Pharmacophore Modeling Using Machine Learning for Screening the Blood–Brain Barrier Permeation of Xenobiotics
Abstract
1. Introduction
2. Materials and Methods
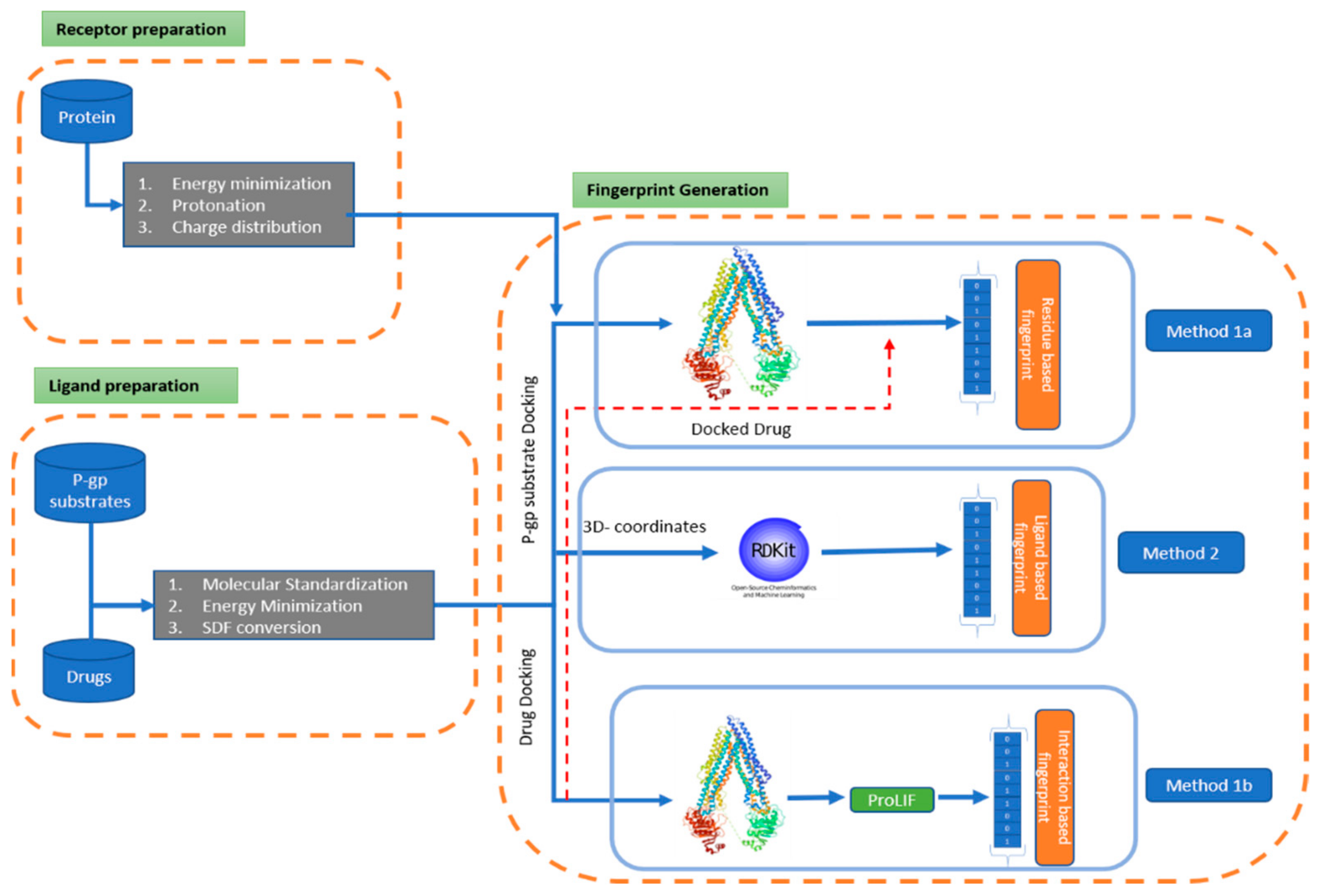
2.1. Overall Methodological Concept
- -
- The collection of chemical data from reviewed literature sources, followed by a filtering and standardization process to obtain a stabilized 3D structure.
- -
- A scaffold of the collected data was generated to analyze the distribution of the core structure of the chemical responsible for permeability.
- -
- Stabilization and hydration of the protein retrieved from the protein data bank (PDB) was undertaken, for docking purposes.
- -
- Three methods were implemented to generate pharmacophore fingerprints; among them, two belong to the receptor-based method and one to the ligand-based method.
- (a)
- The residue-based pharmacophore was generated by docking the P-gp substrates and extracting the most common residues involved in the interaction. The residues were then mapped with the interaction of the drug molecule to generate a 62-bit fingerprint (Figure 2).
- (b)
- The interaction-type pharmacophore was generated using the docked drug data, which was further processed with the proLIF library to generate a 9-bit fingerprint (Figure 3).
- (c)
- The 39,971-bit-long ligand-based pharmacophore fingerprint was generated using Rdkit.
- -
- The generated fingerprint and classical fingerprint were then trained on a classical algorithm, such as Support Vector Machine (SVM), RF (Random Forest), and naïve Bayes, for comparison. A newly developed graph model was also implemented for comparison.


2.2. Data Collection and Scaffold Generation
2.3. P-gp Receptor Preparation
2.4. Ligand Preparation
2.5. Interaction Fingerprint Generation
2.5.1. Receptor-Based Fingerprint
Method DockedFP(1a): Residue-Based Type
Method DockedFP(1b): Interaction-Type-Based Fingerprint
2.5.2. Ligand-Based Generation
Method 2: Rdkit Pharmacophore Fingerprint
2.6. GNN Implementation
2.7. Model Validation
2.8. Evaluation Metrics
3. Results
3.1. Scaffold-Based Chemical Space Analysis
3.2. Classical ML Models
3.3. Comparison with GNN Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BBB | Blood–brain barrier |
| BCRP | Breast cancer resistance protein |
| P-gp | P-glycoprotein |
| CNS | Central nervous system |
| SMILES | Simplified Molecular Input Line Entry System |
| ECFP4 | Extended Connectivity Fingerprint |
| MMFF94 | Merck Molecular Force Field |
| SDF | Structure-Data File |
| TMAP | Tree MAP |
| PDB | Protein Data Bank |
| PDBQT | Protein Data Bank, Partial Charge and Atom Type |
| GNN | Graph Neural Network |
| GCN | Graph Convolution Network |
| GAT | Graph Attention Network |
| ELU | Exponential Linear Unit |
| SVM | Support Vector Machine |
| RF | Random Forest |
| TPSA | Topological Polar Surface Area |
| HBA | Hydrogen Bond Acceptor |
| HBD | Hydrogen Bond Donor |
| RGCN | Relational Graph Convolution Network |
| QSAR | Quantitative structure activity relationship |
| PBPK | Physiologically Based pharmacokinetics |
References
- Daneman, R.; Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [PubMed]
- Deepika, D.; Bravo, N.; Esplugas, R.; Capodiferro, M.; Sharma, R.P.; Schuhmacher, M.; Grimalt, J.O.; Blanco, J.; Kumar, V. Chlorpyrifos, Permethrin and Cyfluthrin effect on cell survival, permeability, and tight junction in an in-vitro model of the Human Blood-Brain Barrier (BBB). NeuroToxicology 2022, 93, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Naito, M.; Ohhara, T.; Sugawara, I.; Tsuruo, T. Functional Involvement of P-Glycoprotein in Blood-Brain-Barrier. J. Biol. Chem. 1992, 267, 20383–20391. [Google Scholar] [CrossRef]
- Doniger, S.; Hofmann, T.; Yeh, J. Predicting CNS Permeability of Drug Molecules: Comparison of Neural Network and Support Vector Machine Algorithms. J. Comput. Biol. 2002, 9, 849–864. [Google Scholar] [CrossRef]
- Martins, I.F.; Teixeira, A.L.; Pinheiro, L.; Falcao, A.O. A Bayesian approach to in Silico blood-brain bar-rier penetration modeling. J. Chem. Inf. Model 2012, 52, 1686–1697. [Google Scholar] [CrossRef]
- Andres, C.; Hutter, M.C. CNS Permeability of Drugs Predicted by a Decision Tree. QSAR Comb. Sci. 2006, 25, 305–309. [Google Scholar] [CrossRef]
- Li, H.; Yap, C.W.; Ung, C.Y.; Xue, Y.; Cao, Z.-W.; Chen, Y.Z. Effect of Selection of Molecular Descriptors on the Prediction of Blood-Brain Barrier Penetrating and Nonpenetrating Agents by Statistical Learning Methods. J. Chem. Inf. Model. 2005, 45, 1376–1384. [Google Scholar] [CrossRef]
- Sato, T.; Honma, T.; Yokoyama, S. Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J. Chem. Inf. Model 2010, 50, 170–185. [Google Scholar] [CrossRef]
- Yang, S.-Y. Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov Today 2010, 15, 444–450. [Google Scholar] [CrossRef]
- Meng, F.; Xi, Y.; Huang, J.; Ayers, P.W. A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. Sci. Data 2021, 8, 1–11. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Bajusz, D.; Rácz, A.; Héberger, K. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations? J. Cheminform. 2015, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Introduction—MolVS 0.1.1 Documentation. Available online: https://molvs.readthedocs.io/en/latest/guide/intro.html (accessed on 23 August 2022).
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 5–6. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef]
- Probst, D.; Reymond, J.-L. Visualization of Very Large High-Dimensional Data Sets as Minimum Spanning Trees. arXiv 2020. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Improving the Physical Realism and Structural Accuracy of Protein Models by a Two-Step Atomic-Level Energy Minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Jouan, E.; Le Vée, M.; Mayati, A.; Denizot, C.; Parmentier, Y.; Fardel, O. Evaluation of P-glycoprotein inhibitory potential using a rhodamine 123 accumulation assay. Pharmaceutics 2016, 8, 12. [Google Scholar] [CrossRef]
- Sharom, F.J.; Liu, R.; Romsicki, Y.; Lu, P. Insights into the Structure and Substrate Interactions of the P-Glycoprotein Multidrug Transporter from Spectroscopic Studies. Biochim. Biophys. Acta (BBA)-Biomembr. 1999, 1461, 327–345. [Google Scholar] [CrossRef]
- Teodori, E.; Dei, S.; Martelli, C.; Scapecchi, S.; Gualtieri, F. The Functions and Structure of ABC Transporters: Implications for the Design of New Inhibitors of Pgp and MRP1 to Control Multidrug Resistance (MDR). Curr. Drug Targets 2006, 7, 893–909. [Google Scholar] [CrossRef] [PubMed]
- Meeko. Available online: https://github.com/forlilab/Meeko (accessed on 23 August 2022).
- Van Rossum, G.; Drake, F.L., Jr. Python Reference Manual; Centrum voor Wiskunde en Informatica Amsterdam: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Bouysset, C.; Fiorucci, S. ProLIF: A library to encode molecular interactions as fingerprints. J. Chemin. 2021, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017, arXiv:1609.02907. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Curran Associates, Inc.: Sydney, Australia, 2019; pp. 8024–8035. Available online: http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf (accessed on 7 August 2022).
- Eastman, P.; Walters, P.; Ramsundar, B.; Pande, V.S. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Mi-Croscopy, Drug Discovery, and More; Beijing O’Reilly: Beijing, China, 2019. [Google Scholar]
- Vapnik, V.N. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wikipedia Contributors. Receiver Operating Characteristic. Wikipedia, Wikimedia Foundation. 20 March 2019. Available online: En.wikipedia.org/wiki/Receiver_operating_characteristic (accessed on 7 August 2022).
- Geldenhuys, W.J.; Mohammad, A.S.; Adkins, C.E.; Lockman, P.R. Molecular determinants of blood-brain barrier permeation. Ther. Deliv. 2015, 6, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-Y.; Pan, J.; Yuan, Y.; Hull, A.-L.; Yang, A.; Zhou, Y. Comparison of prediction models for blood brain barrier permeability and analysis of the molecular descriptors. Pharmazie 2012, 67, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Wu, Z.; Hsieh, C.-Y.; Chen, G.; Liao, B.; Wang, Z.; Shen, C.; Cao, D.; Wu, J.; Hou, T. Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. J. Chemin. 2021, 13, 1–23. [Google Scholar] [CrossRef]
- Ding, Y.; Jiang, X.; Kim, Y. Relational Graph Convolutional Networks for Predicting Blood-Brain Barrier Penetration of Drug Molecules. Available online: https://github.com/dingyan20/BBB-Penetration-Prediction (accessed on 7 August 2022).
- Laroche, C.; Aggarwal, M.; Bender, H.; Benndorf, P.; Birk, B.; Crozier, J.; Negro, G.D.; De Gaetano, F.; Desaintes, C.; Gardner, I.; et al. Finding synergies for 3Rs—Toxicokinetics and read-across: Report from an EPAA partners’ Forum. Regul. Toxicol. Pharmacol. 2018, 99, 5–21. [Google Scholar] [CrossRef]
- Deepika, D.; Sharma, R.P.; Schuhmacher, M.; Kumar, V. An integrative translational framework for chemical induced neurotoxicity—A systematic review. Crit. Rev. Toxicol. 2020, 50, 424–438. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, S. ANN-based Integrated Risk ranking approach: A case study of contaminants of emerging concern of fish and seafood in Europe. Int. J. Environ. Res. Public Health 2021, 18, 1598. [Google Scholar] [CrossRef]
- Balaguer-Trias, J.; Deepika, D.; Schuhmacher, M.; Kumar, V. Impact of Contaminants on Microbiota: Linking the Gut–Brain Axis with Neurotoxicity. Int. J. Environ. Res. Public Health 2022, 19, 1368. [Google Scholar] [CrossRef]


| Models | Features | Accuracy (Train/Test) | Precision (Train/Test) | Recall (Train/Test) | F1 Score (Train/Test) |
|---|---|---|---|---|---|
| Baseline | DockedFP (1a) DockedFP (1b) | 0.50/0.50 0.50/0.50 | 0.60/0.61 0.61/0.61 | 0.48/0.49 0.49/0.49 | 0.53/0.54 0.55/0.55 |
| SVM | ECFP4 fingerprint DockedFP (1a) DockedFP (1b) Rdkit Pharmacoprint ECFP4+ DockedFP (1a) ECFP4+ DockedFP (1b) | 0.92/0.76 0.62/0.61 0.71/0.63 0.88/0.75 0.92/0.76 0.93/0.76 | 0.91/0.77 0.62/0.62 0.70/0.64 0.84/0.75 0.91/0.77 0.92/0.77 | 0.96/0.86 0.93/0.92 0.93/0.87 0.98/0.89 0.96/0.87 0.97/0.86 | 0.94/0.82 0.75/0.74 0.80/0.74 0.90/0.81 0.93/0.82 0.94/0.81 |
| Random Forest * | ECFP4 fingerprint DockedFP (1a) DockedFP (1b) Rdkit Pharmacoprint ECFP4 + DockedFP (1a) ECFP4 + DockedFP (1b) | 1/0.76 0.62/0.61 0.91/0.60 0.99/0.77 1/0.76 1/0.76 | 1/0.76 0.62/62 0.90/0.64 0.99/0.78 1/0.76 1/0.76 | 1/0.86 0.92/0.91 0.95/0.73 0.99/0.84 1/0.87 1/0.87 | 1/0.81 0.75/0.74 0.92/0.69 0.99/0.81 1/0.81 1/0.81 |
| Naïve Byes | ECFP4 fingerprint DockedFP (1a) DockedFP (1b) Rdkit Pharmacoprint ECFP4 + DockedFP (1a) ECFP4 + DockedFP (1b) | 0.76/0.72 0.62/0.62 0.61/0.60 0.72/0.71 0.76/72 0.76/72 | 0.78/0.75 0.62/0.62 0.66/0.64 0.72/0.71 0.78/0.75 0.78/0.75 | 0.83/0.80 0.9/0.9 0.75/0.74 0.90/0.89 0.84/0.8 0.84/0.81 | 0.81/0.78 0.74/0.74 0.70/0.69 0.80/0.79 0.81/0.77 0.81/0.78 |
| Graph Convolution Network (GCN) | Descriptors | 0.81/0.74 | 0.83/0.77 | 0.85/0.80 | 0.84/0.79 |
| Graph Attention Network (GAT) | Descriptors | 0.83/0.74 | 0.87/0.77 | 0.84/0.78 | 0.85/79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Deepika, D.; Kumar, V. Pharmacophore Modeling Using Machine Learning for Screening the Blood–Brain Barrier Permeation of Xenobiotics. Int. J. Environ. Res. Public Health 2022, 19, 13471. https://doi.org/10.3390/ijerph192013471
Kumar S, Deepika D, Kumar V. Pharmacophore Modeling Using Machine Learning for Screening the Blood–Brain Barrier Permeation of Xenobiotics. International Journal of Environmental Research and Public Health. 2022; 19(20):13471. https://doi.org/10.3390/ijerph192013471
Chicago/Turabian StyleKumar, Saurav, Deepika Deepika, and Vikas Kumar. 2022. "Pharmacophore Modeling Using Machine Learning for Screening the Blood–Brain Barrier Permeation of Xenobiotics" International Journal of Environmental Research and Public Health 19, no. 20: 13471. https://doi.org/10.3390/ijerph192013471
APA StyleKumar, S., Deepika, D., & Kumar, V. (2022). Pharmacophore Modeling Using Machine Learning for Screening the Blood–Brain Barrier Permeation of Xenobiotics. International Journal of Environmental Research and Public Health, 19(20), 13471. https://doi.org/10.3390/ijerph192013471








