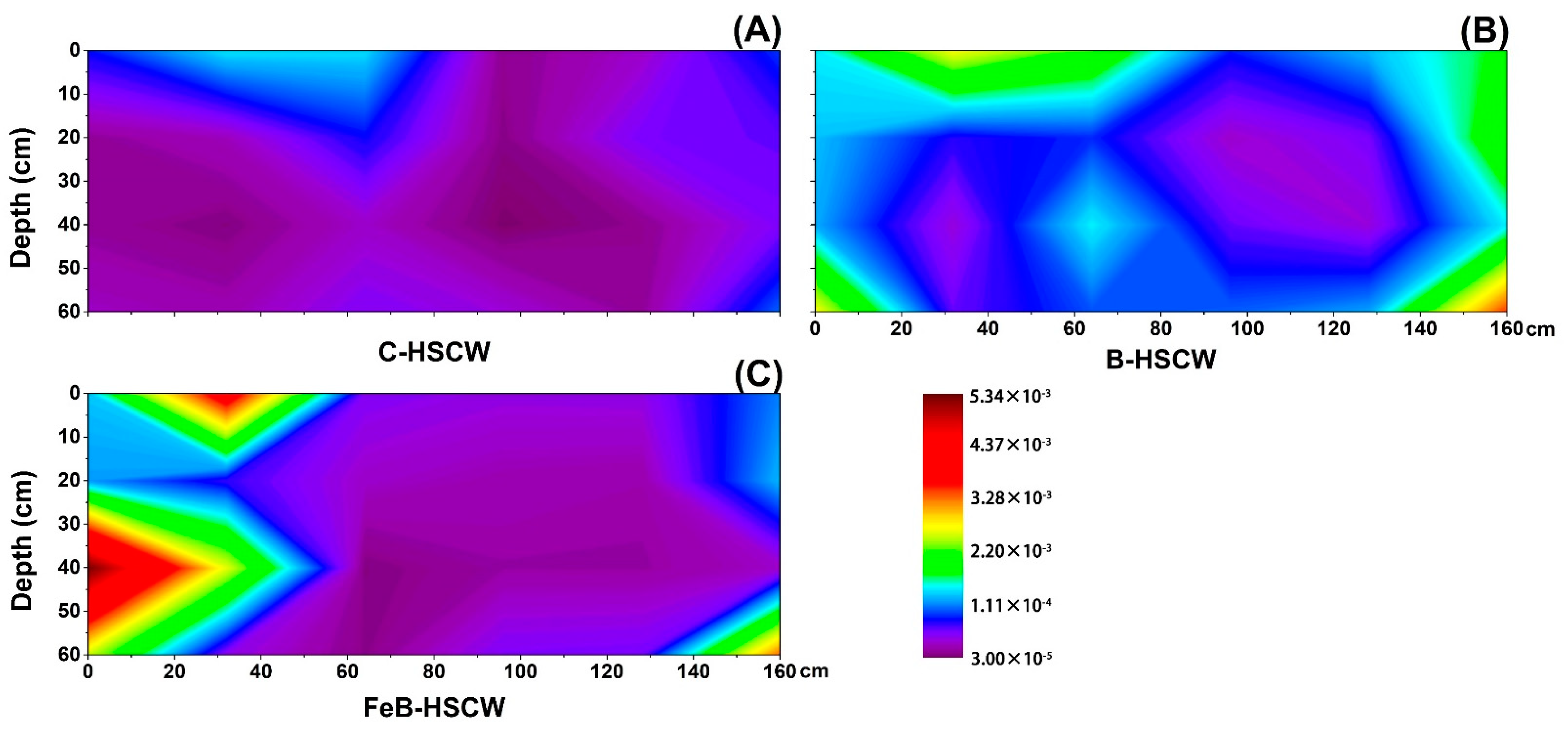
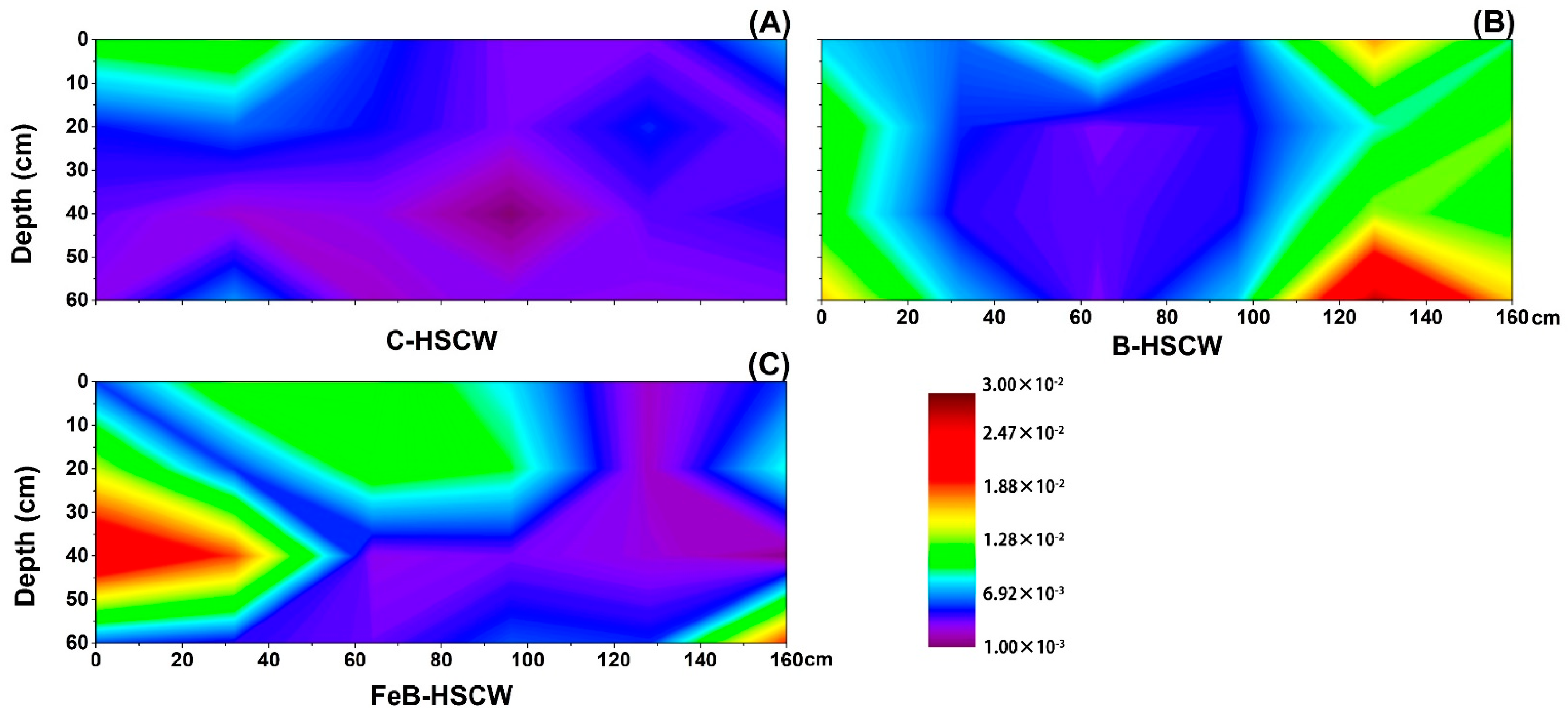
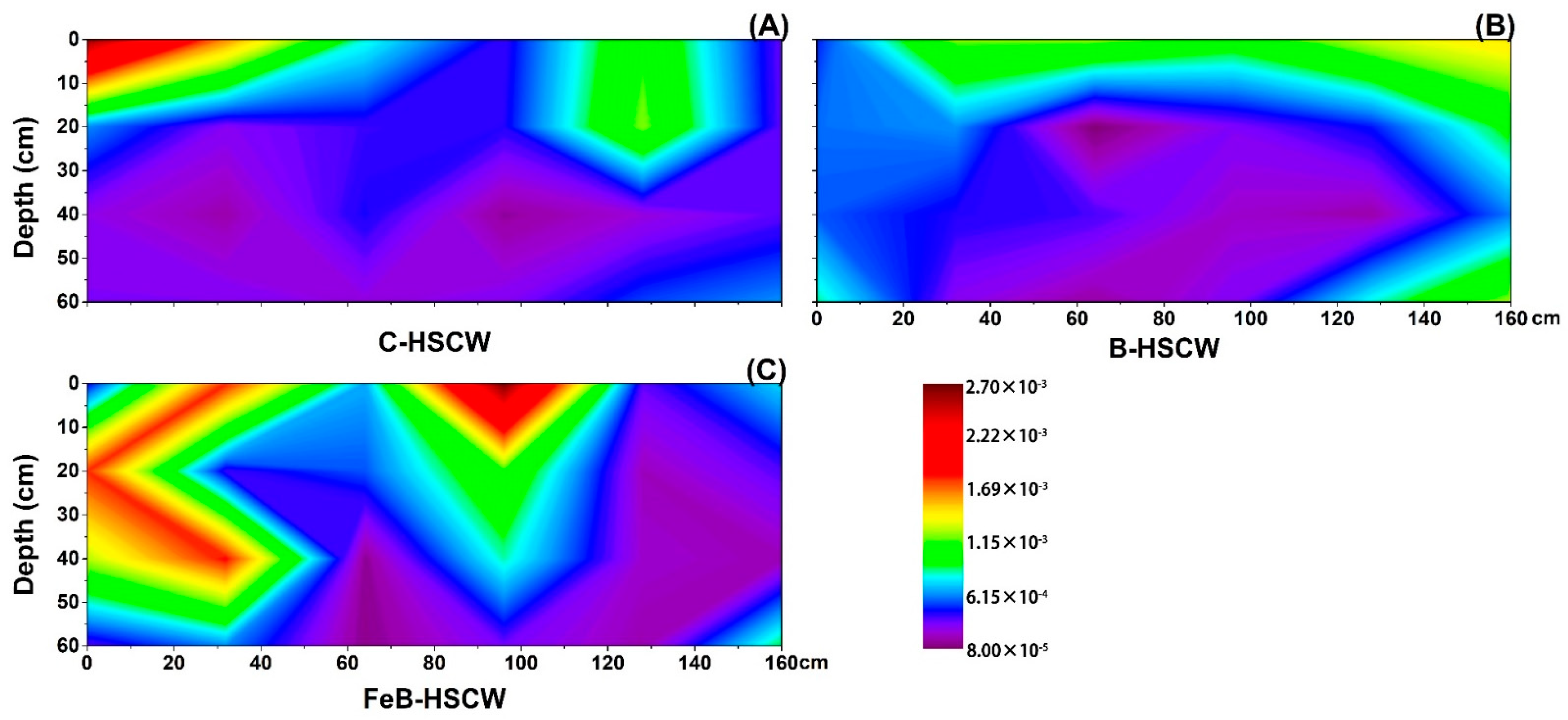
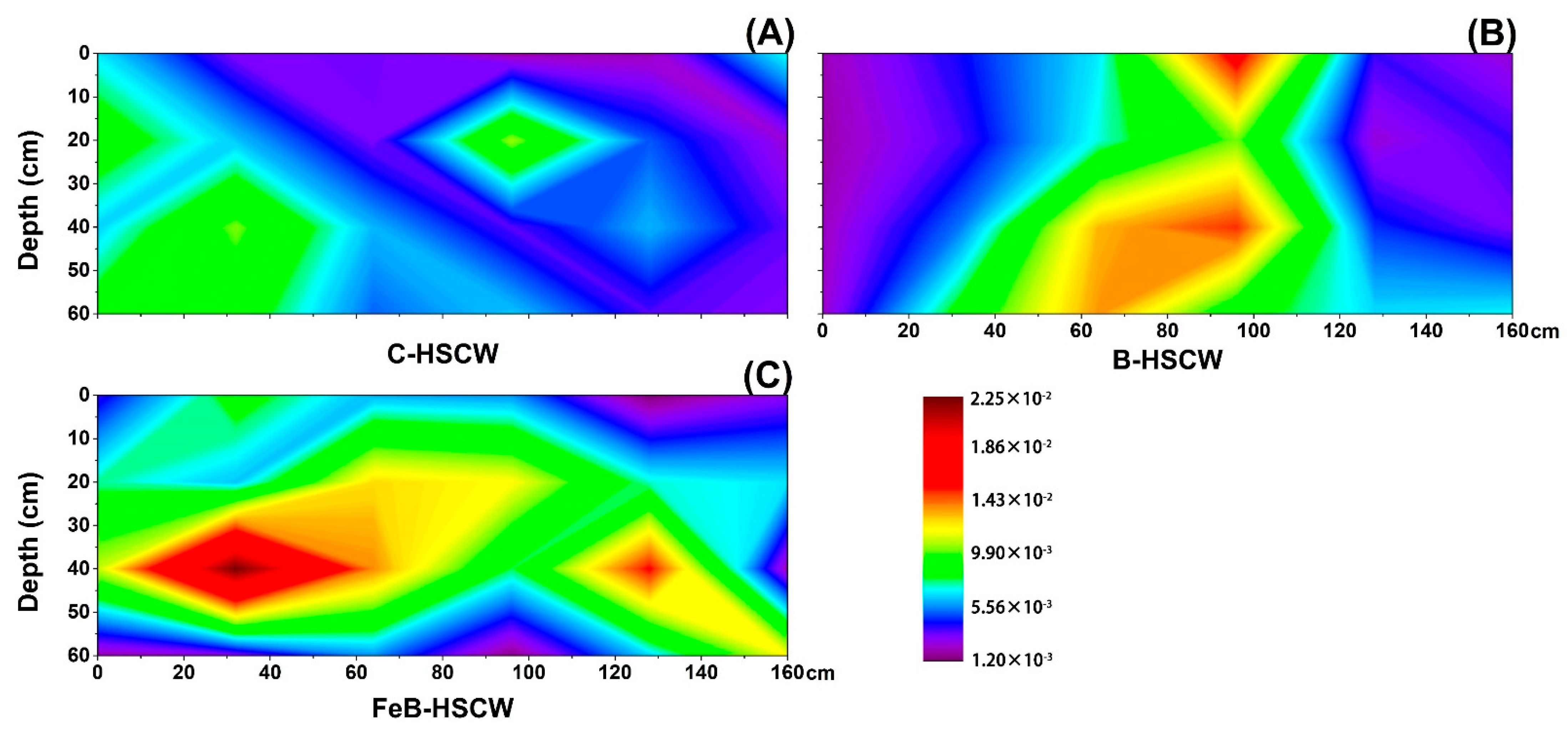


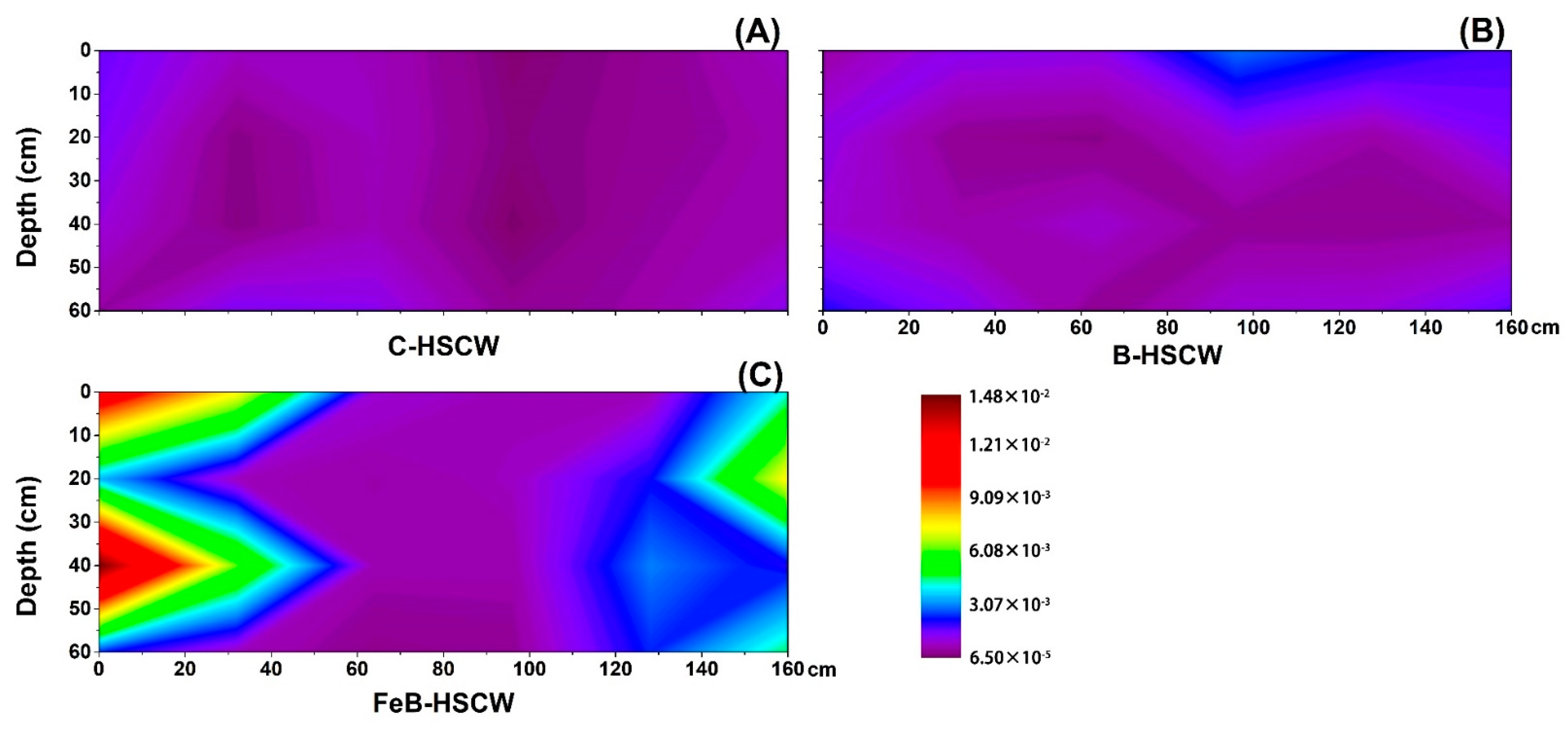
| Estimators | nirS-Denitrifier | nirK-Denitrifier | ||||
|---|---|---|---|---|---|---|
| C-HSCW | B-HSCW | FeB-HSCW | C-HSCW | B-HSCW | FeB-HSCW | |
| Reads | 94707 | 101452 | ||||
| Average Length | 392.39 | 452.089 | ||||
| OTUs | 586 | 674 | 795 | 550 | 630 | 802 |
| Sobs | 394 ± 76.37 | 494 ± 103.24 | 451 ± 49.50 | 376 ± 21.21 | 436.5 ± 2.12 | 444.5 ± 58.69 |
| Shannon | 4.58 ± 0.57 | 4.80 ± 0.54 | 4.68 ± 0.07 | 4.60 ± 0.20 | 4.46 ± 0.31 | 4.80 ± 0.21 |
| Simpson | 0.032 ± 0.016 | 0.024 ± 0.019 | 0.022 ± 0.000 | 0.022± 0.004 | 0.036 ± 0.019 | 0.019 ± 0.006 |
| ACE | 440.29 ± 35.12 | 582.12 ± 115.77 | 523.76 ± 60.54 | 418.83 ± 48.63 | 496.53 ± 13.94 | 487.15 ± 81.17 |
| Chao 1 | 436.49 ± 44.66 | 587.13 ± 131.43 | 523.60 ± 59.32 | 422.62 ± 61.09 | 501.58 ± 10.85 | 480.05 ± 80.34 |
| Coverage | 99.19 ± 0.375% | 98.51 ± 0.304% | 98.74 ± 0.170% | 99.27 ± 0.35% | 99.00 ± 0.12% | 99.21 ± 0.30% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, W.; Yang, L. Community Composition and Spatial Distribution of N-Removing Microorganisms Optimized by Fe-Modified Biochar in a Constructed Wetland. Int. J. Environ. Res. Public Health 2021, 18, 2938. https://doi.org/10.3390/ijerph18062938
Jia W, Yang L. Community Composition and Spatial Distribution of N-Removing Microorganisms Optimized by Fe-Modified Biochar in a Constructed Wetland. International Journal of Environmental Research and Public Health. 2021; 18(6):2938. https://doi.org/10.3390/ijerph18062938
Chicago/Turabian StyleJia, Wen, and Liuyan Yang. 2021. "Community Composition and Spatial Distribution of N-Removing Microorganisms Optimized by Fe-Modified Biochar in a Constructed Wetland" International Journal of Environmental Research and Public Health 18, no. 6: 2938. https://doi.org/10.3390/ijerph18062938
APA StyleJia, W., & Yang, L. (2021). Community Composition and Spatial Distribution of N-Removing Microorganisms Optimized by Fe-Modified Biochar in a Constructed Wetland. International Journal of Environmental Research and Public Health, 18(6), 2938. https://doi.org/10.3390/ijerph18062938





