Metabolomic Alteration in the Plasma of Wild Rodents Environmentally Exposed to Lead: A Preliminary Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rodent Sample Collection
2.2. Identification of Rodent Species
2.3. Estimation of the Age of Rodents
2.4. Extraction and Analysis of Pb
2.5. Metabolome Analysis
2.6. Statistical Analysis
3. Results
3.1. Characteristics and Pb Levels of Animals
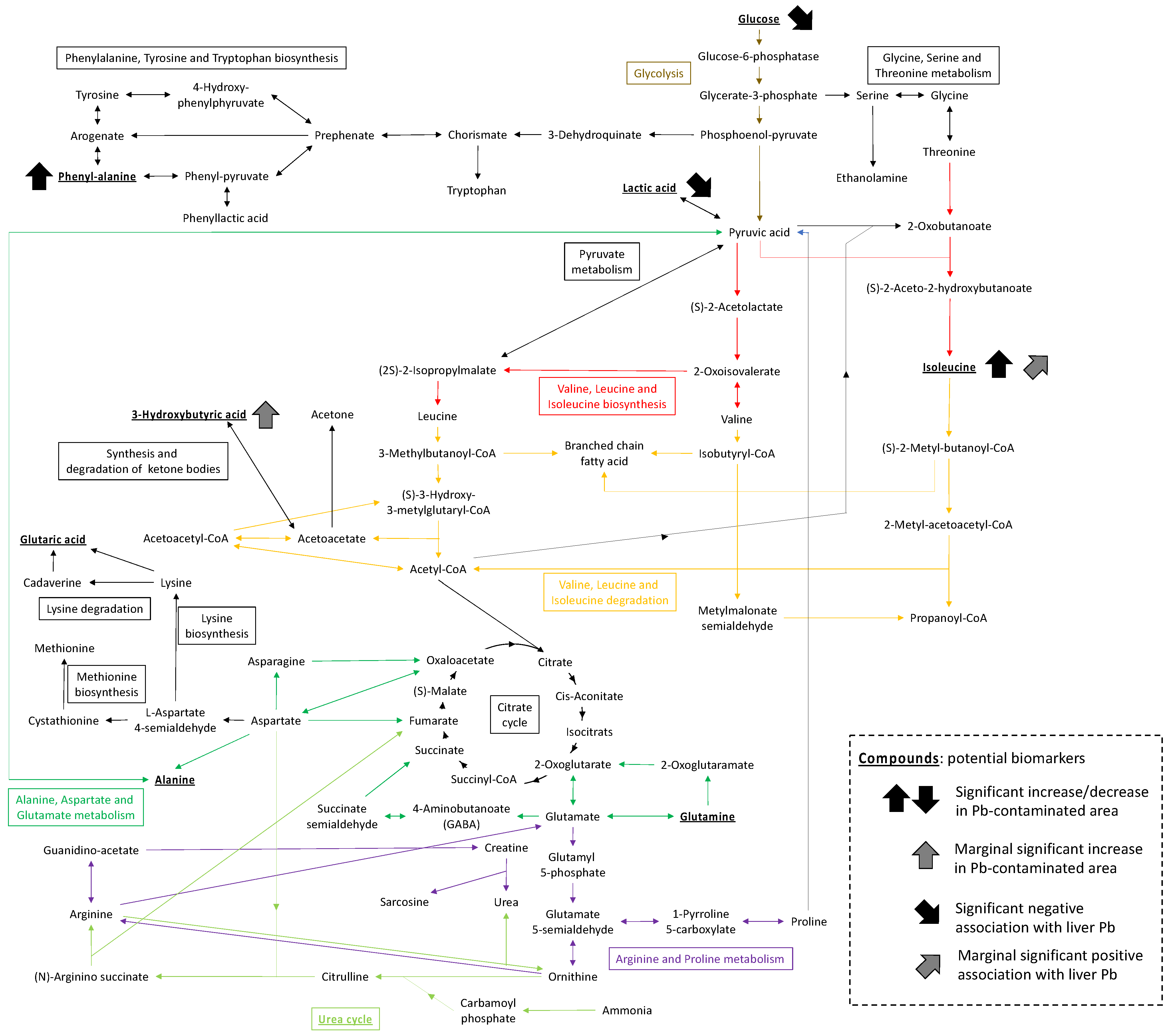
3.2. Metabolome Profiles and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flora, G.; Gupta, D.; Tiwari, A. Toxicity of lead: A review with recent updates. Interdiscip. Toxicol. 2012, 5, 47–58. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Ruckart, P.Z.; Jones, R.L.; Courtney, J.G.; LeBlanc, T.T.; Jackson, W.; Karwowski, M.P.; Cheng, P.Y.; Allwood, P.; Svendsen, E.R.; Breysse, P.N. Update of the Blood Lead Reference Value—United States, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1509–1512. [Google Scholar] [CrossRef]
- García-Sevillano, M.A.; García-Barrera, T.; Gómez-Ariza, J.L. Application of metallomic and metabolomic approaches in exposure experiments on laboratory mice for environmental metal toxicity assessment. Metallomics 2014, 6, 237–248. [Google Scholar] [CrossRef]
- García-Barrera, T.; Rodríguez-Moro, G.; Callejón-Leblic, B.; Arias-Borrego, A.; Gómez-Ariza, J.L. Mass spectrometry based analytical approaches and pitfalls for toxicometabolomics of arsenic in mammals: A tutorial review. Anal. Chim. Acta 2018, 1000, 41–66. [Google Scholar] [CrossRef]
- Kim, K.B.; Um, S.Y.; Chung, M.W.; Jung, S.C.; Oh, J.S.; Kim, S.H.; Na, H.S.; Lee, B.M.; Choi, K.H. Toxicometabolomics approach to urinary biomarkers for mercuric chloride (HgCl2)-induced nephrotoxicity using proton nuclear magnetic resonance (1H NMR) in rats. Toxicol. Appl. Pharmacol. 2010, 249, 114–126. [Google Scholar] [CrossRef]
- Ramirez, T.; Daneshian, M.; Kamp, H.; Bois, F.Y.; Clench, M.R.; Coen, M.; Donley, B.; Fischer, S.M.; Ekman, D.R.; Fabian, E.; et al. Metabolomics in toxicology and preclinical research. Altex 2013, 30, 209. [Google Scholar] [CrossRef] [Green Version]
- Robertson, D.G.; Watkins, P.B.; Reily, M.D. Metabolomics in toxicology: Preclinical and clinical applications. Toxicol. Sci. 2011, 120 (Suppl. 1), S146–S170. [Google Scholar] [CrossRef] [Green Version]
- Dudka, I.; Kossowska, B.; Senhadri, H.; Latajka, R.; Hajek, J.; Andrzejak, R.; Antonowicz-Juchniewicz, J.; Gancarz, R. Metabonomic analysis of serum of workers occupationally exposed to arsenic, cadmium and lead for biomarker research: A preliminary study. Environ. Int. 2014, 68, 71–81. [Google Scholar] [CrossRef]
- Eguchi, A.; Nomiyama, K.; Sakurai, K.; Trang, P.T.K.; Viet, P.H.; Takahashi, S.; Iwata, H.; Tanabe, S.; Todaka, E.; Mori, C. Alterations in urinary metabolomic profiles due to lead exposure from a lead–acid battery recycling site. Environ. Pollut. 2018, 242, 98–105. [Google Scholar] [CrossRef]
- Ladva, C.N.; Golan, R.; Liang, D.; Greenwald, R.; Walker, D.I.; Uppal, K.; Raysoni, A.U.; Tran, V.; Yu, T.; Flanders, W.D.; et al. Particulate metal exposures induce plasma metabolome changes in a commuter panel study. PLoS ONE 2018, 13, e0203468. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xu, X.; He, B.; Guo, J.; Zhao, B.; Zhang, Y.; Zhou, Z.; Zhou, X.; Zhang, R.; Abliz, Z. The impact of chronic environmental metal and benzene exposure on human urinary metabolome among Chinese children and the elderly population. Ecotoxicol. Environ. Saf. 2019, 169, 232–239. [Google Scholar] [CrossRef]
- Mani, M.S.; Joshi, M.B.; Shetty, R.R.; DSouza, V.L.; Swathi, M.; Kabekkodu, S.P.; Dsouza, H.S. Lead exposure induces metabolic reprogramming in rat models. Toxicol. Lett. 2020, 335, 11–27. [Google Scholar] [CrossRef]
- Gao, B.; Chi, L.; Mahbub, R.; Bian, X.; Tu, P.; Ru, H.; Lu, K. Multi-omics reveals that lead exposure disturbs gut microbiome development, key metabolites, and metabolic pathways. Chem. Res. Toxicol. 2017, 30, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, S.M.; Ikenaka, Y.; Hamada, K.; Muzandu, K.; Choongo, K.; Teraoka, H.; Ishizuka, M. Metal and metalloid contamination in roadside soil and wild rats around a Pb–Zn mine in Kabwe, Zambia. Environ. Pollut. 2011, 159, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Doya, R.; Nakayama, S.M.; Nakata, H.; Toyomaki, H.; Yabe, J.; Muzandu, K.; Yohannes, Y.B.; Kataba, A.; Zyambo, G.; Ogawa, T.; et al. Land Use in Habitats Affects Metal Concentrations in Wild Lizards Around a Former Lead Mining Site. Environ. Sci. Technol. 2020, 54, 14474–14481. [Google Scholar] [CrossRef]
- Nakata, H.; Nakayama, S.M.; Yabe, J.; Liazambi, A.; Mizukawa, H.; Darwish, W.S.; Ikenaka, Y.; Ishizuka, M. Reliability of stable Pb isotopes to identify Pb sources and verifying biological fractionation of Pb isotopes in goats and chickens. Environ. Pollut. 2016, 208, 395–403. [Google Scholar] [CrossRef]
- Kataba, A.; Nakayama, S.M.; Nakata, H.; Toyomaki, H.; Yohannes, Y.B.; Yabe, J.; Muzandu, K.; Zyambo, G.; Kubota, A.; Matsukawa, T.; et al. An Investigation of the Wild Rat Crown Incisor as an Indicator of Lead (Pb) Exposure Using Inductively Couple Plasma Mass Spectrometry (ICP-MS) and Laser Ablation ICP-MS. Int. J. Environ. Res. Public Health 2021, 18, 767. [Google Scholar] [CrossRef]
- Nakayama, S.M.; Ikenaka, Y.; Hamada, K.; Muzandu, K.; Choongo, K.; Yabe, J.; Umemura, T.; Ishizuka, M. Accumulation and biological effects of metals in wild rats in mining areas of Zambia. Environ. Monit. Assess. 2013, 185, 4907–4918. [Google Scholar] [CrossRef]
- Nakata, H.; Nakayama, S.M.; Yabe, J.; Muzandu, K.; Toyomaki, H.; Yohannes, Y.B.; Kataba, A.; Zyambo, G.; Ikenaka, Y.; Choongo, K.; et al. Clinical biochemical parameters associated with the exposure to multiple environmental metals in residents from Kabwe, Zambia. Chemosphere 2021, 262, 127788. [Google Scholar] [CrossRef]
- Yabe, J.; Nakayama, S.M.; Ikenaka, Y.; Yohannes, Y.B.; Bortey-Sam, N.; Oroszlany, B.; Muzandu, K.; Choongo, K.; Kabalo, A.N.; Ntapisha, J.; et al. Lead poisoning in children from townships in the vicinity of a lead–zinc mine in Kabwe, Zambia. Chemosphere 2015, 119, 941–947. [Google Scholar] [CrossRef]
- Robins, J.H.; Hingston, M.; Matisoo-Smith, E.; Ross, H.A. Identifying Rattus species using mitochondrial DNA. Mol. Ecol. Notes 2007, 7, 717–729. [Google Scholar] [CrossRef]
- Tanikawa, T. An eye-lens weight curve for determining age in black rats, Rattus rattus. J. Mammal. Soc. Jpn. 1993, 18, 49–51. [Google Scholar]
- Fichet-Calvet, E.; Lecompte, E.; Koivogui, L.; Daffis, S.; Meulen, J.T. Reproductive characteristics of Mastomys natalensis and Lassa virus prevalence in Guinea, West Africa. Vector-Borne Zoonotic Dis. 2008, 8, 41–48. [Google Scholar] [CrossRef]
- Nakata, H.; Nakayama, S.M.; Kataba, A.; Yohannes, Y.B.; Ikenaka, Y.; Ishizuka, M. Evaluation of the ameliorative effect of Spirulina (Arthrospira platensis) supplementation on parameters relating to lead poisoning and obesity in C57BL/6J mice. J. Funct. Foods 2021, 77, 104344. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.N.; Ziegler, A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. arXiv Prepr. 2015, arXiv:1508.04409. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Altmann, A.; Tolosi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef]
- López-Ibáñez, J.; Pazos, F.; Chagoyen, M. MBROLE 2.0—functional enrichment of chemical compounds. Nucleic Acids Res. 2016, 44, W201–W204. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36 (Suppl. 1), D480–D484. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Scalbert, A. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midfold, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Colangelo, P.; Verheyen, E.; Leirs, H.; Tatard, C.; Denys, C.; Dobigny, G.; Duplantier, J.M.; Brouat, C.; Granjon, L.; Lecompte, E. A mitochondrial phylogeographic scenario for the most widespread African rodent, Mastomys natalensis. Biol. J. Linn. Soc. 2013, 108, 901–916. [Google Scholar] [CrossRef] [Green Version]
- Hardin, A.; Nevonen, K.A.; Eckalbar, W.L.; Carbone, L.; Ahituv, N. Comparative Genomic Characterization of the Multimammate Mouse Mastomys coucha. Mol. Biol. Evol. 2019, 36, 2805–2812. [Google Scholar] [CrossRef]
- Lecompte, E.; Aplin, K.; Denys, C.; Catzeflis, F.; Chades, M.; Chevret, P. Phylogeny and biogeography of African Murinae based on mitochondrial and nuclear gene sequences, with a new tribal classification of the subfamily. BMC Evol. Biol. 2008, 8, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.M.; Farag, E.; Mahmoudi, A.; Hassan, M.M.; Atta, M.; Mostafavi, E.; Alnager, I.A.; Farrag, H.A.; Eljack, G.E.A.; Bansal, D.; et al. Morphometric Study of Mus musculus, Rattus norvegicus, and Rattus in Qatar. Animals 2021, 11, 2162. [Google Scholar] [CrossRef]
- Odigie, A.E.; Ekeolu, K.O.; Asemota, D.O.; Uwagie-Ero, E.A.; Aighewi, I.T.; Ighedosa, S.U.; Usifoh, S.F.; Olugasa, B.O.; Asemota, O.; Fagboya, T.E. Comparative non-metric and morphometric analyses of rats at residential halls of the University of Benin campus, Nigeria. J. Infect. Public Health 2018, 11, 412–417. [Google Scholar] [CrossRef]
- Safronetz, D.; Rosenke, K.; Fischer, R.J.; LaCasse, R.A.; Scott, D.P.; Saturday, G.; Hanley, P.W.; Maiga, O.; Sogoba, N.; Schwan, T.G.; et al. Establishment of a genetically confirmed breeding colony of Mastomys natalensis from wild-caught founders from West Africa. Viruses 2021, 13, 590. [Google Scholar] [CrossRef]
- Xia, J.; Lu, L.; Jin, C.; Wang, S.; Zhou, J.; Ni, Y.; Fu, Z.; Jin, Y. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 209, 1–8. [Google Scholar] [CrossRef]
- Price, E.R. The physiology of lipid storage and use in reptiles. Biol. Rev. 2017, 92, 1406–1426. [Google Scholar] [CrossRef]




| Species and Sample Size | Sex (Male, Female) | Body Weight * (g) | Estimated Age ** (day) | Blood Pb ** (µg/dL) | Liver Pb ** (mg/kg, dw) | Kidney Pb *** (mg/kg, dw) | |
|---|---|---|---|---|---|---|---|
| Mutwe Wansofu (polluted site) | Total (N = 18) | 14, 4 | 108 ± 44.3 | 139 ± 107 | 33.6 ± 25.4 | 5.15 ± 3.98 | 14.2 ± 12.2 |
| R. rattus (N = 17) | 14, 3 | 113 ± 40.5 | 190 ± 163 | 33.1 ± 26.0 | 4.36 ± 2.24 | 13.6 ± 12.3 | |
| M. natalensis (N = 1) | 0, 1 | 25.2 | 203 | 42.9 | 15.8 | 24.4 | |
| Kang’omba (control site) | Total (N = 9) | 5, 4 | 49.5 ± 30.1 | 227 ± 136 | 5.62 ± 1.86 | 1.07 ± 0.542 | 4.26 ± 1.03 |
| R. rattus (N = 2) | 1, 1 | 91.3, 107 | 75.9, 234 | 5.57, 6.44 | 0.483, 0.624 | 2.85, 4.83 | |
| M. natalensis (N = 7) | 4, 3 | 35.3 ± 11.5 | 248 ± 142 | 5.50 ± 2.11 | 1.22 ± 0.524 | 4.37 ± 1.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakata, H.; Eguchi, A.; Nakayama, S.M.M.; Yabe, J.; Muzandu, K.; Ikenaka, Y.; Mori, C.; Ishizuka, M. Metabolomic Alteration in the Plasma of Wild Rodents Environmentally Exposed to Lead: A Preliminary Study. Int. J. Environ. Res. Public Health 2022, 19, 541. https://doi.org/10.3390/ijerph19010541
Nakata H, Eguchi A, Nakayama SMM, Yabe J, Muzandu K, Ikenaka Y, Mori C, Ishizuka M. Metabolomic Alteration in the Plasma of Wild Rodents Environmentally Exposed to Lead: A Preliminary Study. International Journal of Environmental Research and Public Health. 2022; 19(1):541. https://doi.org/10.3390/ijerph19010541
Chicago/Turabian StyleNakata, Hokuto, Akifumi Eguchi, Shouta M. M. Nakayama, John Yabe, Kaampwe Muzandu, Yoshinori Ikenaka, Chisato Mori, and Mayumi Ishizuka. 2022. "Metabolomic Alteration in the Plasma of Wild Rodents Environmentally Exposed to Lead: A Preliminary Study" International Journal of Environmental Research and Public Health 19, no. 1: 541. https://doi.org/10.3390/ijerph19010541
APA StyleNakata, H., Eguchi, A., Nakayama, S. M. M., Yabe, J., Muzandu, K., Ikenaka, Y., Mori, C., & Ishizuka, M. (2022). Metabolomic Alteration in the Plasma of Wild Rodents Environmentally Exposed to Lead: A Preliminary Study. International Journal of Environmental Research and Public Health, 19(1), 541. https://doi.org/10.3390/ijerph19010541







