A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster
Abstract
:1. Introduction
Highlights
- Estimates of infectious disease incidence can be time-consuming and tedious;
- Deep learning, in particular RNN-GRU, for automatic data extraction from the free text could be a feasible and timely option;
- Results obtained with MLT were promising, yet, in future development, this text-mining tool should be readily usable by non-technical users as well.
2. Materials and Methods
2.1. Electronic Medical Record Database
2.2. Main Strategy
2.3. Model Choice
2.4. Language Model
2.5. Implementation
- Embeddings: representation of words converted into a syntactic and semantically coherent 300-dimensional structure—input N × 10.000/output N × 10.000 × 300, where N is the number of cases (i.e., one case is the collection of all the HERs of a single child for a given year) in the dataset/minibatch considered.
- Two synchronized layers of bidirectional RNN-GRU modules are composed of 256 nodes each (512 nodes of each layer process the information as a pure sequence summarizing its “meaning”—input N × 10.000 × 300/output N × 512 (output for the first synchronized layer is equal to the input of the second: N × 10.000 × 512))
- Two fully connected layers of 128 nodes each to process the “meaning” vector of information from each record—input N × 512/output N × 128 (output for the first synchronized layer is equal to the input of the second: N × 128).
- Logistic output node: to produce a probability measure for the children being affected by VZV in the corresponding year—input N × 128/output N × 1.
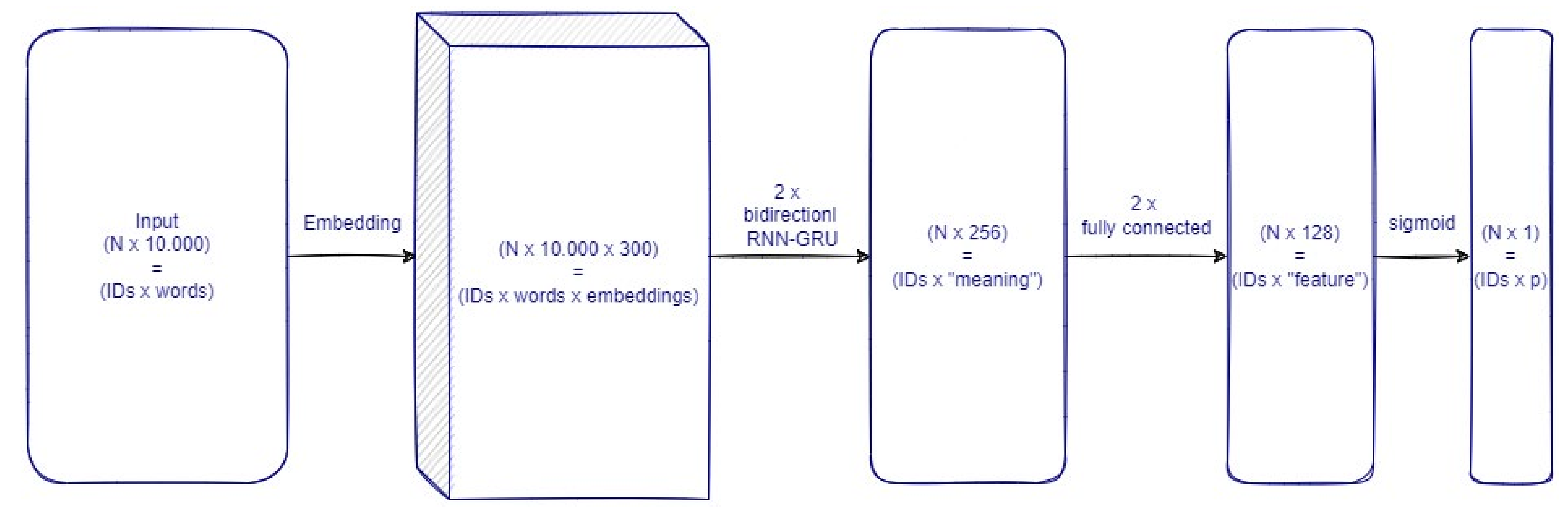
2.6. Training
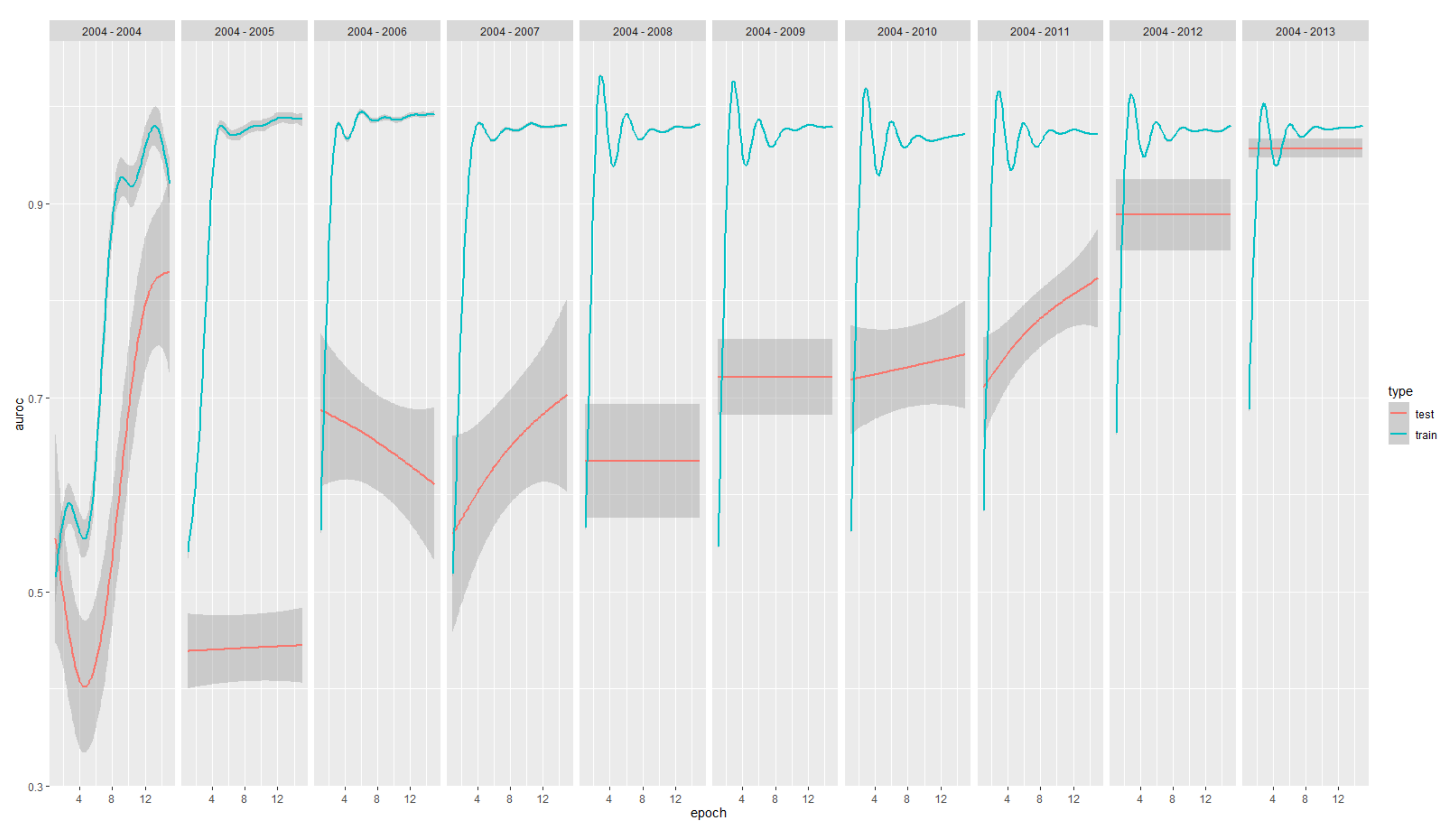
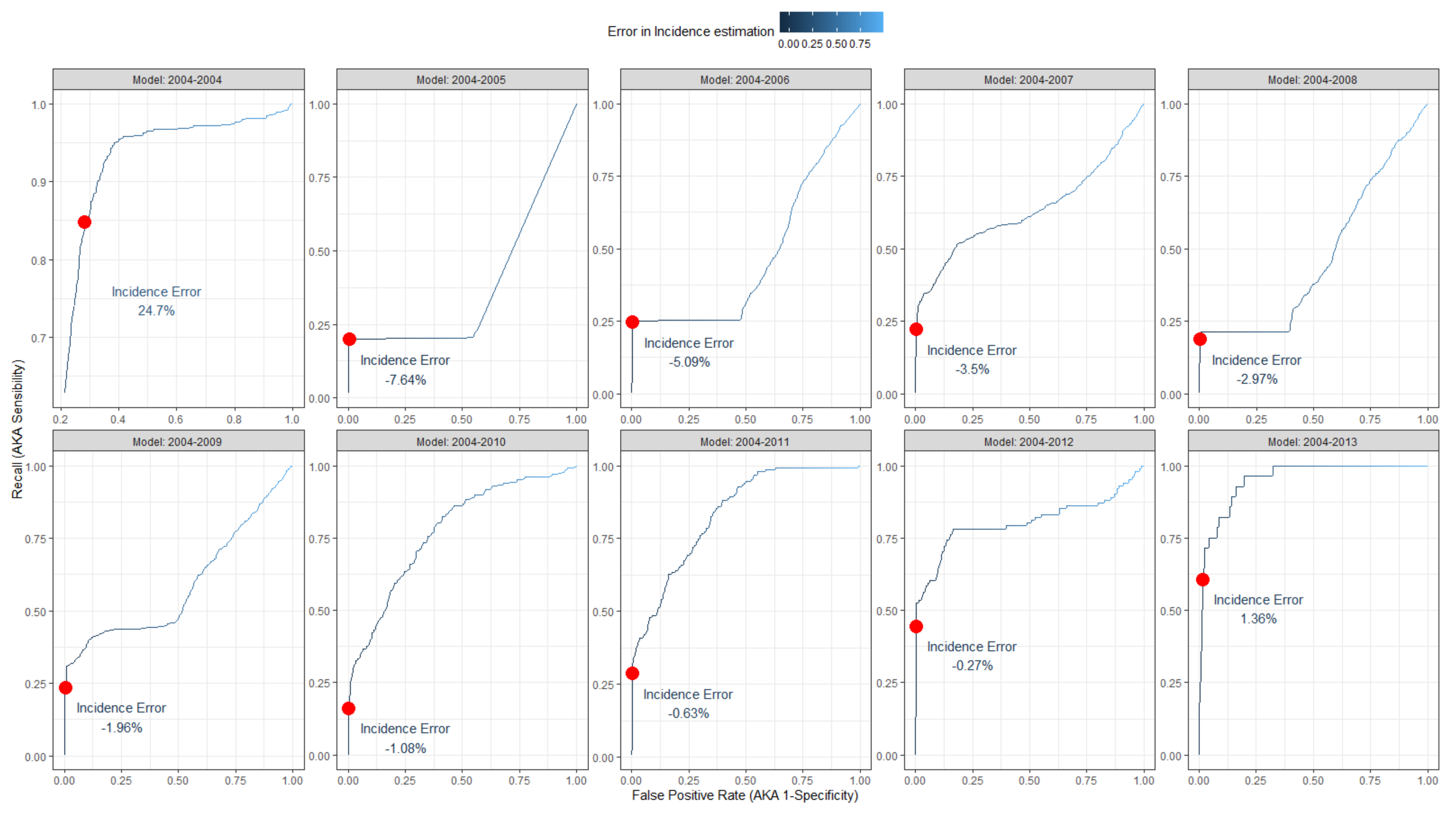
3. Results
Computational Environment
4. Discussion
Limitations
Some Limitations Must Be Acknowledged
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kretzschmar, M.; Mangen, M.J.; Pinheiro, P.; Jahn, B.; Fevre, E.M.; Longhi, S.; Lai, T.; Havelaar, A.H.; Stein, C.; Cassini, A.; et al. New Methodology for Estimating the Burden of Infectious Diseases in Europe. PLoS Med. 2012, 9, e1001205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magill, S.S.; Dumyati, G.; Ray, S.M.; Fridkin, S.K. Evaluating Epidemiology and Improving Surveillance of Infections Associated with Health Care, United States. Emerg. Infect. Dis. 2015, 21, 1537–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baracco, G.J.; Eisert, S.; Saavedra, S.; Hirsch, P.; Marin, M.; Ortega-Sanchez, I.R. Clinical and Economic Impact of Various Strategies for Varicella Immunity Screening and Vaccination of Health Care Personnel. Am. J. Infect. Control 2015, 43, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Damm, O.; Ultsch, B.; Horn, J.; Mikolajczyk, R.T.; Greiner, W.; Wichmann, O. Systematic Review of Models Assessing the Economic Value of Routine Varicella and Herpes Zoster Vaccination in High-Income Countries. BMC Public Health 2015, 15, 533. [Google Scholar] [CrossRef] [Green Version]
- Gabutti, G.; Rota, M.C.; Guido, M.; De Donno, A.; Bella, A.; Ciofi degli Atti, M.L.; Crovari, P. The Epidemiology of Varicella Zoster Virus Infection in Italy. BMC Public Health 2008, 8, 372. [Google Scholar] [CrossRef] [Green Version]
- De Bie, S.; Coloma, P.M.; Ferrajolo, C.; Verhamme, K.M.; Trifiro, G.; Schuemie, M.J.; Straus, S.M.; Gini, R.; Herings, R.; Mazzaglia, G.; et al. The Role of Electronic Healthcare Record Databases in Paediatric Drug Safety Surveillance: A Retrospective Cohort Study. Br. J. Clin. Pharmacol. 2015, 80, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, S.M.; Kaelber, D.C.; Downing, N.L.; Goel, V.V.; Longhurst, C.A. Electronic Health Record-Enabled Research in Children Using the Electronic Health Record for Clinical Discovery. Pediatr. Clin. 2016, 63, 251–268. [Google Scholar] [CrossRef]
- Rosier, A.; Burgun, A.; Mabo, P. Using Regular Expressions to Extract Information on Pacemaker Implantation Procedures from Clinical Reports. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 8–12 November 2008; pp. 81–85. [Google Scholar]
- Nassif, H.; Woods, R.; Burnside, E.; Ayvaci, M.; Shavlik, J.; Page, D. Information Extraction for Clinical Data Mining: A Mammography Case Study. In Proceedings of the 2009 IEEE International Conference on Data Mining Workshops, Miami, FL, USA, 6 December 2009; pp. 37–42. [Google Scholar]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [Green Version]
- Witten, I.H.; Frank, E.; Hall, M.A. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2011. [Google Scholar]
- Lanera, C.; Berchialla, P.; Baldi, I.; Lorenzoni, G.; Tramontan, L.; Scamarcia, A.; Cantarutti, L.; Giaquinto, C.; Gregori, D. Use of Machine Learning Techniques for Case-Detection of Varicella Zoster Using Routinely Collected Textual Ambulatory Records: Pilot Observational Study. JMIR Med. Inform. 2020, 8, e14330. [Google Scholar] [CrossRef]
- Lanera, C.; Berchialla, P.; Sharma, A.; Minto, C.; Gregori, D.; Baldi, I. Screening PubMed Abstracts: Is Class Imbalance Always a Challenge to Machine Learning? Syst. Rev. 2019, 8, 317. [Google Scholar] [CrossRef]
- Hahn, U.; Oleynik, M. Medical Information Extraction in the Age of Deep Learning. Yearb. Med. Inform. 2020, 29, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Jozefowicz, R.; Zaremba, W.; Sutskever, I. An Empirical Exploration of Recurrent Network Architectures. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 2342–2350. [Google Scholar]
- Dona, D.; Mozzo, E.; Scamarcia, A.; Picelli, G.; Villa, M.; Cantarutti, L.; Giaquinto, C. Community-Acquired Rotavirus Gastroenteritis Compared with Adenovirus and Norovirus Gastroenteritis in Italian Children: A Pedianet Study. Int. J. Pediatr. 2016, 2016, 5236243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, E.; Porcu, G.; Hu, T.; Petigara, T.; Senese, F.; Prandi, G.M.; Scamarcia, A.; Cantarutti, L.; Cantarutti, A.; Giaquinto, C. A Retrospective Database Analysis to Estimate the Burden of Acute Otitis Media in Children Aged <15 Years in the Veneto Region (Italy). Children 2022, 9, 436. [Google Scholar] [CrossRef] [PubMed]
- Nicolosi, A.; Sturkenboom, M.; Mannino, S.; Arpinelli, F.; Cantarutti, L.; Giaquinto, C. The Incidence of Varicella: Correction of a Common Error. Epidemiology 2003, 14, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder–Decoder Approaches. In Proceedings of the Proceedings of SSST-8, Eighth Workshop on Syntax, Semantics and Structure in Statistical Translation, Doha, Qatar, 25 October 2014; Association for Computational Linguistics: Doha, Qatar, 2014; pp. 103–111. [Google Scholar]
- Shi, J.; Gao, X.; Kinsman, W.C.; Ha, C.; Gao, G.G.; Chen, Y. DI++: A Deep Learning System for Patient Condition Identification in Clinical Notes. Artif. Intell. Med. 2022, 123, 102224. [Google Scholar] [CrossRef]
- Chae, S.; Kwon, S.; Lee, D. Predicting Infectious Disease Using Deep Learning and Big Data. Int. J. Environ. Res. Public Health 2018, 15, 1596. [Google Scholar] [CrossRef] [Green Version]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching Word Vectors with Subword Information. Trans. Assoc. Comput. Linguist. 2017, 5, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Robbins, H.; Monro, S. A Stochastic Approximation Method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Yao, Y.; Rosasco, L.; Caponnetto, A. On Early Stopping in Gradient Descent Learning. Constr. Approx. 2007, 26, 289–315. [Google Scholar] [CrossRef]
- Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958.
- Habibzadeh, F.; Habibzadeh, P.; Yadollahie, M. On Determining the Most Appropriate Test Cut-off Value: The Case of Tests with Continuous Results. Biochem Med. 2016, 26, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Liu, X. Classification Accuracy and Cut Point Selection. Statist. Med. 2012, 31, 2676–2686. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Holub, K.; Hardy, N.; Kallmes, K. Toward Automated Data Extraction according to Tabular Data Structure: Cross-Sectional Pilot Survey of the Comparative Clinical Literature. JMIR Form. Res. 2021, 5, e33124. [Google Scholar] [CrossRef]
- Dai, H.-J.; Su, C.-H.; Lee, Y.-Q.; Zhang, Y.-C.; Wang, C.-K.; Kuo, C.-J.; Wu, C.-S. Deep Learning-Based Natural Language Processing for Screening Psychiatric Patients. Front. Psychiatry 2021, 11, 533949. [Google Scholar] [CrossRef]
- Ding, Y.; Ma, J.; Luo, X. Applications of Natural Language Processing in Construction. Autom. Constr. 2022, 136, 104169. [Google Scholar] [CrossRef]
- Xu, Q.; Gel, Y.R.; Ramirez Ramirez, L.L.; Nezafati, K.; Zhang, Q.; Tsui, K.-L. Forecasting Influenza in Hong Kong with Google Search Queries and Statistical Model Fusion. PLoS ONE 2017, 12, e0176690. [Google Scholar] [CrossRef] [Green Version]
- Howard, J.; Ruder, S. Universal Language Model Fine-Tuning for Text Classification. arXiv 2018, arXiv:1801.06146. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A Pre-Trained Biomedical Language Representation Model for Biomedical Text Mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef]
- Shi, J.; Liu, S.; Pruitt, L.C.C.; Luppens, C.L.; Ferraro, J.P.; Gundlapalli, A.V.; Chapman, W.W.; Bucher, B.T. Using Natural Language Processing to Improve EHR Structured Data-Based Surgical Site Infection Surveillance. In Proceedings of the AMIA Annual Symposium Proceedings, Washington, DC, USA, 16–20 November 2019; Volume 2019, pp. 794–803. [Google Scholar]
- Mitra, A.; Rawat, B.P.S.; McManus, D.D.; Yu, H. Relation Classification for Bleeding Events from Electronic Health Records Using Deep Learning Systems: An Empirical Study. JMIR Med. Inform. 2021, 9, e27527. [Google Scholar] [CrossRef]

| N | VZV Negative (N = 58,334) | VZV Positive (N = 2325) | |
|---|---|---|---|
| Sex | 60,659 | ||
| Female | 47% (27,340) | 46% (1068) | |
| Male | 53% (30,994) | 54% (1257) | |
| Age [days] | 60,342 | 0.7/2.2/4.28 * | 0.6/1.4/3.1 |
| Years | Training Phase | Testing Phase | |||
|---|---|---|---|---|---|
| Train | Test | Train (#) | Validation (#) | Train (#) | Test (#) |
| 2004 | 2005–2006 | 1588 | 396 | 1984 | 7854 |
| 2004–2005 | 2006–2007 | 4405 | 1099 | 5504 | 9454 |
| 2004–2006 | 2007–2008 | 7873 | 1965 | 9838 | 10,852 |
| 2004–2007 | 2008–2009 | 11,969 | 2389 | 14,958 | 12,020 |
| 2004–2008 | 2009–2010 | 16,555 | 4135 | 20,690 | 13,062 |
| 2004–2009 | 2010–2011 | 21,586 | 5392 | 26,987 | 13,848 |
| 2004–2010 | 2011–2012 | 27,006 | 6746 | 33,752 | 14,139 |
| 2004–2011 | 2012–2013 | 32,666 | 8160 | 40,826 | 14,017 |
| 2004–2012 | 2013–2014 | 38,319 | 9572 | 47,891 | 12,768 |
| 2004–2013 | 2014 | 43,882 | 10,961 | 54,843 | 5816 |
| Model Year | Positives | Negatives | AUC ROC | tp | tn | fp | fn | prec | rec |
|---|---|---|---|---|---|---|---|---|---|
| 2004–2004 | 637 | 1.954 | 0.804 | 540 | 5.180 | 2.037 | 97 | 0.210 | 0.848 |
| 2004–2005 | 172 | 3.348 | 0.385 | 188 | 8.474 | 35 | 757 | 0.843 | 0.199 |
| 2004–2006 | 465 | 3.869 | 0.588 | 194 | 10.024 | 41 | 593 | 0.826 | 0.247 |
| 2004–2007 | 480 | 4.640 | 0.649 | 130 | 11.403 | 33 | 454 | 0.798 | 0.223 |
| 2004–2008 | 307 | 5.425 | 0.582 | 102 | 12.470 | 51 | 439 | 0.667 | 0.189 |
| 2004–2009 | 277 | 6.011 | 0.652 | 98 | 13.386 | 46 | 318 | 0.681 | 0.236 |
| 2004–2010 | 264 | 6.510 | 0.775 | 37 | 13.870 | 40 | 192 | 0.481 | 0.162 |
| 2004–2011 | 152 | 6.922 | 0.835 | 43 | 13.848 | 19 | 107 | 0.694 | 0.287 |
| 2004–2012 | 77 | 6.988 | 0.832 | 45 | 12.645 | 22 | 56 | 0.672 | 0.446 |
| 2004–2013 | 73 | 6.879 | 0.953 | 17 | 5.698 | 90 | 11 | 0.159 | 0.607 |
| Model Year | Years Estimated | Positives | Negatives | Observed Incidence (%) | Estimated Incidence (%) | Estimated Incidence Error (%) |
|---|---|---|---|---|---|---|
| 2004 | 2005–2006 | 637 | 1.954 | 8.11 | 32.8 | 24.7 |
| 2004–2005 | 2006–2007 | 172 | 3.348 | 10 | 2.36 | −7.64 |
| 2004–2006 | 2007–2008 | 465 | 3.869 | 7.25 | 2.17 | −5.09 |
| 2004–2007 | 2008–2009 | 480 | 4.640 | 4.86 | 1.36 | −3.5 |
| 2004–2008 | 2009–2010 | 307 | 5.425 | 4.14 | 1.17 | −2.97 |
| 2004–2009 | 2010–2011 | 277 | 6.011 | 3 | 1.04 | −1.96 |
| 2004–2010 | 2011–2012 | 264 | 6.510 | 1.62 | 0.54 | −1.08 |
| 2004–2011 | 2012–2013 | 152 | 6.922 | 1.07 | 0.44 | −0.63 |
| 2004–2012 | 2013–2014 | 77 | 6.988 | 0.79 | 0.52 | −0.27 |
| 2004–2013 | 2014 | 73 | 6.879 | 0.48 | 1.84 | 1.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanera, C.; Baldi, I.; Francavilla, A.; Barbieri, E.; Tramontan, L.; Scamarcia, A.; Cantarutti, L.; Giaquinto, C.; Gregori, D. A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster. Int. J. Environ. Res. Public Health 2022, 19, 5959. https://doi.org/10.3390/ijerph19105959
Lanera C, Baldi I, Francavilla A, Barbieri E, Tramontan L, Scamarcia A, Cantarutti L, Giaquinto C, Gregori D. A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster. International Journal of Environmental Research and Public Health. 2022; 19(10):5959. https://doi.org/10.3390/ijerph19105959
Chicago/Turabian StyleLanera, Corrado, Ileana Baldi, Andrea Francavilla, Elisa Barbieri, Lara Tramontan, Antonio Scamarcia, Luigi Cantarutti, Carlo Giaquinto, and Dario Gregori. 2022. "A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster" International Journal of Environmental Research and Public Health 19, no. 10: 5959. https://doi.org/10.3390/ijerph19105959
APA StyleLanera, C., Baldi, I., Francavilla, A., Barbieri, E., Tramontan, L., Scamarcia, A., Cantarutti, L., Giaquinto, C., & Gregori, D. (2022). A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster. International Journal of Environmental Research and Public Health, 19(10), 5959. https://doi.org/10.3390/ijerph19105959








