The Detection of SARS-CoV2 Antigen in Wastewater Using an Automated Chemiluminescence Enzyme Immunoassay
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Limit of Detection (LOD)
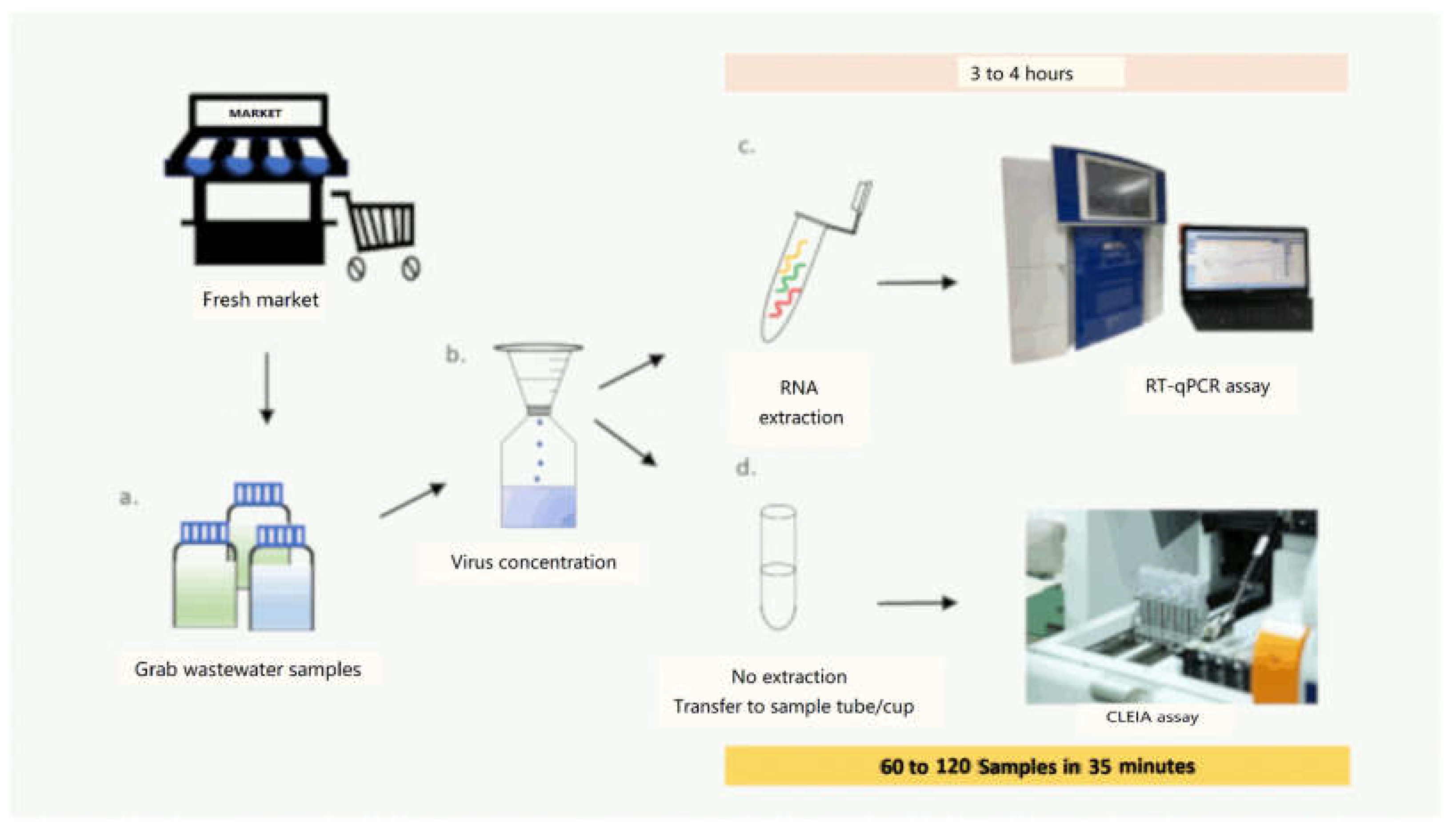
2.1.1. Sample Preparation and Concentration
2.1.2. SARS-CoV-2 Virus Detection and Quantification by RT-qPCR
2.1.3. Detection of SARS-CoV-2 Antigen
2.2. The Field Performance of the SARS-CoV-2 CLEIA
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Mecenas, P.; da Rosa Moreira Bastos, R.T.; Vallinoto, A.C.R.; Normando, D. Effects of temperature and humidity on the spread of COVID-19: A systematic review. PLoS ONE 2020, 15, e0238339. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention National Wastewater Surveillance System (NWSS). Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.html (accessed on 15 December 2021).
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirotsu, Y.; Maejima, M.; Shibusawa, M.; Nagakubo, Y.; Hosaka, K.; Amemiya, K.; Sueki, H.; Hayakawa, M.; Mochizuki, H.; Tsutsui, T.; et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci. Rep. 2020, 10, 18899. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Nagasawa, T.; Ishii, Y.; Yagi, S.; Okuma, S.; Kashiwagi, K.; Maeda, T.; Miyazaki, T.; Yoshizawa, S.; Tateda, K. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2021, 27, 613–616. [Google Scholar] [CrossRef]
- Menchinelli, G.; Bordi, L.; Liotti, F.M.; Palucci, I.; Capobianchi, M.R.; Sberna, G.; Lalle, E.; Romano, L.; De Angelis, G.; Marchetti, S.; et al. Lumipulse G SARS-CoV-2 Ag assay evaluation using clinical samples from different testing groups. Clin. Chem. Lab. Med. 2021, 59, 1468–1476. [Google Scholar] [CrossRef]
- Kobayashi, R.; Murai, R.; Asanuma, K.; Fujiya, Y.; Takahashi, S. Evaluating a novel, highly sensitive, and quantitative reagent for detecting SARS-CoV-2 antigen. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2021, 27, 800–807. [Google Scholar] [CrossRef]
- Ogawa, T.; Fukumori, T.; Nishihara, Y.; Sekine, T.; Okuda, N.; Nishimura, T.; Fujikura, H.; Hirai, N.; Imakita, N.; Kasahara, K. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. J. Clin. Virol. 2020, 131, 104612. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Options for the Use of Rapid Antigen Tests for COVID-19 in the EU/EEA and the UK. 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Options-use-of-rapid-antigen-tests-for-COVID-19_0.pdf (accessed on 15 December 2021).
- Gili, A.; Paggi, R.; Russo, C.; Cenci, E.; Pietrella, D.; Graziani, A.; Stracci, F.; Mencacci, A. Evaluation of Lumipulse® G SARS-CoV-2 antigen assay automated test for detecting SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs for community and population screening. Int. J. Infect. Dis. 2021, 105, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Thongpradit, S.; Prasongtanakij, S.; Srisala, S.; Kumsang, Y.; Chanprasertyothin, S.; Boonkongchuen, P.; Pitidhammabhorn, D.; Manomaipiboon, P.; Somchaiyanon, P.; Chandanachulaka, S.; et al. A Simple Method to Detect SARS-CoV-2 in Wastewater at Low Virus Concentration. J. Environ. Public Health 2022, 2022, 4867626. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Henry, B.M.; Adeli, K.; Plebani, M. Fujirebio Lumipulse SARS-CoV-2 antigen immunoassay: Pooled analysis of diagnostic accuracy. Diagnosis 2022, 9, 149–156. [Google Scholar] [CrossRef]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, U.; Perret-Liaudet, A.; van Waalwijk van Doorn, L.J.C.; Blennow, K.; Chiasserini, D.; Engelborghs, S.; Fladby, T.; Genc, S.; Kruse, N.; Kuiperij, H.B.; et al. A Practical Guide to Immunoassay Method Validation. Front. Neurol. 2015, 6, 179. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Maejima, M.; Shibusawa, M.; Nagakubo, Y.; Hosaka, K.; Amemiya, K.; Sueki, H.; Hayakawa, M.; Mochizuki, H.; Tsutsui, T.; et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020, 99, 397–402. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Maejima, M.; Shibusawa, M.; Natori, Y.; Nagakubo, Y.; Hosaka, K.; Sueki, H.; Amemiya, K.; Hayakawa, M.; Mochizuki, H.; et al. Direct comparison of Xpert Xpress, FilmArray Respiratory Panel, Lumipulse antigen test, and RT-qPCR in 165 nasopharyngeal swabs. BMC Infect. Dis. 2022, 22, 221. [Google Scholar] [CrossRef]
- Ishii, T.; Sasaki, M.; Yamada, K.; Kato, D.; Osuka, H.; Aoki, K.; Morita, T.; Ishii, Y.; Tateda, K. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2021, 27, 915–918. [Google Scholar] [CrossRef]
- Sberna, G.; Basile, F.; Guarino, M.L.; Capobianchi, M.R.; Bordi, L.; Parisi, G. Comparison of Allplex™ SARS-CoV-2 Assay, Easy SARS-CoV-2 WE and Lumipulse quantitative SARS-CoV-2 antigen test performance using automated systems for the diagnosis of COVID-19. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 113, 113–115. [Google Scholar] [CrossRef]
- Yin, N.; Debuysschere, C.; Daubie, V.; Hildebrand, M.; Martin, C.; Curac, S.; Ponthieux, F.; Payen, M.-C.; Vandenberg, O.; Hallin, M. Evaluation and Modelling of the Performance of an Automated SARS-CoV-2 Antigen Assay According to Sample Type, Target Population and Epidemic Trends. Diagnostics 2022, 12, 447. [Google Scholar] [CrossRef]
- Osterman, A.; Iglhaut, M.; Lehner, A.; Späth, P.; Stern, M.; Autenrieth, H.; Muenchhoff, M.; Graf, A.; Krebs, S.; Blum, H.; et al. Comparison of four commercial, automated antigen tests to detect SARS-CoV-2 variants of concern. Med. Microbiol. Immunol. 2021, 210, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Gandolfo, C.; Morecchiato, F.; Pistello, M.; Rossolini, G.M.; Cusi, M.G. Detection of SARS-CoV-2 N protein allelic variants by rapid high-throughput CLEIA antigen assay. J. Clin. Virol. 2021, 142, 104942. [Google Scholar] [CrossRef] [PubMed]
- Amoah, I.D.; Mthethwa, N.P.; Pillay, L.; Deepnarain, N.; Pillay, K.; Awolusi, O.O.; Kumari, S.; Bux, F. RT-LAMP: A Cheaper, Simpler and Faster Alternative for the Detection of SARS-CoV-2 in Wastewater. Food Environ. Virol. 2021, 13, 447–456. [Google Scholar] [CrossRef]
- Daigle, J.; Racher, K.; Hazenberg, J.; Yeoman, A.; Hannah, H.; Duong, D.; Mohammed, U.; Spreitzer, D.; Gregorchuk, B.S.J.; Head, B.M.; et al. A Sensitive and Rapid Wastewater Test for SARS-CoV-2 and Its Use for the Early Detection of a Cluster of Cases in a Remote Community. Appl. Environ. Microbiol 2022, 88, e0174021. [Google Scholar] [CrossRef] [PubMed]
| Spike Cell | CLEIA | RT-qPCR | ||||
|---|---|---|---|---|---|---|
| Antigen Concentrations (pg/mL) | Interpretation | N (Mean Ct ± SD) | ORF1ab (Mean Ct ± SD) | S (Mean Ct ± SD) | Interpretation | |
| 107 | 2399.67 | Positive | 33.04 ± 0.65 | 33.02 ± 0.45 | 23.04 ± 3.56 | Positive |
| 106 | 396.08 | Positive | 23.25 ± 0.02 | 22.81 ± 0.13 | 23.45 ± 1.86 | Positive |
| 105 | 37.56 | Positive | 26.16 ± 0.08 | 25.97 ± 0.17 | 29.39 ± 2.93 | Positive |
| 104 | 3.63 | Positive | 29.34 ± 0.26 | 28.84 ± 0.19 | 26.84 ± 1.66 | Positive |
| 103 | 0.39 | Negative | 32.52 ± 0.24 | 31.56 ± 0.39 | 23.96 ± 1.97 | Positive |
| 102 | 0.01 | Negative | 36.05 ± 0.96 | 29.56 ± 0.00 | 24.40 ± 2.48 | Positive |
| 101 | 0.01 | Negative | 37.27 ± 1.44 | 31.97 ± 5.79 | 24.19 ± 6.02 | Inconclusive |
| No spike | UD | Negative | UD | UD | UD | Negative |
| Sample | RT-qPCR (Ct) | CLEIA | ||||
|---|---|---|---|---|---|---|
| N | ORF1ab | S | Interpretation | Antigen Concentration (pg/mL) | Interpretation | |
| PP1 | 34.21 | 31.89 | 33.98 | Positive | 17.2 | Positive |
| PP2 | 32.24 | 30.94 | 34.85 | Positive | 9.68 | Positive |
| PP3 | UD | 33.35 | UD | Positive | 30.78 | Positive |
| PP4 | UD | 33.83 | 36.97 | Positive | 11.76 | Positive |
| PP5 | UD | UD | UD | Negative | 0.39 | Negative |
| PP6 | UD | UD | UD | Negative | 0.44 | Negative |
| PP7 | 33.39 | 30.46 | 31.32 | Positive | 1.9 | Positive |
| PP8 | 33.96 | 31.77 | 33.92 | Positive | 38.46 | Positive |
| SC1 | UD | 36.51 | UD | Positive | 13.5 | Positive |
| SC2 | 36.41 | 34.81 | UD | Positive | 8.52 | Positive |
| SC3 | UD | UD | UD | Negative | 0.27 | Negative |
| RS1 | UD | UD | UD | Negative | 0.9 | Negative |
| RS2 | UD | UD | UD | Negative | 7.19 | Positive |
| RS3 | UD | UD | UD | Negative | 1.83 | Positive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thongpradit, S.; Prasongtanakij, S.; Srisala, S.; Chanprasertyothin, S.; Pasomsub, E.; Ongphiphadhanakul, B. The Detection of SARS-CoV2 Antigen in Wastewater Using an Automated Chemiluminescence Enzyme Immunoassay. Int. J. Environ. Res. Public Health 2022, 19, 7783. https://doi.org/10.3390/ijerph19137783
Thongpradit S, Prasongtanakij S, Srisala S, Chanprasertyothin S, Pasomsub E, Ongphiphadhanakul B. The Detection of SARS-CoV2 Antigen in Wastewater Using an Automated Chemiluminescence Enzyme Immunoassay. International Journal of Environmental Research and Public Health. 2022; 19(13):7783. https://doi.org/10.3390/ijerph19137783
Chicago/Turabian StyleThongpradit, Supranee, Somsak Prasongtanakij, Supanart Srisala, Suwannee Chanprasertyothin, Ekawat Pasomsub, and Boonsong Ongphiphadhanakul. 2022. "The Detection of SARS-CoV2 Antigen in Wastewater Using an Automated Chemiluminescence Enzyme Immunoassay" International Journal of Environmental Research and Public Health 19, no. 13: 7783. https://doi.org/10.3390/ijerph19137783
APA StyleThongpradit, S., Prasongtanakij, S., Srisala, S., Chanprasertyothin, S., Pasomsub, E., & Ongphiphadhanakul, B. (2022). The Detection of SARS-CoV2 Antigen in Wastewater Using an Automated Chemiluminescence Enzyme Immunoassay. International Journal of Environmental Research and Public Health, 19(13), 7783. https://doi.org/10.3390/ijerph19137783






