Epidemiological Patterns of Cannabis- and Substance- Related Congenital Uronephrological Anomalies in Europe: Geospatiotemporal and Causal Inferential Study
Abstract
:1. Introduction
1.1. Background
1.2. Cannabinoid Genotoxicity
1.3. Genotoxic Mechanisms
1.4. Exponential Dose-Response Effect Curve
1.5. Lessons from VACTERL Syndrome
1.6. Study Questions
2. Methods
2.1. Data
2.2. National Assignment
2.3. Derived Data
2.4. Data Imputation
2.5. Statistics
2.6. Covariate Selection
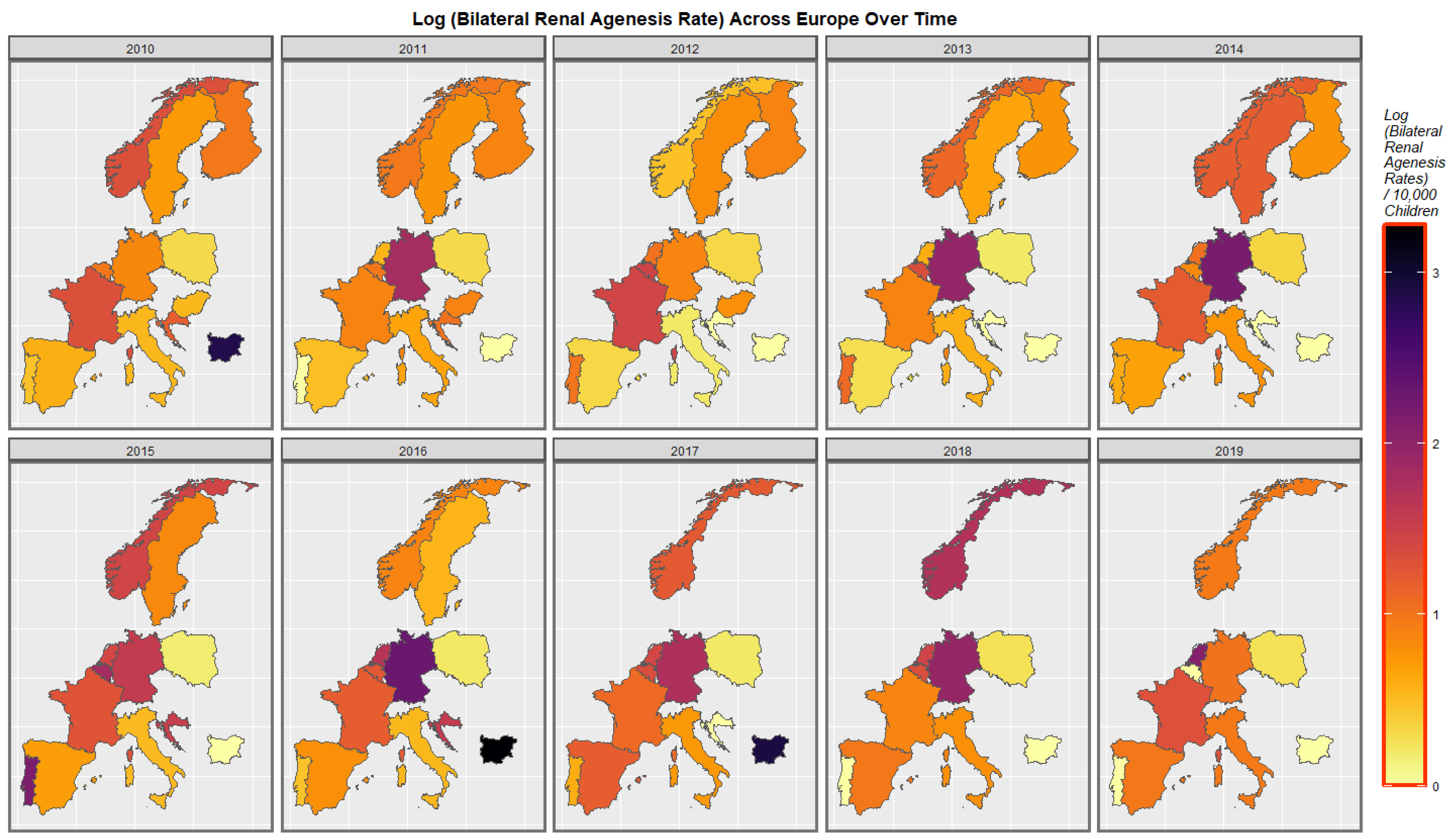
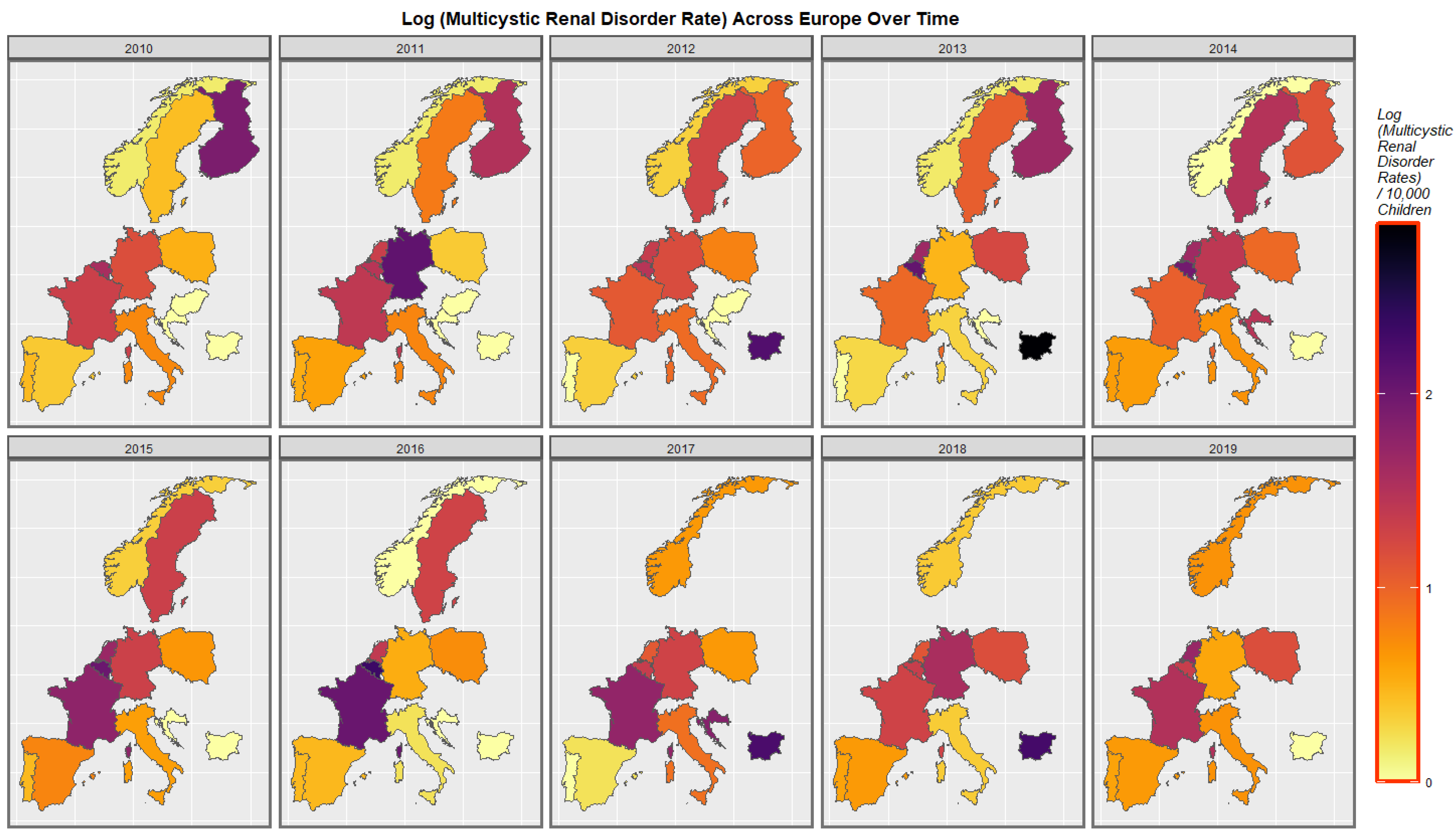
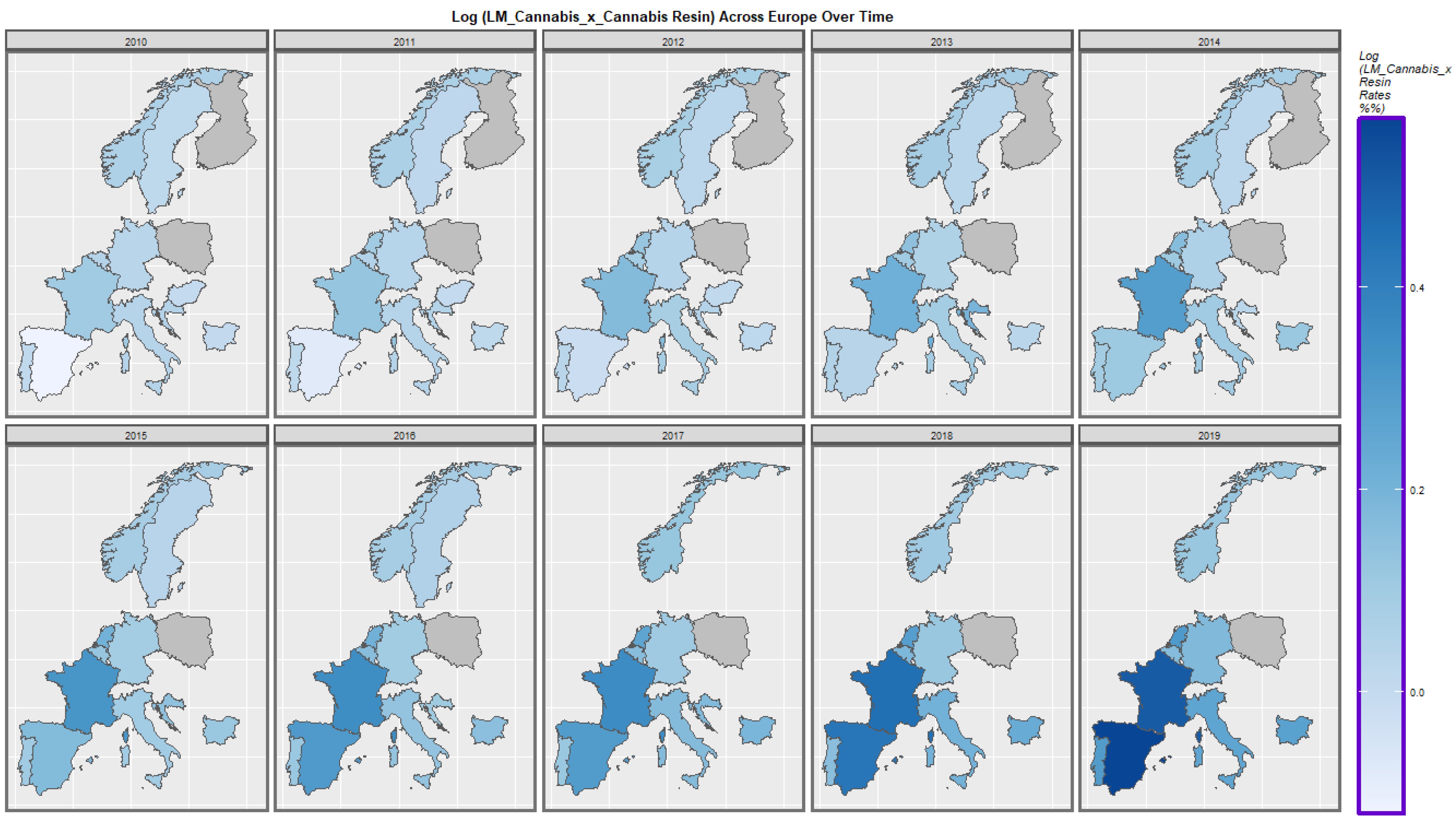
2.7. Panel and Geospatial Analysis
2.8. Causal Inference
2.9. Data Availability
2.10. Ethics
3. Results
3.1. Input Data
3.2. Bivariate Analysis
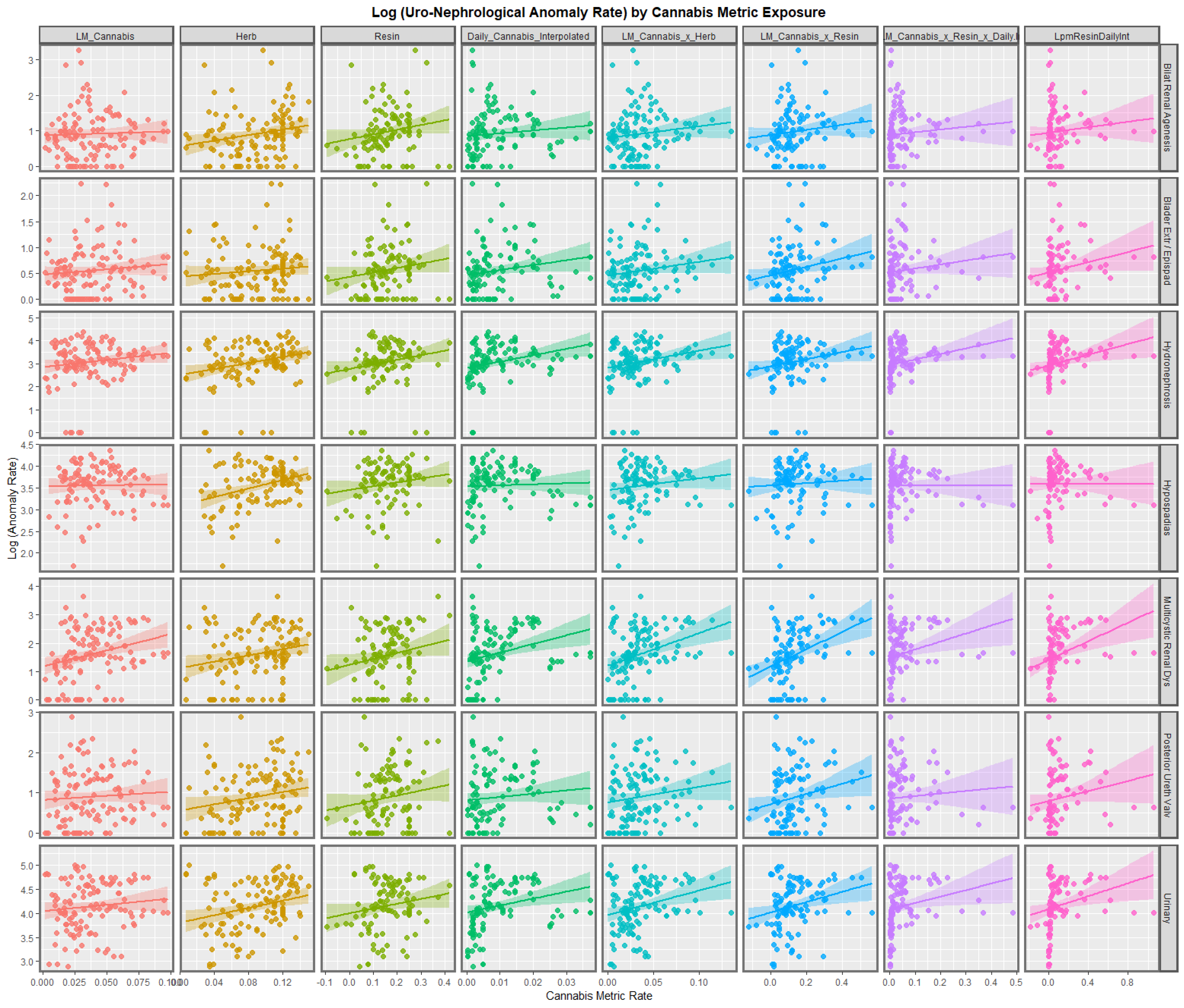


3.2.1. Bivariate Graphs

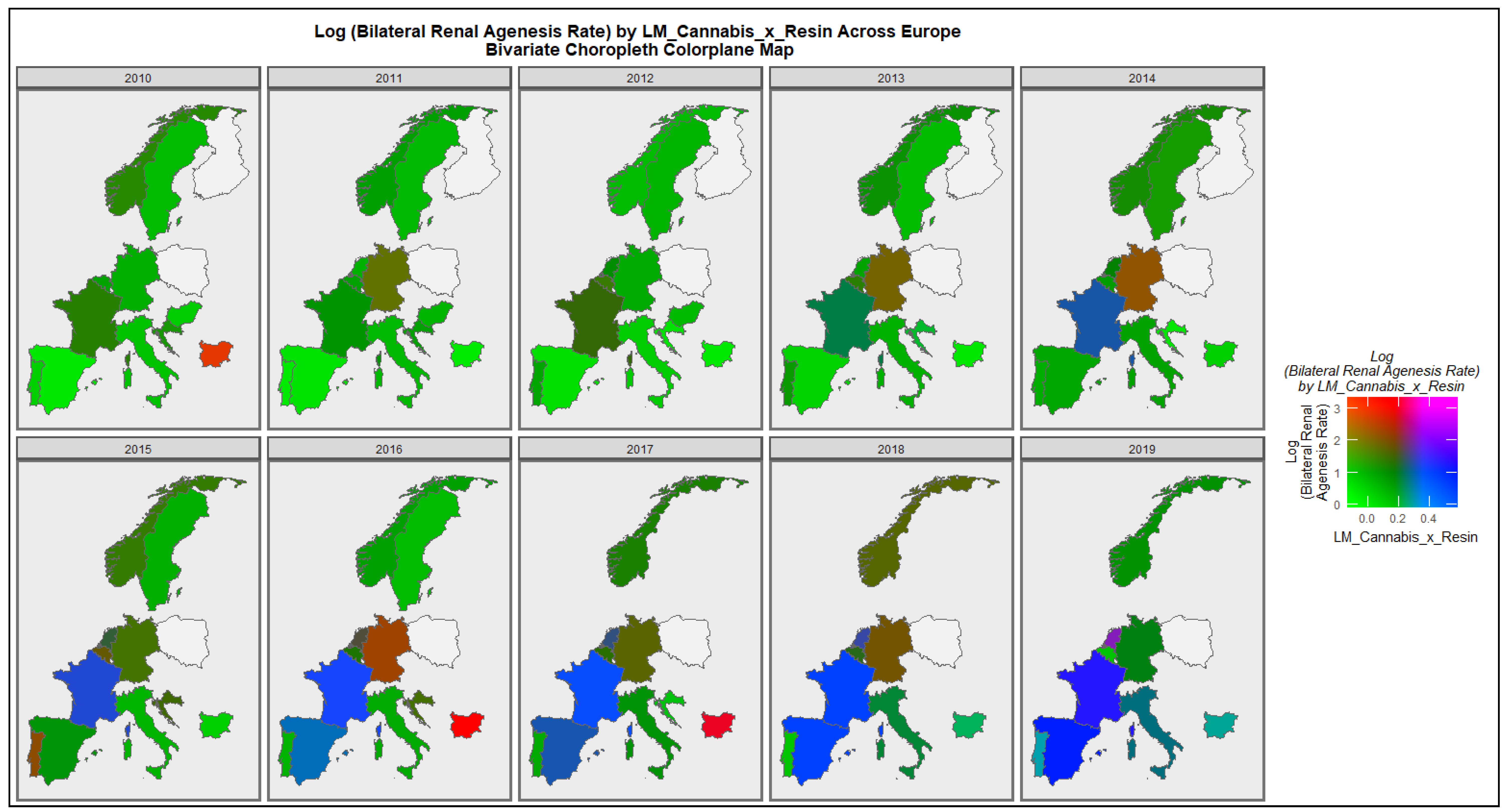
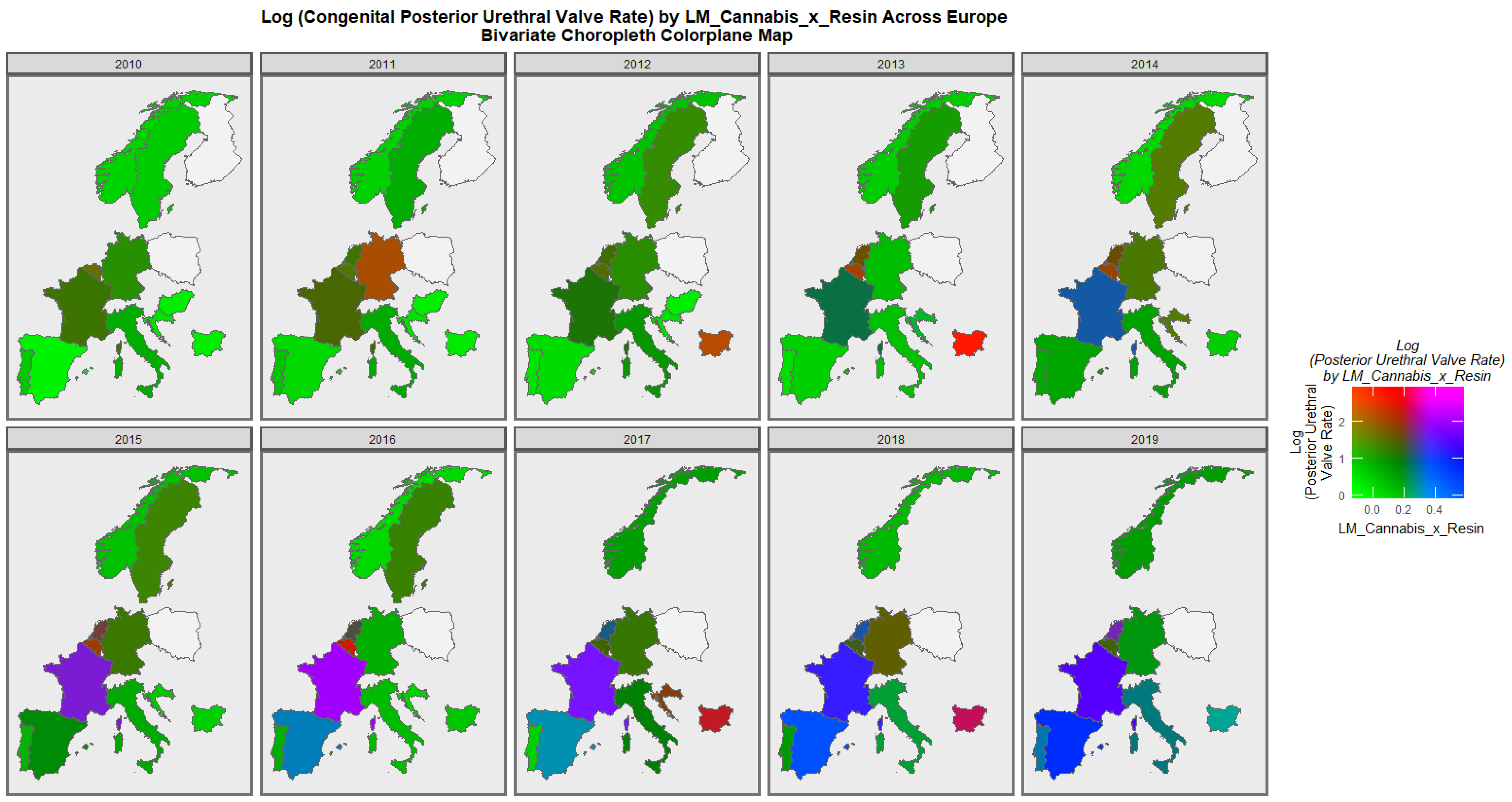
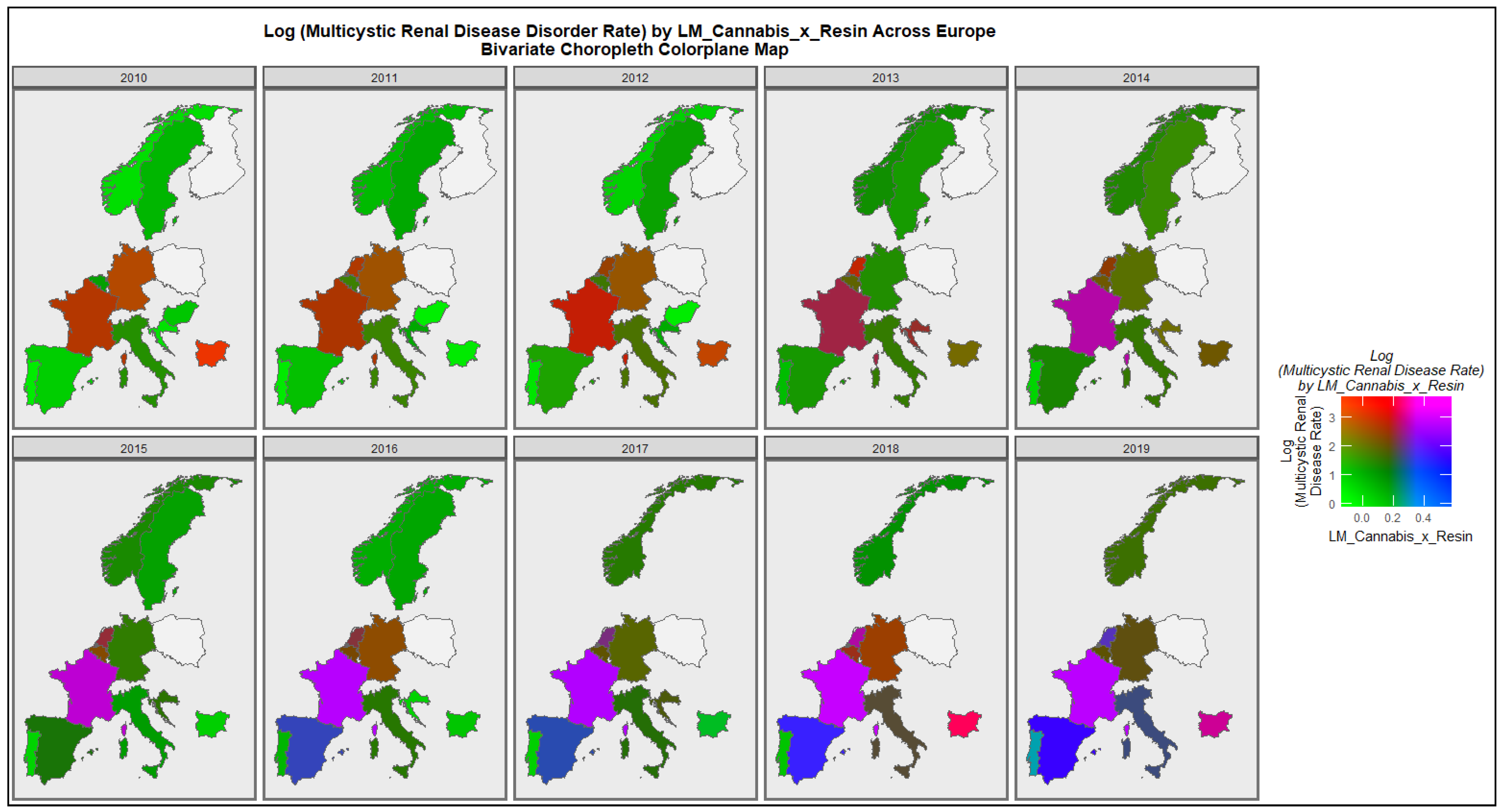
3.2.2. Bivariate Overview
3.2.3. Bivariate Linear Regressions
3.3. Covariate Selection
3.4. Multivariable Regressions
3.4.1. IPW Panel Regression
3.4.2. Geospatial Regression
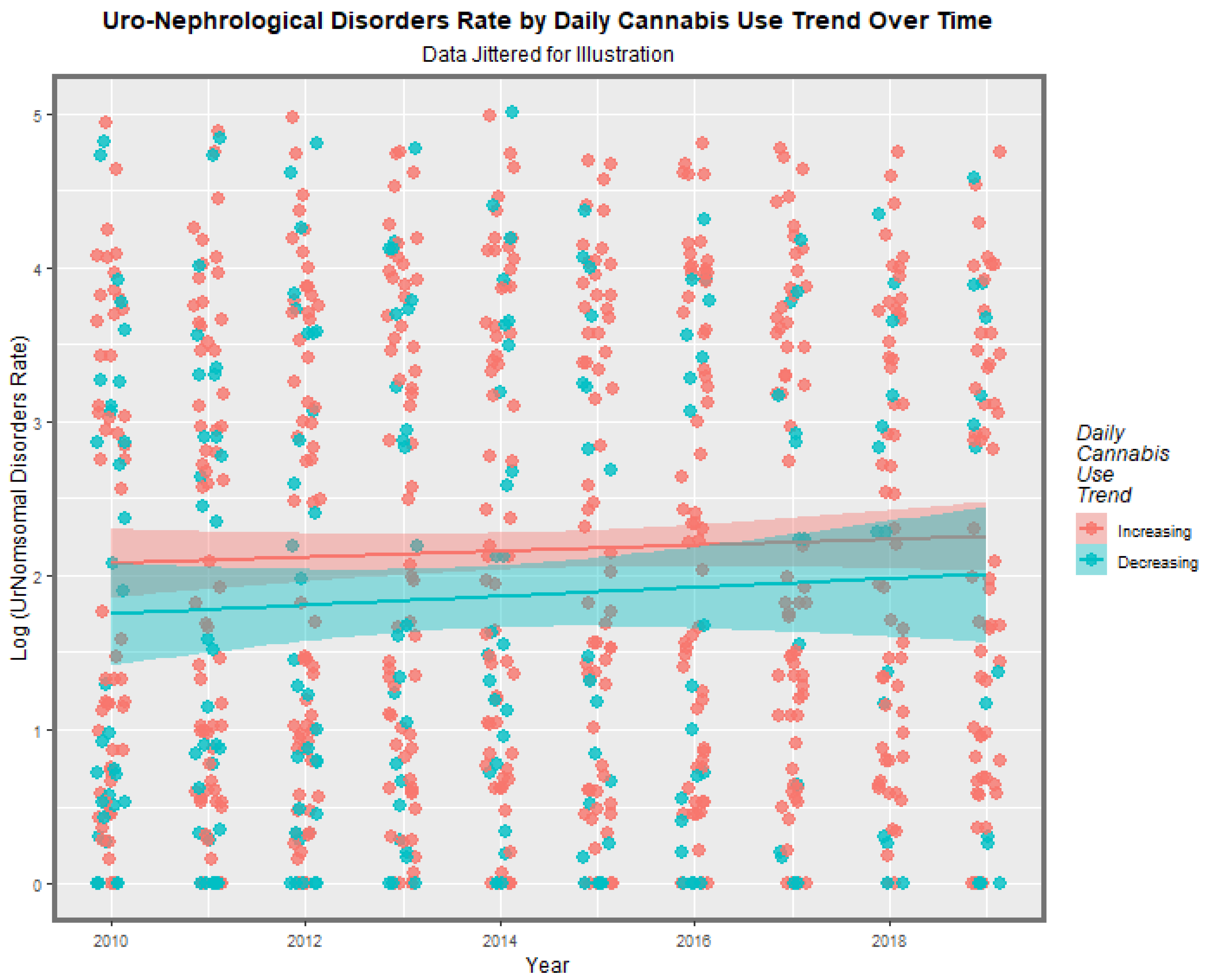
3.5. Causal Inference
E-Values

| Anomaly | Substance | Mean Anomaly Rate | Estimate | Std.Error | Sigma | t_statistic | p-Value | E-Value Estimate | E-Value Lower Bound |
|---|---|---|---|---|---|---|---|---|---|
| Multicystic Renal Dys | Daily.Interpol. | 3.4655 | 30.4447 | 9.5953 | 0.9007 | 3.1729 | 0.0019 | 4.57 × 1013 | 2.66 × 105 |
| Hydronephrosis | Daily.Interpol. | 13.8455 | 27.8771 | 8.7897 | 0.8250 | 3.1716 | 0.0020 | 4.52 × 1013 | 2.62 × 105 |
| Urinary | Daily.Interpol. | 35.4475 | 14.3684 | 5.3583 | 0.5029 | 2.6815 | 0.0084 | 3.90 × 1011 | 2.27 × 103 |
| Multicystic Renal Dys | LMCannabis_Herb | 3.4655 | 11.4522 | 3.0267 | 0.8688 | 3.7837 | 2.43 × 10−4 | 3.24 × 105 | 656.0696 |
| Hypospadias | Herb | 19.2037 | 4.9196 | 1.3727 | 0.4546 | 3.5838 | 5.15 × 10−4 | 3.78 × 104 | 174.7051 |
| Multicystic Renal Dys | LM_Cannabis | 3.4655 | 11.4477 | 3.6514 | 0.8837 | 3.1352 | 0.0022 | 2.63 × 105 | 167.9388 |
| Urinary | LMCannabis_Herb | 35.4475 | 5.0871 | 1.7232 | 0.4947 | 2.9521 | 0.0038 | 2.32 × 104 | 46.5273 |
| Hydronephrosis | Daily.Interpol. | 13.8455 | 6.5820 | 2.1094 | 0.8202 | 3.1204 | 0.0023 | 2.97 × 103 | 30.0042 |
| Urinary | Herb | 35.4475 | 3.8164 | 1.2706 | 0.4941 | 3.0036 | 0.0032 | 2.26 × 103 | 22.7009 |
| Hydronephrosis | LMCannabis_Herb | 13.8455 | 7.3128 | 2.8950 | 0.8310 | 2.5260 | 0.0128 | 6.01 × 103 | 11.6605 |
| Multicystic Renal Dys | LMCannabis_Resin | 3.4655 | 3.0558 | 0.7366 | 0.8820 | 4.1488 | 6.81 × 10−5 | 46.3050 | 10.0602 |
| Multicystic Renal Dys | Herb | 3.4655 | 5.5861 | 2.3083 | 0.8976 | 2.4200 | 0.0170 | 575.8297 | 5.3720 |
| Bilat Renal Agenesis | Herb | 1.3617 | 3.9363 | 1.6424 | 0.6386 | 2.3967 | 0.0181 | 545.3091 | 5.0547 |
| Multicystic Renal Dys | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 3.4655 | 2.8187 | 1.0685 | 0.9122 | 2.6379 | 0.0095 | 32.7720 | 3.5552 |
| Posterior Ureth Valv | Herb | 1.3062 | 3.8400 | 1.7052 | 0.6630 | 2.2519 | 0.0261 | 388.4327 | 3.4114 |
| Multicystic Renal Dys | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.4655 | 1.6437 | 0.4955 | 0.9272 | 3.3171 | 0.0013 | 9.5088 | 3.2871 |
| Hydronephrosis | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 13.8455 | 2.4167 | 0.9825 | 0.8388 | 2.4597 | 0.0154 | 27.0122 | 2.8132 |
| Urinary | LMCannabis_Resin | 35.4475 | 1.0733 | 0.4108 | 0.4919 | 2.6129 | 0.0103 | 14.0498 | 2.6800 |
| Hydronephrosis | Cocaine | 13.8455 | 0.4916 | 0.0962 | 0.7729 | 5.1079 | 1.24 × 10−6 | 2.9663 | 2.2125 |
| Urinary | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 35.4475 | 1.3384 | 0.5944 | 0.5074 | 2.2517 | 0.0263 | 21.5361 | 2.0829 |
| Urinary | Cocaine | 35.4475 | 0.2657 | 0.0590 | 0.4738 | 4.5041 | 1.56 × 10−5 | 2.7191 | 2.0031 |
| Urinary | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 35.4475 | 0.6738 | 0.2664 | 0.4985 | 2.5291 | 0.0130 | 6.2987 | 1.9731 |
| Multicystic Renal Dys | Cocaine | 3.4655 | 0.4660 | 0.1063 | 0.8533 | 4.3857 | 2.50 × 10−5 | 2.6723 | 1.9630 |
| Hydronephrosis | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 13.8455 | 1.1656 | 0.4627 | 0.8659 | 2.5189 | 0.0134 | 6.2645 | 1.9582 |
| Hypospadias | Cocaine | 19.2037 | 0.2230 | 0.0608 | 0.4534 | 3.6679 | 3.86 × 10−4 | 2.5043 | 1.7674 |
| Posterior Ureth Valv | LMCannabis_Resin | 1.3062 | 1.2729 | 0.5745 | 0.6880 | 2.2154 | 0.0289 | 10.2449 | 1.7331 |
| Hydronephrosis | LMCannabis_Resin | 13.8455 | 1.5445 | 0.7215 | 0.8639 | 2.1407 | 0.0346 | 9.6488 | 1.5671 |
| Bladder Extr/Epispad | LMCannabis_Resin | 0.6657 | 0.8171 | 0.3848 | 0.4608 | 2.1235 | 0.0361 | 9.5157 | 1.5282 |
| Hydronephrosis | Resin | 13.8455 | 1.9876 | 0.9624 | 0.8652 | 2.0652 | 0.0414 | 15.6631 | 1.4781 |
| Urinary | Amphetamine | 35.4475 | 0.1765 | 0.0622 | 0.4959 | 2.8377 | 0.0053 | 2.1095 | 1.4480 |
| Bladder Extr/Epispad | Cocaine | 0.6657 | 0.1511 | 0.0548 | 0.4405 | 2.7545 | 0.0068 | 2.0738 | 1.4166 |
| Hydronephrosis | Amphetamine | 13.8455 | 0.2789 | 0.1039 | 0.8283 | 2.6856 | 0.0083 | 2.0566 | 1.3940 |
| Hypospadias | Amphetamine | 19.2037 | 0.1577 | 0.0603 | 0.4667 | 2.6142 | 0.0103 | 2.0598 | 1.3754 |
| Multicystic Renal Dys | Resin | 3.4655 | 2.1084 | 1.0383 | 0.9334 | 2.0306 | 0.0448 | 15.1060 | 1.3692 |
| Bilat Renal Agenesis | Cocaine | 1.3617 | 0.2017 | 0.0793 | 0.6368 | 2.5443 | 0.0122 | 2.0018 | 1.3404 |
| Posterior Ureth Valv | Annual_Alcohol | 1.3062 | 0.0958 | 0.0325 | 0.6537 | 2.9450 | 0.0039 | 1.5462 | 1.2646 |
| Multicystic Renal Dys | Amphetamine | 3.4655 | 0.2537 | 0.1129 | 0.9005 | 2.2464 | 0.0265 | 1.9067 | 1.2203 |
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Herb.THC + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.06 (0.04, 0.07) | 8.09 × 10−12 | rho | −0.5677 | 2.99 × 10−6 |
| Alcohol | 0.03 (0, 0.06) | 0.0394 | lambda | 0.4206 | 1.45 × 10−5 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −4.7 (−5.97, −3.43) | 4.05 × 10−13 | |||
| LM.Cannabis_x_Herb.THC | 14.1 (10.53, 17.67) | 7.84 × 10−15 | |||
| Cocaine | 0.23 (0.12, 0.33) | 2.82 × 10−5 | |||
| Income | 0 (0, 0) | 1.23 × 10−14 | |||
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | −0.04 (−0.07, −0.01) | 0.0067 | phi | 6.1554 | 0.0464 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −11.75 (−15.66, −7.83) | 4.18 × 10−9 | rho | −0.5643 | 8.32 × 10−7 |
| Daily.Interpol. | 19.94 (2.57, 37.3) | 0.024429 | lambda | 0.5731 | 6.79 × 10−10 |
| Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.51 (0.34, 0.67) | 3.07 × 10−9 | |||
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 53.87 (26.64, 81.11) | 0.0001 | |||
| Tobacco: LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −2.33 (−3.51, −1.14) | 0.0001 | |||
| 1 Lag | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.04 (0.02, 0.07) | 0.0003 | rho | 0.2013 | 0.497 |
| Daily.Interpol. | −31.9 (−53.26, −10.54) | 0.0033 | lambda | 0.0696 | 0.782 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.85 (0.18, 1.53) | 0.0130 | |||
| Alcohol | 0.07 (0.02, 0.12) | 0.0106 | |||
| Cocaine | 0.52 (0.28, 0.75) | 1.52 × 10−5 | |||
| Income | 0 (0, 0) | 0.0001 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.12 (0.06, 0.18) | 9.17 × 10−5 | psi | 0.5495 | 4.92 × 10−10 |
| LM.Cannabis_x_Resin.THC | 1.17 (0.11, 2.23) | 0.0310 | rho | −0.5777 | 0.000168 |
| Income | 0 (0, 0) | 0.0001 | lambda | 0.5010 | 0.000692 |
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.12 (0.06, 0.18) | 9.17 × 10−5 | psi | 0.5495 | 4.92 × 10−10 |
| LM.Cannabis_x_Resin.THC | 1.17 (0.11, 2.23) | 0.0310 | rho | −0.5777 | 0.000168 |
| Income | 0 (0, 0) | 0.0001 | lambda | 0.5010 | 0.000692 |
| 2 Lags | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC + Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Alcohol + + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.06 (0.02, 0.1) | 0.0060 | psi | 0.5834 | 1.62 × 10−9 |
| LM.Cannabis_x_Resin.THC | 3.9 (1.97, 5.84) | 7.72 × 10−5 | rho | 0.7044 | 3.78 × 10−16 |
| Daily.Interpol. | 74.8 (28.64, 120.96) | 0.0015 | lambda | −0.6093 | 7.22 × 10−08 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −7.05 (−12.74, −1.36) | 0.0152 | |||
| Amphetamines | −0.27 (−0.53, 0) | 0.0493 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Resin + Daily.Interpol. + Herb + Amphetamines + Cocaine + Income | |||||
| Alcohol | 0.13 (0.07, 0.18) | 3.62 × 10−6 | rho | 0.6238 | 6.72 × 10−10 |
| Income | 0 (0, 0) | 1.10 × 10−8 | lambda | −0.5490 | 1.92 × 10−6 |
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Herb + Amphetamines + Cocaine + Income | |||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 4.06 (0.57, 7.55) | 0.0228 | rho | 0.6314 | 2.39 × 10−11 |
| Alcohol | 0.14 (0.09, 0.2) | 4.66 × 10−7 | lambda | −0.5346 | 1.34 × 10−6 |
| Income | 0 (0, 0) | 1.49 × 10−8 | |||
| LM.Cannabis_x_Herb.THC: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −0.15 (−0.3, −0.01) | 0.0327 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Amphetamines + Cocaine + Income | |||||
| LM.Cannabis_x_Resin.THC | 2.33 (0.83, 3.83) | 0.0023 | rho | 0.5579 | 6.16 × 10−5 |
| Alcohol | 0.18 (0.11, 0.24) | 6.12 × 10−8 | lambda | −0.4473 | 0.00217 |
| Cocaine | 0.58 (0.26, 0.9) | 0.0004 | |||
| Resin | 0 (0, 0) | 0.0033 | |||
| Tobacco: Daily.Interpol. | −1.87 (−2.9, −0.84) | 0.0004 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Herb + Amphetamines + Cocaine + Income | |||||
| Tobacco | −0.04 (−0.07, 0) | 0.0283 | psi | 0.4594 | 3.24 × 10−7 |
| Daily.Interpol. | 67.26 (23.46, 111.06) | 0.0026 | rho | 0.4513 | 0.00694 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −9.26 (−16.96, −1.56) | 0.0185 | lambda | −0.5073 | 0.00149 |
| Herb | 12.02 (6.1, 17.94) | 6.95 × 10−5 | |||
| Interactive | |||||
| Rate ~ Tobacco * Daily.Interpol. + Resin + Herb + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | −0.05 (−0.09, −0.01) | 0.0064 | psi | 0.5095 | 1.42 × 10−9 |
| Herb | 9.93 (4.34, 15.53) | 0.0005 | rho | 0.4324 | 0.0151 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −5.24 (−9.59, −0.89) | 0.0182 | lambda | −0.4830 | 0.00603 |
| Tobacco: Daily.Interpol. | 2.9 (1.29, 4.51) | 0.0004 | |||
| 1 Lag | |||||
| Rate ~ Tobacco + Herb * Resin + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. * Daily.Interpol. + Herb + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Herb | 6.01 (2.26, 9.76) | 0.0017 | psi | 0.5117 | 1.30E−9 |
| Cocaine | 0.35 (0.1, 0.59) | 0.0058 | rho | −0.5342 | 0.00382 |
| lambda | 0.4013 | 0.0324 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * Herb + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + Resin + Alcohol + Amphetamines + Cocaine + Income | |||||
| Resin | −2.46 (−4.02, −0.9) | 0.0020 | psi | 0.8338 | < 2.22 × 10−16 |
| Tobacco: Herb | −0.25 (−0.42, −0.08) | 0.0033 | rho | 0.6650 | 6.20 × 10−13 |
| lambda | −0.5945 | 5.88 × 10−7 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Daily.Interpol. + Resin + Herb + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.04 (0.02, 0.06) | 0.0001 | psi | 0.2311 | 0.0177 |
| Alcohol | 0.15 (0.08, 0.22) | 9.98 × 10−6 | rho | −0.5195 | 6.58 × 10−5 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −10.18 (−14.3, −6.06) | 1.29 × 10−6 | lambda | 0.5252 | 2.31 × 10−5 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.09 (1.55, 4.64) | 8.82 × 10−5 | |||
| Herb | 4.77 (1.87, 7.67) | 0.0013 | |||
| Amphetamines | −0.19 (−0.36, −0.03) | 0.0218 | |||
| Cocaine | 0.51 (0.3, 0.71) | 1.54 × 10−6 | |||
| Interactive | |||||
| Rate ~ Tobacco * Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Herb + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.04 (0.02, 0.06) | 0.0001 | psi | 0.2311 | 0.0177 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −10.18 (−14.3, −6.06) | 1.29 × 10−6 | rho | −0.5195 | 6.58 × 10−5 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.09 (1.55, 4.64) | 8.82 × 10−5 | lambda | 0.5252 | 2.33 × 10−5 |
| Herb | 4.77 (1.87, 7.67) | 0.0013 | |||
| Alcohol | 0.15 (0.08, 0.22) | 9.98 × 10−6 | |||
| Amphetamines | −0.19 (−0.36, −0.03) | 0.0218 | |||
| Cocaine | 0.51 (0.3, 0.71) | 1.54 × 10−6 | |||
| 1 Lag | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Herb.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.07 (0.02, 0.11) | 0.0027 | psi | 0.3419 | 0.0013 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 2.65 (0.59, 4.71) | 0.0114 | rho | −0.4716 | 0.00201 |
| Daily.Interpol. | 91.2 (37.3, 145.1) | 0.0009 | lambda | 0.5018 | 0.000476 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −18.6 (−28.09, −9.11) | 0.0001 | |||
| LM.Cannabis_x_Herb.THC | 23.1 (11.16, 35.04) | 0.0002 | |||
| Cocaine | −0.57 (−0.97, −0.16) | 0.0057 | |||
| Income | 0 (0, 0) | 0.0036 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Herb.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.06 (0.01, 0.1) | 0.0241 | psi | 0.3206 | 0.00367 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.06 (0.45, 5.67) | 0.0217 | rho | −0.3717 | 0.06365 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −7.99 (−13.97, −2.01) | 0.0087 | lambda | 0.4126 | 0.0323 |
| Anomaly | Term | p-Value | E-Value Estimate | Lower Bound E-Value |
|---|---|---|---|---|
| Urinary | Additive | |||
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0012 | 9.03 | 3.23 | |
| LM.Cannabis_x_Herb.THC | 1.91 × 10−5 | 6.69 × 108 | 1.22 × 105 | |
| Interactive | ||||
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0165 | 15.85 | 2.38 | |
| LM.Cannabis_x_Resin.THC | 0.0079 | 67.00 | 4.73 | |
| 2 Lags | ||||
| Tobacco: Resin | 6.32 × 10−5 | 2.86 | 2.02 | |
| Multicystic Renal Disease | Additive | |||
| LM.Cannabis_x_Resin.THC | 2.61 × 10−8 | 21.28 | 9.43 | |
| Interactive | ||||
| Daily.Interpol. | 0.0498 | 6.23 × 1063 | 13.74 | |
| Tobacco: Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0063 | 1.61 × 105 | 56.63 | |
| 2 Lags | ||||
| Daily.Interpol. | 0.0433 | 1.18 × 10103 | 1.15 × 105 | |
| LM.Cannabis_x_Resin.THC | 0.0097 | 606.01 | 8.35 | |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0269 | 59.95 | 2.51 | |
| Bilateral Renal Agenesis | Additive | |||
| Resin | 0.0273 | 6.58 | 1.62 | |
| Daily.Interpol. | 0.0003 | 1.21 × 1017 | 1.95 × 108 | |
| Interactive | ||||
| Daily.Interpol. | 5.10 × 10−8 | 3.41 × 10122 | 3.30 × 1081 | |
| 2 Lags | ||||
| LM.Cannabis_x_Resin.THC | 4.60 × 10−15 | 42.81 | 22.36 | |
| Hydronephrosis | Additive | |||
| Daily.Interpol. | 5.45 × 10−6 | 5.81 × 1028 | 1.24 × 1017 | |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0004 | 7.24 | 3.13 | |
| Herb | 0.0002 | 6.62 × 104 | 303.56 | |
| Interactive | ||||
| Daily.Interpol. | 0.0038 | 1.45 × 1091 | 1.36 × 1031 | |
| Resin | 0.0046 | 2.73 × 102 | 9.28 | |
| Herb | 1.75 × 10−7 | 3.51 × 109 | 2.01 × 106 | |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0040 | 11.7 | 3.06 | |
| 2 Lags | ||||
| Herb | 1.87 × 10−5 | 2.33 × 107 | 2.03 × 104 | |
| Posterior Urethral Valve | Additive | |||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 2.25 × 10−7 | 10.52 | 5.47 | |
| Interactive | ||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.38 × 10−6 | 2.65 | 2.02 | |
| LM.Cannabis_x_Resin.THC | 2.66 × 10−10 | 43.56 | 17.85 | |
| 2 Lags | ||||
| Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0481 | 8.15 × 104 | 1.89 |
| Anomaly | Term | p-Value | E-Value Estimate | Lower Bound E-Value |
|---|---|---|---|---|
| Urinary | Additive | |||
| LM.Cannabis_x_Herb.THC | 7.84 × 10−15 | 2.18 × 1013 | 1.14 × 1010 | |
| Interactive | ||||
| Daily.Interpol. | 0.0244 | 5.96 × 1017 | 285.07 | |
| Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.07 × 10−9 | 5.01 | 3.38 | |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0001 | 3.34 × 1047 | 4.98 × 1023 | |
| 1 Lag | ||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0130 | 8.58 | 2.10 | |
| Multicystic Renal Disease | Additive | |||
| LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 | |
| Interactive | ||||
| LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 | |
| 2 Lags | ||||
| LM.Cannabis_x_Resin.THC | 7.72 × 10−5 | 6.62 × 103 | 119.49 | |
| Daily.Interpol. | 0.0015 | 6.01 × 1067 | 1.67 × 1026 | |
| Bilateral Renal Agenesis | Interactive | |||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0228 | 3.54 × 103 | 5.18 | |
| 2 Lags | ||||
| LM.Cannabis_x_Resin.THC | 0.0023 | 162.57 | 9.09 | |
| Hydronephrosis | Additive | |||
| Daily.Interpol. | 0.0026 | 82.56 | 7.02 | |
| Herb | 6.95 × 10−5 | 4.51 × 108 | 3.52 × 104 | |
| Interactive | ||||
| Herb | 0.0005 | 6.89 × 106 | 1.46 × 103 | |
| Tobacco: Daily.Interpol. | 0.0004 | 161.31 | 13.62 | |
| 1 Lag | ||||
| Herb | 0.0017 | 1.23 × 105 | 1258.11 | |
| Posterior Urethral Valve | Additive | |||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10−5 | 954.95 | 43.51 | |
| Herb | 0.0013 | 2.74 × 105 | 83.66 | |
| Interactive | ||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10−5 | 955.32 | 43.52 | |
| Herb | 0.0013 | 2.73 × 104 | 83.68 | |
| 1 Lag | ||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0114 | 252.91 | 5.45 | |
| Daily.Interpol. | 0.0009 | 5.23 × 1072 | 1.08 × 1030 | |
| LM.Cannabis_x_Herb.THC | 0.0002 | 4.09 × 1018 | 1.39 × 109 | |
| 2 Lags | ||||
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0217 | 365.38 | 3.75 |
| No. | Anomaly | Regression | Model Type | Term | p-Value | E-Value Estimate | Lower Bound E-Value |
|---|---|---|---|---|---|---|---|
| 1 | Bilateral Renal Agenesis | Panel | Interactive | Daily.Interpol. | 5.10 × 10−8 | 3.41 × 10122 | 3.30 × 1081 |
| 2 | Hydronephrosis | Panel | Interactive | Daily.Interpol. | 0.0038 | 1.45 × 1091 | 1.36 × 1031 |
| 3 | Posterior Urethral Valve | Spatial | 1 Lag | Daily.Interpol. | 0.0009 | 5.23 × 1072 | 1.08 × 1030 |
| 4 | Multicystic Renal Disease | Spatial | 2 Lags | Daily.Interpol. | 0.0015 | 6.01 × 1067 | 1.67 × 1026 |
| 5 | Urinary | Spatial | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0001 | 3.34 × 1047 | 4.98 × 1023 |
| 6 | Hydronephrosis | Panel | Additive | Daily.Interpol. | 5.45 × 10−6 | 5.81 × 1028 | 1.24 × 1017 |
| 7 | Urinary | Spatial | Additive | LM.Cannabis_x_Herb.THC | 7.84 × 10−15 | 2.18 × 1013 | 1.14 × 1010 |
| 8 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Herb.THC | 0.0002 | 4.09 × 1018 | 1.39 × 109 |
| 9 | Bilateral Renal Agenesis | Panel | Additive | Daily.Interpol. | 0.0003 | 1.21 × 1017 | 1.95 × 108 |
| 10 | Hydronephrosis | Panel | Interactive | Herb | 1.75 × 10−7 | 3.51 × 109 | 2.01 × 106 |
| 11 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC | 1.91 × 10−5 | 6.69 × 108 | 1.22 × 105 |
| 12 | Multicystic Renal Disease | Panel | 2 Lags | Daily.Interpol. | 0.0433 | 1.18 × 10103 | 1.15 × 105 |
| 13 | Hydronephrosis | Spatial | Additive | Herb | 6.95 × 10−5 | 4.51 × 108 | 3.52 × 104 |
| 14 | Hydronephrosis | Panel | 2 Lags | Herb | 1.87 × 10−5 | 2.33 × 107 | 2.03 × 104 |
| 15 | Hydronephrosis | Spatial | Interactive | Herb | 0.0005 | 6.89 × 106 | 1.46 × 103 |
| 16 | Hydronephrosis | Spatial | 1 Lag | Herb | 0.0017 | 1.23 × 105 | 1258.11 |
| 17 | Hydronephrosis | Panel | Additive | Herb | 0.0002 | 6.62 × 104 | 303.56 |
| 18 | Urinary | Spatial | Interactive | Daily.Interpol. | 0.0244 | 5.96 × 1017 | 285.07 |
| 19 | Multicystic Renal Disease | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | 7.72 × 10−5 | 6.62 × 103 | 119.49 |
| 20 | Posterior Urethral Valve | Spatial | Interactive | Herb | 0.0013 | 2.73 × 104 | 83.68 |
| 21 | Posterior Urethral Valve | Spatial | Additive | Herb | 0.0013 | 2.74 × 105 | 83.66 |
| 22 | Multicystic Renal Disease | Panel | Interactive | Tobacco: Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0063 | 1.61 × 105 | 56.63 |
| 23 | Posterior Urethral Valve | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10-5 | 955.32 | 43.52 |
| 24 | Posterior Urethral Valve | Spatial | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10−5 | 954.95 | 43.51 |
| 25 | Bilateral Renal Agenesis | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | 4.60 × 10−15 | 42.81 | 22.36 |
| 26 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC | 2.66 × 10−10 | 43.56 | 17.85 |
| 27 | Multicystic Renal Disease | Panel | Interactive | Daily.Interpol. | 0.0498 | 6.23 × 1063 | 13.74 |
| 28 | Hydronephrosis | Spatial | Interactive | Tobacco: Daily.Interpol. | 0.0004 | 161.31 | 13.62 |
| 29 | Multicystic Renal Disease | Panel | Additive | LM.Cannabis_x_Resin.THC | 2.61 × 10−8 | 21.28 | 9.43 |
| 30 | Hydronephrosis | Panel | Interactive | Resin | 0.0046 | 273.00 | 9.28 |
| 31 | Bilateral Renal Agenesis | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | 0.0023 | 162.57 | 9.09 |
| 32 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | 0.0097 | 606.01 | 8.35 |
| 33 | Hydronephrosis | Spatial | Additive | Daily.Interpol. | 0.0026 | 82.56 | 7.02 |
| 34 | Posterior Urethral Valve | Panel | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 2.25 × 10−7 | 10.52 | 5.47 |
| 35 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0114 | 252.91 | 5.45 |
| 36 | Bilateral Renal Agenesis | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0228 | 3540.00 | 5.18 |
| 37 | Urinary | Panel | Interactive | LM.Cannabis_x_Resin.THC | 0.0079 | 67.00 | 4.73 |
| 38 | Posterior Urethral Valve | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0217 | 365.38 | 3.75 |
| 39 | Urinary | Spatial | Interactive | Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.07 × 10−9 | 5.01 | 3.38 |
| 40 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0012 | 9.03 | 3.23 |
| 41 | Hydronephrosis | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0004 | 7.24 | 3.13 |
| 42 | Hydronephrosis | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0040 | 11.7 | 3.06 |
| 43 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0269 | 59.95 | 2.51 |
| 44 | Urinary | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0165 | 15.85 | 2.38 |
| 45 | Urinary | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0130 | 8.58 | 2.10 |
| 46 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.38 × 10−6 | 2.65 | 2.02 |
| 47 | Urinary | Panel | 2 Lags | Tobacco: Resin | 6.32 × 10−5 | 2.86 | 2.02 |
| 48 | Posterior Urethral Valve | Panel | 2 Lags | Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0481 | 81500.00 | 1.89 |
| 49 | Bilateral Renal Agenesis | Panel | Additive | Resin | 0.0273 | 6.58 | 1.62 |
| 50 | Multicystic Renal Disease | Spatial | Additive | LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 |
| 51 | Multicystic Renal Disease | Spatial | Interactive | LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 |
| No. | Anomaly | Regression | Model Type | Term | p-Value | E-Value Estimate | Lower Bound E-Value |
|---|---|---|---|---|---|---|---|
| 1 | Bilateral Renal Agenesis | Panel | Interactive | Daily.Interpol. | 5.10 × 10−8 | 3.41 × 10122 | 3.30 × 1081 |
| 2 | Bilateral Renal Agenesis | Panel | Additive | Daily.Interpol. | 0.0003 | 1.21 × 1017 | 1.95 × 108 |
| 3 | Bilateral Renal Agenesis | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | 4.60 × 10−15 | 42.81 | 22.36 |
| 4 | Bilateral Renal Agenesis | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | 0.0023 | 162.57 | 9.09 |
| 5 | Bilateral Renal Agenesis | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0228 | 3540.00 | 5.18 |
| 6 | Bilateral Renal Agenesis | Panel | Additive | Resin | 0.0273 | 6.58 | 1.62 |
| 7 | Hydronephrosis | Panel | Interactive | Daily.Interpol. | 0.0038 | 1.45 × 1091 | 1.36 × 1031 |
| 8 | Hydronephrosis | Panel | Additive | Daily.Interpol. | 5.45 × 10-6 | 5.81 × 1028 | 1.24 × 1017 |
| 9 | Hydronephrosis | Panel | Interactive | Herb | 1.75 × 10−7 | 3.51 × 109 | 2.01 × 106 |
| 10 | Hydronephrosis | Spatial | Additive | Herb | 6.95 × 10−5 | 4.51 × 108 | 3.52 × 104 |
| 11 | Hydronephrosis | Panel | 2 Lags | Herb | 1.87 × 10−5 | 2.33 × 107 | 2.03 × 104 |
| 12 | Hydronephrosis | Spatial | Interactive | Herb | 0.0005 | 6.89 × 106 | 1.46 × 103 |
| 13 | Hydronephrosis | Spatial | 1 Lag | Herb | 0.0017 | 1.23 × 105 | 1258.11 |
| 14 | Hydronephrosis | Panel | Additive | Herb | 0.0002 | 6.62 × 104 | 303.56 |
| 15 | Hydronephrosis | Spatial | Interactive | Tobacco: Daily.Interpol. | 0.0004 | 161.31 | 13.62 |
| 16 | Hydronephrosis | Panel | Interactive | Resin | 0.0046 | 273.00 | 9.28 |
| 17 | Hydronephrosis | Spatial | Additive | Daily.Interpol. | 0.0026 | 82.56 | 7.02 |
| 18 | Hydronephrosis | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0004 | 7.24 | 3.13 |
| 19 | Hydronephrosis | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0040 | 11.7 | 3.06 |
| 20 | Multicystic Renal Disease | Spatial | 2 Lags | Daily.Interpol. | 0.0015 | 6.01 × 1067 | 1.67 × 1026 |
| 21 | Multicystic Renal Disease | Panel | 2 Lags | Daily.Interpol. | 0.0433 | 1.18 × 10103 | 1.15 × 105 |
| 22 | Multicystic Renal Disease | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | 7.72 × 10−5 | 6.62 × 103 | 119.49 |
| 23 | Multicystic Renal Disease | Panel | Interactive | Tobacco: Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0063 | 1.61 × 105 | 56.63 |
| 24 | Multicystic Renal Disease | Panel | Interactive | Daily.Interpol. | 0.0498 | 6.23 × 1063 | 13.74 |
| 25 | Multicystic Renal Disease | Panel | Additive | LM.Cannabis_x_Resin.THC | 2.61 × 10−8 | 21.28 | 9.43 |
| 26 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | 0.0097 | 606.01 | 8.35 |
| 27 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0269 | 59.95 | 2.51 |
| 28 | Multicystic Renal Disease | Spatial | Additive | LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 |
| 29 | Multicystic Renal Disease | Spatial | Interactive | LM.Cannabis_x_Resin.THC | 0.0310 | 9.68 | 1.60 |
| 30 | Posterior Urethral Valve | Spatial | 1 Lag | Daily.Interpol. | 0.0009 | 5.23 × 1072 | 1.08 × 1030 |
| 31 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Herb.THC | 0.0002 | 4.09 × 1018 | 1.39 × 109 |
| 32 | Posterior Urethral Valve | Spatial | Interactive | Herb | 0.0013 | 2.73 × 104 | 83.68 |
| 33 | Posterior Urethral Valve | Spatial | Additive | Herb | 0.0013 | 2.74 × 105 | 83.66 |
| 34 | Posterior Urethral Valve | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10−5 | 955.32 | 43.52 |
| 35 | Posterior Urethral Valve | Spatial | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 8.82 × 10−5 | 954.95 | 43.51 |
| 36 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC | 2.66 × 10−10 | 43.56 | 17.85 |
| 37 | Posterior Urethral Valve | Panel | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 2.25 × 10−7 | 10.52 | 5.47 |
| 38 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0114 | 252.91 | 5.45 |
| 39 | Posterior Urethral Valve | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0217 | 365.38 | 3.75 |
| 40 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.38 × 10−6 | 2.65 | 2.02 |
| 41 | Posterior Urethral Valve | Panel | 2 Lags | Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0481 | 81500.00 | 1.89 |
| 42 | Urinary | Spatial | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0001 | 3.34 × 1047 | 4.98 × 1023 |
| 43 | Urinary | Spatial | Additive | LM.Cannabis_x_Herb.THC | 7.84 × 10−15 | 2.18 × 1013 | 1.14 × 1010 |
| 44 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC | 1.91 × 10−5 | 6.69 × 108 | 1.22 × 105 |
| 45 | Urinary | Spatial | Interactive | Daily.Interpol. | 0.0244 | 5.96 × 1017 | 285.07 |
| 46 | Urinary | Panel | Interactive | LM.Cannabis_x_Resin.THC | 0.0079 | 67.00 | 4.73 |
| 47 | Urinary | Spatial | Interactive | Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 3.07 × 10−9 | 5.01 | 3.38 |
| 48 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0012 | 9.03 | 3.23 |
| 49 | Urinary | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.0165 | 15.85 | 2.38 |
| 50 | Urinary | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 0.0130 | 8.58 | 2.10 |
| 51 | Urinary | Panel | 2 Lags | Tobacco: Resin | 6.32 × 10−5 | 2.86 | 2.02 |
| Anomaly | Number | Mean Minimum E-Value | Median Minimum E-Value | Min Minimum E-Value | Max Minimum E-Value | Mean E-Value Estimate | Median E-Value Estimate | Min E-Value Estimate | Max E-Value Estimate |
|---|---|---|---|---|---|---|---|---|---|
| Hydronephrosis | 13 | 1.05 × 1030 | 1258.11 | 3.06 | 1.36 × 1031 | 1.12 × 1090 | 1.23 × 105 | 7.24 | 1.45 × 1091 |
| Posterior Urethral Valve | 12 | 9.00 × 1028 | 30.68 | 1.89 | 1.08 × 1030 | 4.36 × 1071 | 955.14 | 2.65 | 5.23 × 1072 |
| Bilateral Renal Agenesis | 6 | 5.50 × 1080 | 15.73 | 1.62 | 3.30 × 1081 | 5.68 × 10121 | 1.85 × 103 | 6.58 | 3.41 × 10122 |
| Multicystic Renal Disease | 10 | 1.67 × 1025 | 11.59 | 1.6 | 1.67 × 1026 | 1.18 × 10102 | 3.61 × 103 | 9.68 | 1.18 × 10103 |
| Urinary | 10 | 4.98 × 1022 | 4.055 | 2.02 | 4.98 × 1023 | 3.34 × 1046 | 41.43 | 2.86 | 3.34 × 1047 |
| No. | Anomaly | Regression | Model Type | Term | Group | p-Value | E-Value Estimate | Lower Bound E-Value |
|---|---|---|---|---|---|---|---|---|
| 1 | Bilateral Renal Agenesis | Panel | Interactive | Daily.Interpol. | Daily | 5.10 × 10−8 | 3.41 × 10122 | 3.30 × 1081 |
| 2 | Hydronephrosis | Panel | Interactive | Daily.Interpol. | Daily | 0.0038 | 1.45 × 1091 | 1.36 × 1031 |
| 3 | Posterior Urethral Valve | Spatial | 1 Lag | Daily.Interpol. | Daily | 0.0009 | 5.23 × 1072 | 1.08 × 1030 |
| 4 | Multicystic Renal Disease | Spatial | 2 Lags | Daily.Interpol. | Daily | 0.0015 | 6.01 × 1067 | 1.67 × 1026 |
| 5 | Hydronephrosis | Panel | Additive | Daily.Interpol. | Daily | 5.45 × 10−6 | 5.81 × 1028 | 1.24 × 1017 |
| 6 | Bilateral Renal Agenesis | Panel | Additive | Daily.Interpol. | Daily | 0.0003 | 1.21 × 1017 | 1.95 × 108 |
| 7 | Multicystic Renal Disease | Panel | 2 Lags | Daily.Interpol. | Daily | 0.0433 | 1.18 × 10103 | 1.15 × 105 |
| 8 | Urinary | Spatial | Interactive | Daily.Interpol. | Daily | 0.0244 | 5.96 × 1017 | 285.07 |
| 9 | Multicystic Renal Disease | Panel | Interactive | Daily.Interpol. | Daily | 0.0498 | 6.23 × 1063 | 13.74 |
| 10 | Hydronephrosis | Spatial | Additive | Daily.Interpol. | Daily | 0.0026 | 82.56 | 7.02 |
| 11 | Posterior Urethral Valve | Panel | 2 Lags | Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Daily | 0.0481 | 81500.00 | 1.89 |
| 12 | Hydronephrosis | Panel | Interactive | Herb | Herb | 1.75 × 10−7 | 3.51 × 109 | 2.01 × 106 |
| 13 | Hydronephrosis | Spatial | Additive | Herb | Herb | 6.95 × 10−5 | 4.51 × 108 | 3.52 × 104 |
| 14 | Hydronephrosis | Panel | 2 Lags | Herb | Herb | 1.87 × 10−5 | 2.33 × 107 | 2.03 × 104 |
| 15 | Hydronephrosis | Spatial | Interactive | Herb | Herb | 0.0005 | 6.89 × 106 | 1.46 × 103 |
| 16 | Hydronephrosis | Spatial | 1 Lag | Herb | Herb | 0.0017 | 1.23 × 105 | 1258.11 |
| 17 | Hydronephrosis | Panel | Additive | Herb | Herb | 0.0002 | 6.62 × 104 | 303.56 |
| 18 | Posterior Urethral Valve | Spatial | Interactive | Herb | Herb | 0.0013 | 2.73 × 104 | 83.68 |
| 19 | Posterior Urethral Valve | Spatial | Additive | Herb | Herb | 0.0013 | 2.74 × 105 | 83.66 |
| 20 | Urinary | Spatial | Additive | LM.Cannabis_x_Herb.THC | Herb | 7.84 × 10−15 | 2.18 × 1013 | 1.14 × 1010 |
| 21 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Herb.THC | Herb | 0.0002 | 4.09 × 1018 | 1.39 × 109 |
| 22 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC | Herb | 1.91 × 10−5 | 6.69 × 108 | 1.22 × 105 |
| 23 | Urinary | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | Herb | 0.0012 | 9.03 | 3.23 |
| 24 | Hydronephrosis | Panel | Additive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | Herb | 0.0004 | 7.24 | 3.13 |
| 25 | Hydronephrosis | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | Herb | 0.0040 | 11.7 | 3.06 |
| 26 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | Herb | 0.0269 | 59.95 | 2.51 |
| 27 | Urinary | Panel | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | Herb | 0.0165 | 15.85 | 2.38 |
| 28 | Urinary | Spatial | Interactive | LM.Cannabis_x_Herb.THC_x_Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Herb | 0.0001 | 3.34 × 1047 | 4.98 × 1023 |
| 29 | Multicystic Renal Disease | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | Resin | 7.72 × 10−5 | 6.62 × 103 | 119.49 |
| 30 | Bilateral Renal Agenesis | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | Resin | 4.60 × 10−15 | 42.81 | 22.36 |
| 31 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC | Resin | 2.66 × 10−10 | 43.56 | 17.85 |
| 32 | Multicystic Renal Disease | Panel | Additive | LM.Cannabis_x_Resin.THC | Resin | 2.61 × 10−8 | 21.28 | 9.43 |
| 33 | Bilateral Renal Agenesis | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC | Resin | 0.0023 | 162.57 | 9.09 |
| 34 | Multicystic Renal Disease | Panel | 2 Lags | LM.Cannabis_x_Resin.THC | Resin | 0.0097 | 606.01 | 8.35 |
| 35 | Urinary | Panel | Interactive | LM.Cannabis_x_Resin.THC | Resin | 0.0079 | 67.00 | 4.73 |
| 36 | Multicystic Renal Disease | Spatial | Additive | LM.Cannabis_x_Resin.THC | Resin | 0.0310 | 9.68 | 1.60 |
| 37 | Multicystic Renal Disease | Spatial | Interactive | LM.Cannabis_x_Resin.THC | Resin | 0.0310 | 9.68 | 1.60 |
| 38 | Posterior Urethral Valve | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 8.82 × 10−5 | 955.32 | 43.52 |
| 39 | Posterior Urethral Valve | Spatial | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 8.82 × 10−5 | 954.95 | 43.51 |
| 40 | Posterior Urethral Valve | Panel | Additive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 2.25 × 10−7 | 10.52 | 5.47 |
| 41 | Posterior Urethral Valve | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 0.0114 | 252.91 | 5.45 |
| 42 | Bilateral Renal Agenesis | Spatial | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 0.0228 | 3540.00 | 5.18 |
| 43 | Posterior Urethral Valve | Spatial | 2 Lags | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 0.0217 | 365.38 | 3.75 |
| 44 | Urinary | Spatial | 1 Lag | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 0.0130 | 8.58 | 2.10 |
| 45 | Posterior Urethral Valve | Panel | Interactive | LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 3.38 × 10−6 | 2.65 | 2.02 |
| 46 | Hydronephrosis | Panel | Interactive | Resin | Resin | 0.0046 | 273.00 | 9.28 |
| 47 | Bilateral Renal Agenesis | Panel | Additive | Resin | Resin | 0.0273 | 6.58 | 1.62 |
| 48 | Hydronephrosis | Spatial | Interactive | Tobacco: Daily.Interpol. | Daily | 0.0004 | 161.31 | 13.62 |
| 49 | Multicystic Renal Disease | Panel | Interactive | Tobacco: Daily.Interpol.: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 0.0063 | 1.61 × 10+5 | 56.63 |
| 50 | Urinary | Spatial | Interactive | Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | Resin | 3.07 × 10−9 | 5.01 | 3.38 |
| 51 | Urinary | Panel | 2 Lags | Tobacco: Resin | Resin | 6.32 × 10−5 | 2.86 | 2.02 |
| Group | Number | Mean Minimum E-Value | Median Minimum E-Value | Minimum Minimum E-Value | Maximum Minimum E-Value | Mean E-Value Estimate | Median E-Value Estimate | Minimum E-Value Estimate | Maximum E-Value Estimate |
|---|---|---|---|---|---|---|---|---|---|
| Daily | 12 | 2.75 × 1080 | 9.76 × 107 | 1.89 | 3.30 × 1081 | 2.84 × 10121 | 3.12 × 1063 | 82.56 | 3.41 × 10122 |
| Herb | 17 | 2.93 × 1022 | 1258.11 | 2.38 | 4.98 × 1023 | 1.96 × 1046 | 2.74 × 105 | 7.24 | 3.34 × 1047 |
| Resin | 22 | 17.20 | 5.46 | 1.6 | 119.49 | 7952.74 | 55.28 | 2.65 | 1.61 × 105 |
| Comparison | W-Statistic | Alternative | p-Value |
|---|---|---|---|
| mEV, Daily_v_Herb | 132 | two.sided | 0.1947 |
| mEV, Daily_v_Resin | 223 | two.sided | 0.0011 |
| mEV, Herb_v_Resin | 292 | two.sided | 0.0031 |
| EVEstimate, Daily_v_Herb | 165 | two.sided | 0.0043 |
| EVEstimate, Daily_v_Resin | 243 | two.sided | 6.82 × 10−5 |
| EVEstimate, Herb_v_Resin | 297 | two.sided | 0.0019 |
4. Discussion
4.1. Main Results
4.2. Detailed Main Results
4.3. Choice of Anomalies
4.4. Qualitative Causal Inference
4.5. Quantitative Causal Inference
4.6. Mechanisms
4.7. Cannabinoid Inhibition of Embryonic Morphogens
4.8. Epigenomic Control of Uronephrological Gene Expression
4.9. Nephrogenic Genes in Renal Organoids
| Nearest Gene Name | Chromosome Number | Nearest Gene Number | Dependency Status | Functional Annotation | Page | Distance from Nearest Gene | Relative Position | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|---|
| JAG1 | 20 | ENSG00000101384 | Dependence | Notch Ligand | 36 | 0 | Intron | 1.31 × 10−6 | 0.007992 |
| NOTCH2 | 1 | ENSG00000134250 | Dependence | Notch Pathway | 55 | 0 | Intron | 3.07 × 10−6 | 0.007992 |
| NOTCH1 | 9 | ENSG00000101384 | Dependence | Notch Pathway | 56 | 0 | Intron | 3.29 × 10−6 | 0.012443 |
| NOTCH2 | 1 | ENSG00000134250 | Dependence | Notch Pathway | 64 | 0 | Intron | 4.19 × 10−6 | 0.013994 |
| NOTCH3 | 19 | ENSG00000074181 | Dependence | Notch Pathway | 86 | 3797 | Downstream | 7.65 × 10−6 | 0.018677 |
| NOTCH2NLC | 1 | ENSG00000286219 | Withdrawal | Notch Pathway | 165 | 0 | Intron | 2.85 × 10−6 | 0.012636 |
| RBPJ | 4 | ENSG00000168214 | Dependence | Notch Pathway | 107 | 0 | Intron | 1.17 × 10−5 | 0.022698 |
| RBPJP2 | 9 | ENSG00000274181 | Dependence | Notch Pathway | 121 | 19867 | Upstream | 1.50 × 10−5 | 0.025700 |
| RBPJ | 4 | ENSG00000168214 | Withdrawal | Notch Pathway | 125 | 0 | Intron | 1.34 × 10−8 | 0.000859 |
| Nearest Gene Name | Chromosome Number | Nearest Gene Number | Dependency Status | Functional Annotation | Page | Function | Number Genes Identified | p-Value | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Skin Lesion | 325 | Notch Processing | 115 | 1.65 × 10−6 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Skin cancer | 325 | Notch Processing | 113 | 4.79 × 10−6 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Cutaneous melanoma | 326 | Notch Processing | 110 | 7.71 × 10−6 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Solid Organ cancer | 327 | Notch Processing | 150 | 9.16 × 10−6 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Cancer | 329 | Notch Processing | 151 | 4.32 × 10−5 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Melanoma | 329 | Notch Processing | 115 | 6.21 × 10−5 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Solid Organ cancer | 335 | Notch Processing | 149 | 1.63 × 10−4 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Protein processing | 353 | Notch Processing | 4 | 4.69 × 10−3 | |
| PSENEN | 19 | ENSG00000205155 | Withdrawal | Organismal death | 356 | Notch Processing | 39 | 6.70 × 10−3 |
| Nearest Gene Name | Chromosome Number | Nearest Gene Number | Dependency Status | Functional Annotation | Page | Distance from Nearest Gene | Relative Position | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|---|
| PTCH1 | 9 | ENSG00000185920 | Dependence | Shh Receptor | 58 | 0 | Intron | 3.46 × 10−6 | 0.012789 |
| PTCHD1-AS | X | ENSG00000233067 | Dependence | lnc Promoter/enhancer | 91 | 0 | Intron | 8.61 × 10−6 | 0.019678 |
| PTCHD1-AS | X | ENSG00000233067 | Withdrawal | lnc Promoter/enhancer | 129 | 0 | Intron | 8.21 × 10−8 | 0.002096 |
| PTCHD4 | 6 | ENSG00000244694 | Withdrawal | Shh Receptor; Otopalatodigital syndrome | 138 | 0 | Intron | 4.21 × 10−7 | 0.005104 |
| PTCH1 | 9 | ENSG00000185920 | Withdrawal | Shh Receptor | 185 | 0 | Intron | 5.80 × 10−6 | 0.017679 |
| SUFU | 16 | ENSG00000161996 | Withdrawal | Hedgehog Inhibitor | 207 | 0 | Exon | 1.01 × 10−5 | 0.022942 |
| Gli3 | 7 | ENSG00000106571 | Dependence | Shh mediator | 78 | 81,232 | Downstream | 6.35 × 10−6 | 0.017090 |
| Gli3 | 7 | ENSG00000106571 | Dependence | Shh mediator | 99 | 0 | Intron | 1.00 × 10−5 | 0.021181 |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Shh mediator | 124 | 20,318 | Downstream | 8.23 × 10−9 | 0.000646 |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Shh mediator | 182 | 0 | Intron | 5.28 × 10−6 | 0.001687 |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Shh mediator | 231 | 0 | Intron | 1.62 × 10−5 | 0.028539 |
| Nearest Gene Name | Chromosome Number | Nearest Gene Number | Dependency Status | Functional Annotation | Page | Function | Number Genes Identified | p-Value | |
| PTCH1 | 9 | ENSG00000185920 | KEGG Pathway | Notch Processing | 237 | Notch Processing | 31 | 0.044117 | |
| PTCH1 | 9 | ENSG00000185920 | KEGG Pathway | Skin cancer | 238 | Notch Processing | 54 | 0.067770 | |
| PSENEN | 19 | ENSG00000185920 | Withdrawal | Cutaneous melanoma | 326 | Notch Processing | 110 | 0.000008 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Skin lesion | 325 | Notch TF | 115 | 1.65 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Head & Neck SCC | 325 | Notch TF | 53 | 3.59 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Skin cancer | 325 | Notch TF | 113 | 4.79 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Lung adenocarcinoma | 325 | Notch TF | 42 | 5.84 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Cancer | 325 | Notch TF | 149 | 7.17 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Large bowel cancer | 326 | Notch TF | 120 | 7.45 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Cutaneous melanoma | 326 | Notch TF | 110 | 7.71 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Highg grade astrocytoma | 326 | Notch TF | 82 | 8.42 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Abdominal adenocarcinoma | 326 | Notch TF | 135 | 8.46 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Solid cancer | 327 | Notch TF | 150 | 9.16 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Haed and Neck cancer | 327 | Notch TF | 137 | 9.54 × 10−6 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Sensory development | 327 | Notch TF | 18 | 1.30 × 10−5 | |
| Gli3 | 7 | ENSG00000106571 | Withdrawal | Carcinoma | 327 | Notch TF | 148 | 1.38 × 10−5 |
| Nearest Gene Name | Chromosome Number | Nearest Gene Number | Dependency Status | Functional Annotation | Page | Distance from Nearest Gene | Relative Position | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|---|
| CCDC178 | 18 | ENSG00000166960 | Dependence | Coiled coil domain containing | 21 | 0 | Intron | 3.78 × 10−7 | 0.004344 |
| CCDC178 | 18 | ENSG00000166960 | Dependence | Coiled coil domain containing | 47 | 0 | Intron | 2.29 × 10−6 | 0.010512 |
| CCDC171 | 9 | ENSG00000164989 | Dependence | Coiled coil domain containing | 53 | 0 | Intron | 2.88 × 10−6 | 0.011635 |
| CCDC178 | 18 | ENSG00000166960 | Dependence | Coiled coil domain containing | 119 | 0 | Intron | 1.46 × 10−5 | 0.025394 |
| CCDC171 | 9 | ENSG00000164989 | Withdrawal | Coiled coil domain containing | 359 | 0 | Intron | 8.36 × 10−8 | 0.002109 |
| CCDC170 | 6 | ENSG00000120262 | Withdrawal | Coiled coil domain containing | 136 | 0 | Intron | 3.29 × 10−7 | 0.004420 |
| CCDC175 | 14 | ENSG00000151838 | Withdrawal | Coiled coil domain containing | 148 | 0 | Intron | 1.11 × 10−6 | 0.008146 |
| CCDC171 | 9 | ENSG00000164989 | Withdrawal | Coiled coil domain containing | 169 | 57,410 | Downstream | 3.38 × 10−6 | 0.013764 |
| CCDC171 | 9 | ENSG00000164989 | Withdrawal | Coiled coil domain containing | 170 | 0 | Intron | 3.50 × 10−6 | 0.139199 |
| CCDC178 | 18 | ENSG00000166960 | Withdrawal | Coiled coil domain containing | 186 | 0 | Intron | 5.94 × 10−6 | 0.017905 |
| CCDC171 | 9 | ENSG00000164989 | Withdrawal | Coiled coil domain containing | 189 | 0 | Intron | 6.67 × 10−6 | 0.019063 |
| CCDC171 | 9 | ENSG00000164989 | Withdrawal | Coiled coil domain containing | 203 | 0 | Intron | 9.26 × 10−6 | 0.022131 |
| MYH7 | 14 | ENSG00000092054 | Dependence | Myosin heavy chain 7B | 81 | 12,605 | Upstream | 6.91 × 10−6 | 0.017781 |
| ROCK1 | 18 | ENSG00000067900 | Dependence | Rho associated Coiled Coil Kinase | 20 | 2,527,461 | Downstream | 3.17 × 10−7 | 0.003991 |
| ROCK1 | 18 | ENSG00000067900 | Dependence | Rho associated Coiled Coil Kinase | 20 | 0 | Intron | 5.41 × 10−6 | 0.015884 |
| ROCK1 | 18 | ENSG00000067900 | Dependence | Rho associated Coiled Coil Kinase | 20 | 411,279 | Downstream | 4.72 × 10−6 | 0.016044 |
| EPCAM-DT | 2 | ENSG00000234690 | Dependence | Epithelial Cell adhesion molecule—divergent transcript | 221 | 0 | Intron | 1.33 × 10−5 | 0.025950 |
| BMPR2 | 1 | ENSG00000204217 | Dependence | BMP Receptor 2 | 12 | 0 | Intron | 8.38 × 10−8 | 0.001951 |
| BMPR1B | 4 | ENSG00000138696 | Dependence | BMP Receptor 1B | 25 | 0 | Intron | 5.68 × 10−7 | 0.005306 |
| BMPR2 | 4 | ENSG00000138696 | Dependence | BMP Receptor 1B | 35 | 0 | Intron | 1.22 × 10−6 | 0.007711 |
| BMPER | 7 | ENSG00000164619 | Dependence | BMP Endothelial regulator | 42 | 0 | Intron | 1.83 × 10−6 | 0.009422 |
| BMP6 | 6 | ENSG00000153162 | Dependence | BMP 6 | 56 | 0 | Intron | 3.18 × 10−6 | 0.012217 |
| BMPR1B | 4 | ENSG00000138696 | Dependence | BMP Receptor 1B | 70 | 0 | Intron | 5.19 × 10−6 | 0.015541 |
| BMPR1A | 10 | ENSG00000107779 | Dependence | BMP Receptor 1A | 96 | 0 | Intron | 9.50 × 10−6 | 0.020621 |
| BMPR1B | 4 | ENSG00000138696 | Dependence | BMP Receptor 1B | 98 | 0 | Intron | 9.94 × 10−6 | 0.021066 |
| BMPER | 7 | ENSG00000164619 | Dependence | BMP Endothelial regulator | 113 | 0 | Intron | 1.32 × 10−5 | 0.024145 |
| BMPR1B | 4 | ENSG00000138696 | Withdrawal | BMP Receptor 1B | 124 | 0 | Intron | 7.92 × 10−9 | 0.000631 |
| WNT7B | 22 | ENSG00000188064 | Dependence | Wnt family member 7B | 74 | 0 | Intron | 5.78 × 10−6 | 0.016375 |
| WNT7B | 22 | ENSG00000188064 | Withdrawal | Wnt family member 7B | 161 | 0 | Intron | 2.38 × 10−6 | 0.011749 |
| WNT7A | 3 | ENSG00000154764 | Dependence | Wnt family member 7A | 119 | 0 | Intron | 1.47 × 10−5 | 0.025468 |
| WNT7A | 3 | ENSG00000154764 | Withdrawal | Wnt family member 7A | 123 | 0 | Intron | 4.13 × 10−9 | 0.000453 |
| WNT2 | 7 | ENSG00000105989 | Dependence | Wnt family member 2 | 26 | 0 | Intron | 1.02 × 10−6 | 0.007849 |
| WNT5A | 3 | ENSG00000114251 | Withdrawal | Wnt family member 5A | 150 | 26,665 | Downstream | 1.25 × 10−6 | 0.008600 |
| WNT7A | 3 | ENSG00000154764 | Dependence | Wnt family member 7A | 150 | 13,677 | Upstream | 1.27 × 10−6 | 0.008678 |
| WNT7B | 22 | ENSG00000188064 | Withdrawal | Wnt family member 7B | 161 | 0 | Intron | 2.38 × 10−6 | 0.011749 |
| WNT9B | 17 | ENSG00000158955 | Withdrawal | Wnt family member 9B | 211 | 0 | Intron | 1.10 × 10−5 | 0.023817 |
| WNT16 | 7 | ENSG00000002745 | Withdrawal | Wnt family member 16 | 227 | 0 | Intron | 1.50 × 10−5 | 0.027474 |
4.10. Exponential Genotoxic Effects
4.11. Strengths and Limitations
4.12. Generalizability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reece, A.S.; Hulse, G.K. Geotemporospatial and Causal Inferential Epidemiological Overview and Survey of USA Cannabis, Cannabidiol and Cannabinoid Genotoxicity Expressed in Cancer Incidence 2003–2017: Part 1–Continuous Bivariate Analysis. Arch. Public Health 2022, 80, 99–133. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Geotemporospatial and Causal Inferential Epidemiological Overview and Survey of USA Cannabis, Cannabidiol and Cannabinoid Genotoxicity Expressed in Cancer Incidence 2003–2017: Part 2–Categorical Bivariate Analysis and Attributable Fractions. Arch. Public Health 2022, 80, 100–135. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Geotemporospatial and Causal Inferential Epidemiological Overview and Survey of USA Cannabis, Cannabidiol and Cannabinoid Genotoxicity Expressed in Cancer Incidence 2003–2017: Part 3–Spatiotemporal, Multivariable and Causal Inferential Pathfinding and Exploratory Analyses of Prostate and Ovarian Cancers. Arch. Public Health 2022, 80, 100–136. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Cannabinoid exposure as a major driver of pediatric acute lymphoid Leukaemia rates across the USA: Combined geospatial, multiple imputation and causal inference study. BMC Cancer 2021, 21, 984–1017. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. A geospatiotemporal and causal inference epidemiological exploration of substance and cannabinoid exposure as drivers of rising US pediatric cancer rates. BMC Cancer 2021, 21, 197–230. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Contemporary epidemiology of rising atrial septal defect trends across USA 1991-2016: A combined ecological geospatiotemporal and causal inferential study. BMC Pediatrics 2020, 20, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Geotemporospatial and causal inference epidemiological analysis of US survey and overview of cannabis, cannabidiol and cannabinoid genotoxicity in relation to congenital anomalies 2001–2015. BMC Pediatrics 2022, 22, 47–124. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Broad Spectrum epidemiological contribution of cannabis and other substances to the teratological profile of northern New South Wales: Geospatial and causal inference analysis. BMC Pharmacol. Toxicol. 2020, 21, 75–103. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Causal inference multiple imputation investigation of the impact of cannabinoids and other substances on ethnic differentials in US testicular cancer incidence. BMC Pharmacol. Toxicol. 2021, 22, 40–71. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Co-occurrence across time and space of drug- and cannabinoid- exposure and adverse mental health outcomes in the National Survey of Drug Use and Health: Combined geotemporospatial and causal inference analysis. BMC Public Health 2020, 20, 1655–1669. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Cannabis Genotoxicity and Cancer Incidence: A Highly Concordant Synthesis of European and USA Datasets. In Cannabis, Cannabinoids and Endocannabinoids. Volume 1; Preedy, V., Patel, V., Eds.; Elsevier: London, UK, 2022; in press. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Cannabinoid Genotoxicity and Congenital Anomalies: A Convergent Synthesis of European and USA Datasets. In Cannabis, Cannabinoids and Endocannabinoids. Volume 1; Preedy, V., Patel, V., Eds.; Elsevier: London, UK, 2022; in press. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Cannabis Teratology Explains Current Patterns of Coloradan Congenital Defects: The Contribution of Increased Cannabinoid Exposure to Rising Teratological Trends. Clin. Pediatrics 2019, 58, 1085–1123. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Epidemiological Associations of Various Substances and Multiple Cannabinoids with Autism in USA. Clin. Pediatrics Open Access 2019, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Reece, A.S.; Hulse, G.K. Cannabinoid- and Substance- Relationships of European Congenital Anomaly Patterns: A Space-Time Panel Regression and Causal Inferential Study. Environ. Epigenetics 2022, 8, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Geospatiotemporal and Causal Inference Study of Cannabis and Other Drugs as Risk Factors for Female Breast Cancer USA 2003-2017. Environ. Epigenetics 2022, 2022, 1–22. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Cannabis Consumption Patterns Explain the East-West Gradient in Canadian Neural Tube Defect Incidence: An Ecological Study. Glob. Pediatrics Health 2019, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Reece, A.S.; Hulse, G.K. Canadian Cannabis Consumption and Patterns of Congenital Anomalies: An Ecological Geospatial Analysis. J. Addict. Med. 2020, 14, e195–e210. [Google Scholar] [CrossRef]
- Forrester, M.B.; Merz, R.D. Risk of selected birth defects with prenatal illicit drug use, Hawaii, 1986–2002. J. Toxicol. Environ. Health 2007, 70, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Quadruple convergence—Rising cannabis prevalence, intensity, concentration and use disorder treatment. Lancet Reg. Health Eur. 2021, 10, 100245–100246. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Epidemiological Overview of Cannabis- and Substance- Carcinogenesis in Europe: A Lagged Causal Inferential Panel Regression Modelling and Marginal Effects Study. 2022; manuscript submitted. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Cannabis in Pregnancy—Rejoinder, Exposition and Cautionary Tales. Psychiatr. Times 2020, 37. Available online: https://www.psychiatrictimes.com/view/cannabis-pregnancy-rejoinder-exposition-cautionary-tales (accessed on 18 September 2022).
- Reece, A.S.; Hulse, G.K. Epidemiological Overview of Multidimensional Chromosomal and Genome Toxicity of Cannabis Exposure in Congenital Anomalies and Cancer Development. Sci. Rep. 2021, 11, 13892–13912. [Google Scholar]
- Reece, A.S. Rapid Response: Cannabinoid Genotoxic Trifecta—Cancerogenesis, Clinical Teratogenesis and Cellular Ageing. Br. Med. J. 2022, 376, n3114. Available online: https://www.bmj.com/content/376/bmj.n3114/rr-1 (accessed on 18 September 2022).
- Reece, A.S. Limblessness: Cannabinoids Inhibit Key Embryonic Morphogens both Directly and Epigenomically. Br. Med. J. 2022, 376, n3114. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Geospatiotemporal and Causal Inferential Epidemiological Survey and Exploration of Cannabinoid- and Substance- Related Carcinogenesis in USA 2003–2017. In Epidemiology of Cannabis: Genotoxicity and Neurotoxicity, Epigenomics and Aging; Elsevier: New York, NY, USA; Volume 1, in press.
- Reece, A.S.; Hulse, G.K. European Epidemiological Patterns of Cannabis- and Substance- Related Congenital Body Wall Anomalies: Geospatiotemporal and Causal Inferential Study. Intern. J. Environ. Res. Public Health 2022, 19, 9027. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Cannabis- and Substance- Related Epidemiological Patterns of Chromosomal Congenital Anomalies in Europe: Geospatiotemporal and Causal Inferential Study. Int. J. Environ. Res. Public Health 2022, 19, 11208. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. State Trends of Cannabis Liberalization as a Causal Driver of Increasing Testicular Cancer Rates across the USA. Int. J. Environ. Res. Public Health 2022, 19, 12759. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Epidemiology of Δ8THC-Related Carcinogenesis in USA: A Panel Regression and Causal Inferential Study. Int. J. Environ. Res. Public Health 2022, 19, 7726. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Epidemiological association of cannabinoid- and drug- exposures and sociodemographic factors with limb reduction defects across USA 1989–2016: A geotemporospatial study. Spat. Spatio-Temporal Epidemiol. 2022, 41, 100480–100490. [Google Scholar] [CrossRef]
- Manthey, J.; Freeman, T.P.; Kilian, C.; Lopez-Pelayo, H.; Rehm, J. Public health monitoring of cannabis use in Europe: Prevalence of use, cannabis potency, and treatment rates. Lancet Reg. Health Eur. 2021, 10, 100227–200237. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Epigenomic and Other Evidence for Cannabis-Induced Aging Contextualized in a Synthetic Epidemiologic Overview of Cannabinoid-Related Teratogenesis and Cannabinoid-Related Carcinogenesis. Mendeley Data, 2022; manuscript submitted. [Google Scholar]
- Reece, A.S.; Norman, A.; Hulse, G.K. Cannabis exposure as an interactive cardiovascular risk factor and accelerant of organismal ageing: A longitudinal study. BMJ Open 2016, 6, e011891–e011901. [Google Scholar] [CrossRef] [Green Version]
- Reece, A.S.; Hulse, G.K. Extending the “Paracentral Dogma” of biology with the metabolome: Implications for understanding genomic-glycomic-metabolic-epigenomic synchronization. Engineering 2022, in press. [CrossRef]
- Reece, A.S.; Hulse, G.K. Cannabis, Cannabidiol, Cannabinoids and Multigenerational Policy. Engineering 2022, in press. [CrossRef]
- Huang, H.F.S.; Nahas, G.G.; Hembree, W.C. Effects of Marijuana Inhalation on Spermatogenesis of the Rat. In Marijuana in Medicine; Nahas, G.G., Sutin, K.M., Harvey, D.J., Agurell, S., Eds.; Human Press: Totowa, NJ, USA; New York, NY, USA, 1999; Volume 1, pp. 359–366. [Google Scholar]
- Zimmerman, A.M.; Zimmerman, S.; Raj, A.Y. Effects of Cannabinoids on Spermatogensis in Mice. In Marijuana and Medicine; Nahas, G.G., Sutin, K.M., Harvey, D.J., Agurell, S., Eds.; Human Press: Totowa, NJ, USA; New York, NY, USA, 1999; Volume 1, pp. 347–358. [Google Scholar]
- Morishima, A. Effects of cannabis and natural cannabinoids on chromosomes and ova. NIDA Res. Monogr. 1984, 44, 25–45. [Google Scholar] [PubMed]
- Rossato, M.; Ion Popa, F.; Ferigo, M.; Clari, G.; Foresta, C. Human sperm express cannabinoid receptor Cb1, the activation of which inhibits motility, acrosome reaction, and mitochondrial function. J. Clin. Endocrinol. Metab. 2005, 90, 984–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossato, M.; Pagano, C.; Vettor, R. The cannabinoid system and male reproductive functions. J. Neuroendocrinol. 2008, 20 (Suppl. 1), 90–93. [Google Scholar] [CrossRef] [PubMed]
- Chioccarelli, T.; Cacciola, G.; Altucci, L.; Lewis, S.E.; Simon, L.; Ricci, G.; Ledent, C.; Meccariello, R.; Fasano, S.; Pierantoni, R.; et al. Cannabinoid receptor 1 influences chromatin remodeling in mouse spermatids by affecting content of transition protein 2 mRNA and histone displacement. Endocrinology 2010, 151, 5017–5029. [Google Scholar] [CrossRef]
- Russo, C.; Ferk, F.; Mišík, M.; Ropek, N.; Nersesyan, A.; Mejri, D.; Holzmann, K.; Lavorgna, M.; Isidori, M.; Knasmüller, S. Low doses of widely consumed cannabinoids (cannabidiol and cannabidivarin) cause DNA damage and chromosomal aberrations in human-derived cells. Arch. Toxicol. 2019, 93, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Stenchever, M.A.; Kunysz, T.J.; Allen, M.A. Chromosome breakage in users of marihuana. Am. J. Obstet. Gynecol. 1974, 118, 106–113. [Google Scholar] [CrossRef]
- Leuchtenberger, C.; Leuchtenberger, R. Morphological and cytochemical effects of marijuana cigarette smoke on epithelioid cells of lung explants from mice. Nature 1971, 234, 227–229. [Google Scholar] [CrossRef]
- McClintock, B. The Production of Homozygous Deficient Tissues with Mutant Characteristics by Means of the Aberrant Mitotic Behavior of Ring-Shaped Chromosomes. Genetics 1938, 23, 315–376. [Google Scholar] [CrossRef]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef]
- DiNieri, J.A.; Wang, X.; Szutorisz, H.; Spano, S.M.; Kaur, J.; Casaccia, P.; Dow-Edwards, D.; Hurd, Y.L. Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol. Psychiatry 2011, 70, 763–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szutorisz, H.; DiNieri, J.A.; Sweet, E.; Egervari, G.; Michaelides, M.; Carter, J.M.; Ren, Y.; Miller, M.L.; Blitzer, R.D.; Hurd, Y.L. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology 2014, 39, 1315–1323. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.T.; Szutorisz, H.; Garg, P.; Martin, Q.; Landry, J.A.; Sharp, A.J.; Hurd, Y.L. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated with Cross-Generational Effects of Adolescent THC Exposure. Neuropsychopharmacology 2015, 40, 2993–3005. [Google Scholar] [CrossRef] [Green Version]
- Szutorisz, H.; Hurd, Y.L. Epigenetic Effects of Cannabis Exposure. Biol. Psychiatry 2016, 79, 586–594. [Google Scholar] [CrossRef] [Green Version]
- Szutorisz, H.; Hurd, Y.L. High times for cannabis: Epigenetic imprint and its legacy on brain and behavior. Neurosci. Biobehav. Rev. 2018, 85, 93–101. [Google Scholar] [CrossRef]
- Ellis, R.J.; Bara, A.; Vargas, C.A.; Frick, A.L.; Loh, E.; Landry, J.; Uzamere, T.O.; Callens, J.E.; Martin, Q.; Rajarajan, P.; et al. Prenatal Δ(9)-Tetrahydrocannabinol Exposure in Males Leads to Motivational Disturbances Related to Striatal Epigenetic Dysregulation. Biol. Psychiatry 2021, 92, 127–138. [Google Scholar] [CrossRef]
- Murphy, S.K.; Itchon-Ramos, N.; Visco, Z.; Huang, Z.; Grenier, C.; Schrott, R.; Acharya, K.; Boudreau, M.H.; Price, T.M.; Raburn, D.J.; et al. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 2018, 13, 1208–1221. [Google Scholar] [CrossRef] [Green Version]
- Schrott, R.; Murphy, S.K.; Modliszewski, J.L.; King, D.E.; Hill, B.; Itchon-Ramos, N.; Raburn, D.; Price, T.; Levin, E.D.; Vandrey, R.; et al. Refraining from use diminishes cannabis-associated epigenetic changes in human sperm. Environ. Epigenetics 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Blevins, R.D.; Regan, J.D. delta-9-Tetrahydrocannabinol: Effect on macromolecular synthesis in human and other mammalian cells. Arch. Toxicol. 1976, 35, 127–135. [Google Scholar] [CrossRef] [PubMed]
- McClean, D.K.; Zimmerman, A.M. Action of delta 9-tetrahydrocannabinol on cell division and macromolecular synthesis in division-synchronized protozoa. Pharmacology 1976, 14, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Nahas, G.G.; Morishima, A.; Desoize, B. Effects of cannabinoids on macromolecular synthesis and replication of cultured lymphocytes. Fed. Proc. 1977, 36, 1748–1752. [Google Scholar] [PubMed]
- Mon, M.J.; Jansing, R.L.; Doggett, S.; Stein, J.L.; Stein, G.S. Influence of delta9-tetrahydrocannabinol on cell proliferation and macromolecular biosynthesis in human cells. Biochem. Pharmacol. 1978, 27, 1759–1765. [Google Scholar] [CrossRef]
- Mon, M.J.; Haas, A.E.; Stein, J.L.; Stein, G.S. Influence of psychoactive and nonpsychoactive cannabinoids on cell proliferation and macromolecular biosynthesis in human cells. Biochem. Pharmacol. 1981, 30, 31–43. [Google Scholar] [CrossRef]
- Mon, M.J.; Haas, A.E.; Stein, J.L.; Stein, G.S. Influence of psychoactive and nonpsychoactive cannabinoids on chromatin structure and function in human cells. Biochem. Pharmacol. 1981, 30, 45–58. [Google Scholar] [CrossRef]
- Yang, X.; Hegde, V.L.; Rao, R.; Zhang, J.; Nagarkatti, P.S.; Nagarkatti, M. Histone modifications are associated with Delta9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J. Biol. Chem. 2014, 289, 18707–18718. [Google Scholar] [CrossRef] [PubMed]
- Mehrnoush, V.; De Lima, S.G.; Kotb, A.; Hyndman, M.E. The association of bladder cancer and Cannabis: A systematic review. Arch. Ital. Urol. Androl. 2022, 94, 248–251. [Google Scholar] [CrossRef]
- Payne, K.S.; Mazur, D.J.; Hotaling, J.M.; Pastuszak, A.W. Cannabis and Male Fertility: A Systematic Review. J. Urol. 2019, 202, 674–681. [Google Scholar] [CrossRef] [Green Version]
- Busch, F.W.; Seid, D.A.; Wei, E.T. Mutagenic activity of marihuana smoke condensates. Cancer Lett. 1979, 6, 319–324. [Google Scholar] [CrossRef]
- Zimmerman, A.M.; Raj, A.Y. Influence of cannabinoids on somatic cells in vivo. Pharmacology 1980, 21, 277–287. [Google Scholar] [CrossRef]
- Tahir, S.K.; Zimmerman, A.M. Influence of marihuana on cellular structures and biochemical activities. Pharmacol. Biochem. Behav. 1991, 40, 617–623. [Google Scholar] [CrossRef]
- Vela, G.; Martín, S.; García-Gil, L.; Crespo, J.A.; Ruiz-Gayo, M.; Fernández-Ruiz, J.J.; Garcia-Lecumberri, C.; Pelaprat, D.; Fuentes, J.A.; Ramos, J.A.; et al. Maternal exposure to delta9-tetrahydrocannabinol facilitates morphine self-administration behavior and changes regional binding to central mu opioid receptors in adult offspring female rats. Brain Res. 1998, 807, 101–109. [Google Scholar] [CrossRef]
- Koller, V.J.; Auwarter, V.; Grummt, T.; Moosmann, B.; Misik, M.; Knasmuller, S. Investigation of the in vitro toxicological properties of the synthetic cannabimimetic drug CP-47,497-C8. Toxicol. Appl. Pharmacol. 2014, 277, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Koller, V.J.; Ferk, F.; Al-Serori, H.; Mišík, M.; Nersesyan, A.; Auwärter, V.; Grummt, T.; Knasmuller, S. Genotoxic properties of representatives of alkylindazoles and aminoalkyl-indoles which are consumed as synthetic cannabinoids. Food Chem. Toxicol. 2015, 80, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Price, P.J.; Suk, W.A.; Spahn, G.J.; Freeman, A.E. Transformation of Fischer rat embryo cells by the combined action of murine leukemia virus and (-)-trans- 9 -tetrahydrocannabinol. Proc. Soc. Exp. Biol. Med. 1972, 140, 454–456. [Google Scholar] [CrossRef]
- Hölzel, B.N.; Pfannkuche, K.; Allner, B.; Allner, H.T.; Hescheler, J.; Derichsweiler, D.; Hollert, H.; Schiwy, A.; Brendt, J.; Schaffeld, M.; et al. Following the adverse outcome pathway from micronucleus to cancer using H2B-eGFP transgenic healthy stem cells. Arch. Toxicol. 2020, 94, 3265–3280. [Google Scholar] [CrossRef] [PubMed]
- Tahir, S.K.; Trogadis, J.E.; Stevens, J.K.; Zimmerman, A.M. Cytoskeletal organization following cannabinoid treatment in undifferentiated and differentiated PC12 cells. Biochem. Cell Biol. 1992, 70, 1159–1173. [Google Scholar] [CrossRef]
- Sarafian, T.A.; Kouyoumjian, S.; Khoshaghideh, F.; Tashkin, D.P.; Roth, M.D. Delta 9-tetrahydrocannabinol disrupts mitochondrial function and cell energetics. Am. J. Physiol. 2003, 284, L298–L306. [Google Scholar]
- Sarafian, T.A.; Habib, N.; Oldham, M.; Seeram, N.; Lee, R.P.; Lin, L.; Tashkin, D.P.; Roth, M.D. Inhaled marijuana smoke disrupts mitochondrial energetics in pulmonary epithelial cells in vivo. Am. J. Physiol. 2006, 290, L1202–L1209. [Google Scholar] [CrossRef]
- Morimoto, S.; Tanaka, Y.; Sasaki, K.; Tanaka, H.; Fukamizu, T.; Shoyama, Y.; Shoyama, Y.; Taura, F. Identification and characterization of cannabinoids that induce cell death through mitochondrial permeability transition in Cannabis leaf cells. J. Biol. Chem. 2007, 282, 20739–20751. [Google Scholar] [CrossRef] [Green Version]
- Fisar, Z.; Singh, N.; Hroudova, J. Cannabinoid-induced changes in respiration of brain mitochondria. Toxicol. Lett. 2014, 231, 62–71. [Google Scholar] [CrossRef]
- Singh, N.; Hroudova, J.; Fisar, Z. Cannabinoid-Induced Changes in the Activity of Electron Transport Chain Complexes of Brain Mitochondria. J. Mol. Neurosci. 2015, 56, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Gant, J. Scientists are baffled by spatter of babies born without hands or arms in France, as investigation fails to discover a cause. In Daily Mail; Daily Mail: London, UK, 2019. [Google Scholar]
- Agence France-Presse in Paris. France-Presse in Paris. France to investigate cause of upper limb defects in babies. In The Guardian; The Guardian: London, UK, 2018. [Google Scholar]
- Willsher, K. Baby arm defects prompt nationwide investigation in France. In Guardian; The Guardian: London, UK, 2018. [Google Scholar]
- Babies Born with Deformed Hands Spark Investigation in Germany. Available online: https://edition.cnn.com/2019/09/16/health/hand-deformities-babies-gelsenkirchen-germany-intl-scli-grm/index.html (accessed on 18 September 2022).
- VACTERL Association. Available online: https://www.gosh.nhs.uk/conditions-and-treatments/conditions-we-treat/vacterl-association-0/ (accessed on 1 January 2022).
- Reece, A.S.; Hulse, G.K. Epidemiological Patterns of Cannabis- and Substance- Related General Congenital Anomalies Across Europe 2010–2019: Space-Time and Causal Inference Study. 2022; manuscript submitted. [Google Scholar]
- Eurocat Data: Prevalence Charts and Tables. Available online: https://eu-rd-platform.jrc.ec.europa.eu/eurocat/eurocat-data/prevalence_en (accessed on 1 January 2022).
- Global Health Observatory. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/total-(recorded-unrecorded)-alcohol-per-capita-(15-)-consumption (accessed on 1 October 2021).
- European Monitoring Centre for Drugs and Drug Addiction (EMCDDA). Statistical Bulletin 2021—Prevalence of Drug Use. Available online: https://www.emcdda.europa.eu/data/stats2021/gps_en (accessed on 1 October 2021).
- The World Bank. Crude Data: Adjusted Net National Income per Capita (Current US$). Available online: https://data.worldbank.org/indicator/NY.ADJ.NNTY.PC.CD (accessed on 1 October 2021).
- R: A Language and Environment for Statistical Computing. Available online: https://cran.r-project.org/ (accessed on 1 October 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; Francios, R.; Groelmund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686–1691. [Google Scholar] [CrossRef] [Green Version]
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Viridis: Default Color Maps from ‘matplotlib’. Available online: https://CRAN.R-project.org/package=viridis (accessed on 1 October 2021).
- Colorplaner: Ggplot2 Extension to Visualize Two Variables Per Color Aesthetic Through Colorspace Projection. Available online: https://github.com/wmurphyrd/colorplaner (accessed on 1 October 2021).
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Team, R.C. Linear and Nonlinear Mixed Effects Models. R Package Version 2020, 3, 1–89. [Google Scholar]
- Broom.Mixed: Tidying Methods for Mixed Models. Available online: http://github.com/bbolker/broom.mixed (accessed on 1 October 2021).
- Broom: Convert Statistical Objects into Tidy Tibbles. Available online: https://CRAN.R-project.org/package=broom (accessed on 1 October 2021).
- Leeper, T.J. Margins: Marginal Effects for Model Objects; Leeper, T.J., Ed.; R package version 0.3.26; Central R Archive Network: Houston, TX, USA, 2021; Volume 1, pp. 1–36. [Google Scholar]
- Wright, M.N.; Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Greenwell, B.M.; Boehmke, B.C. Variable Importance Plots—An Introduction to the vip Package. R J. 2021, 12, 343–366. [Google Scholar] [CrossRef]
- Package ‘plm’. Available online: https://cran.r-project.org/web/packages/plm/plm.pdf (accessed on 1 October 2021).
- Bivand, R.; Anselin, L.; Berke, O.; Bernat, A.; Carvalho, M.; Chun, Y.; Dormann, C.; Dray, S.; Halbersma, R.; Lewis-Koh, N.; et al. The spdep Package; CRAN (Central R-Archive Network): Trieste, Italy, 2007; pp. 1–143. [Google Scholar]
- Millo, G.; Piras, G. splm: Spatial Panel Data Models in R. J. Stat. Softw. 2012, 47, 1–38. [Google Scholar] [CrossRef]
- Millo, G.; Piras, G. Package ‘splm’; CRAN (Central R-Archive Network): Trieste, Italy, 2018; pp. 1–27. Available online: https://cran.r-project.org/web/packages/splm/splm.pdf (accessed on 1 October 2021).
- Croissant, Y.; Millo, G. Panel Data Econometrics with R; John Wiley and Sons: Oxford, UK, 2019. [Google Scholar]
- Wal, W.; Geskus, R. ipw: An R Package for Inverse Probability Weighting. J. Stat. Softw. 2011, 43, 1–23. [Google Scholar] [CrossRef] [Green Version]
- VanderWeele, T.J.; Ding, P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef] [Green Version]
- VanderWeele, T.J.; Martin, J.N.; Mathur, M.B. E-values and incidence density sampling. Epidemiology 2020, 31, e51–e52. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Mathur, M.B. Commentary: Developing best-practice guidelines for the reporting of E-values. Int. J. Epidemiol. 2020, 49, 1495–1497. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Ding, P.; Mathur, M. Technical Considerations in the Use of the E-Value. J. Causal Inference 2019, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pearl, J.; Mackaenzie, D. The Book of Why. The New Science of Cause and Effect; Basic Books: New York, NY, USA, 2019; Volume 1. [Google Scholar]
- Package ‘EValue’. Available online: https://cran.r-project.org/web/packages/EValue/EValue.pdf (accessed on 1 October 2021).
- Hill, A.B. The Environment and Disease: Association or Causation? Proc. R Soc. Med. 1965, 58, 295–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reece, A.S.; Hulse, G.K. Effect of Cannabis Legalization on US Autism Incidence and Medium Term Projections. Clin. Pediatrics Open Access 2019, 4, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Reece, A.S. Rapid Response: Known Cannabis Teratogenicity Needs to be Carefully Considered. BMJ 2018, 362, k3357. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Impacts of cannabinoid epigenetics on human development: Reflections on Murphy et al. ‘cannabinoid exposure and altered DNA methylation in rat and human sperm’ epigenetics. Epigenetics 2019, 14, 1041–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fish, E.W.; Murdaugh, L.B.; Zhang, C.; Boschen, K.E.; Boa-Amponsem, O.; Mendoza-Romero, H.N.; Tarpley, M.; Chdid, L.; Mukhopadhyay, S.; Cole, G.J.; et al. Cannabinoids exacerbate alcohol teratogenesis by a CB1-hedgehog interaction. Sci. Rep. 2019, 9, 16057–16075. [Google Scholar] [CrossRef] [Green Version]
- Aguado, T.; Romero, E.; Monory, K.; Palazuelos, J.; Sendtner, M.; Marsicano, G.; Lutz, B.; Guzmán, M.; Galve-Roperh, I. The CB1 cannabinoid receptor mediates excitotoxicity-induced neural progenitor proliferation and neurogenesis. J. Biol. Chem. 2007, 282, 23892–23898. [Google Scholar] [CrossRef]
- Williams, E.J.; Walsh, F.S.; Doherty, P. The FGF receptor uses the endocannabinoid signaling system to couple to an axonal growth response. J. Cell Biol. 2003, 160, 481–486. [Google Scholar] [CrossRef] [Green Version]
- Birerdinc, A.; Jarrar, M.; Stotish, T.; Randhawa, M.; Baranova, A. Manipulating molecular switches in brown adipocytes and their precursors: A therapeutic potential. Prog. Lipid Res. 2013, 52, 51–61. [Google Scholar] [CrossRef]
- Richard, D.; Picard, F. Brown fat biology and thermogenesis. Front. Biosci. 2011, 16, 1233–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.R.; Yang, Y.; Ward, R.; Gao, L.; Liu, Y. Orexin receptors: Multi-functional therapeutic targets for sleeping disorders, eating disorders, drug addiction, cancers and other physiological disorders. Cell. Signal. 2013, 25, 2413–2423. [Google Scholar] [CrossRef] [PubMed]
- Fraher, D.; Ellis, M.K.; Morrison, S.; McGee, S.L.; Ward, A.C.; Walder, K.; Gibert, Y. Lipid Abundance in Zebrafish Embryos Is Regulated by Complementary Actions of the Endocannabinoid System and Retinoic Acid Pathway. Endocrinology 2015, 156, 3596–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kučukalić, S.; Ferić Bojić, E.; Babić, R.; Avdibegović, E.; Babić, D.; Agani, F.; Jakovljević, M.; Kučukalić, A.; Bravo Mehmedbašić, A.; Šabić Džananović, E.; et al. Genetic susceptibility to posttraumatic stress disorder: Analyses of the oxytocin receptor, retinoic acid receptor-related orphan receptor A and cannabinoid receptor 1 genes. Psychiatr. Danub. 2019, 31, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, W.I. Retinoic acids and hepatic stellate cells in liver disease. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. 2), 75–79. [Google Scholar] [CrossRef]
- Ungricht, R.; Guibbal, L.; Lasbennes, M.C.; Orsini, V.; Beibel, M.; Waldt, A.; Cuttat, R.; Carbone, W.; Basler, A.; Roma, G.; et al. Genome-wide screening in human kidney organoids identifies developmental and disease-related aspects of nephrogenesis. Cell Stem Cell 2022, 29, 160–175.e167. [Google Scholar] [CrossRef]
- Carlson, B.M. Human Embryology and Developmental Biology; Elsevier: Philadelphia, PA, USA, 2019. [Google Scholar]

| No. | E-Value Estimate | Lower Bound E-Value |
|---|---|---|
| 1 | 3.41 × 10122 | 3.30 × 1081 |
| 2 | 1.18 × 10103 | 1.36 × 1031 |
| 3 | 1.45 × 1091 | 1.08 × 1030 |
| 4 | 5.23 × 1072 | 1.67 × 1026 |
| 5 | 6.01 × 1067 | 4.98 × 1023 |
| 6 | 6.23 × 1063 | 1.24 × 1017 |
| 7 | 3.34 × 1047 | 1.14 × 1010 |
| 8 | 5.81 × 1028 | 1.39 × 109 |
| 9 | 4.09 × 1018 | 1.95 × 108 |
| 10 | 5.96 × 1017 | 2.01 × 106 |
| 11 | 1.21 × 1017 | 1.22 × 105 |
| 12 | 2.18 × 1013 | 1.15 × 105 |
| 13 | 3.51 × 109 | 3.52 × 104 |
| 14 | 6.69 × 108 | 2.03 × 104 |
| 15 | 4.51 × 108 | 1.46 × 103 |
| 16 | 2.33 × 107 | 1258.11 |
| 17 | 6.89 × 106 | 303.56 |
| 18 | 2.74 × 105 | 285.07 |
| 19 | 1.61 × 105 | 119.49 |
| 20 | 1.23 × 105 | 83.68 |
| 21 | 8.150 × 104 | 83.66 |
| 22 | 6.62 × 104 | 56.63 |
| 23 | 2.73 × 104 | 43.52 |
| 24 | 6.62 × 103 | 43.51 |
| 25 | 3540.00 | 22.36 |
| 26 | 955.32 | 17.85 |
| 27 | 954.95 | 13.74 |
| 28 | 606.01 | 13.62 |
| 29 | 365.38 | 9.43 |
| 30 | 273.00 | 9.28 |
| 31 | 252.91 | 9.09 |
| 32 | 162.57 | 8.35 |
| 33 | 161.31 | 7.02 |
| 34 | 82.56 | 5.47 |
| 35 | 67.00 | 5.45 |
| 36 | 59.95 | 5.18 |
| 37 | 43.56 | 4.73 |
| 38 | 42.81 | 3.75 |
| 39 | 21.28 | 3.38 |
| 40 | 15.85 | 3.23 |
| 41 | 11.7 | 3.13 |
| 42 | 10.52 | 3.06 |
| 43 | 9.68 | 2.51 |
| 44 | 9.68 | 2.38 |
| 45 | 9.03 | 2.10 |
| 46 | 8.58 | 2.02 |
| 47 | 7.24 | 2.02 |
| 48 | 6.58 | 1.89 |
| 49 | 5.01 | 1.62 |
| 50 | 2.86 | 1.60 |
| 51 | 2.65 | 1.60 |
| Functional Annotation | Status | Page | Number Genes Identified | p-Value |
|---|---|---|---|---|
| Congenital Anomalies | ||||
| Congenital anomalies of kidney and urinary tract | Withdrawal | 351 | 9 | 0.003850 |
| Familial congenital anomalies of kidney and urinary tract | Withdrawal | 352 | 8 | 0.004400 |
| Urological diseases | Withdrawal | 357 | 1 | 0.007010 |
| Renal agenesis | Withdrawal | 311 | 5 | 0.000429 |
| Renal tubular morphology | Withdrawal | 313 | 8 | 0.000606 |
| Renal morphology & development | Withdrawal | 322 | 16 | 0.002160 |
| Morphology Urinary system | Withdrawal | 322 | 17 | 0.002200 |
| Renal lesions | Withdrawal | 322 | 48 | 0.002200 |
| Renal tubular abnormality | Withdrawal | 323 | 7 | 0.002550 |
| Urinary tract cancer | Withdrawal | 320 | 84 | 0.001890 |
| Male Cancers | ||||
| Male genital neoplasm | Withdrawal | 341 | 60 | 0.000675 |
| Male genital neoplasm | Dependence | 279 | 153 | 1.14 × 10−8 |
| Malignant male genital neoplasm | Dependence | 279 | 151 | 1.60 × 10−8 |
| Prostate cancer | Dependence | 279 | 148 | 2.88 × 10−8 |
| Prostate cancer | Withdrawal | 340 | 59 | 5.33 × 10−4 |
| Prostate cancer cell adhesion | Withdrawal | 357 | 1 | 0.007010 |
| Female Cancers | ||||
| Female genital neoplasm | Withdrawal | 349 | 78 | 0.003030 |
| Female genital neoplasm | Withdrawal | 350 | 79 | 0.003460 |
| Female genital adenocarcinoma | Dependence | 272 | 197 | 1.84 × 10−11 |
| Female genital carcinoma | Dependence | 273 | 215 | 2.05 × 10−11 |
| Female genital carcinoma | Dependence | 276 | 276 | 8.95 × 10−11 |
| Nearest Gene Name | Page | Functional Annotation | Number Genes Identified | p-Value |
|---|---|---|---|---|
| WNT3A | 239 | Head and neck cancer | 356 | 7.73 × 10−20 |
| WNT8B | 239 | Head and neck cancer | 356 | 7.73 × 10−20 |
| WNT3A | 239 | Head and neck cancer | 342 | 7.74 × 10−20 |
| WNT8B | 239 | Head and neck cancer | 342 | 7.74 × 10−20 |
| WNT3A | 240 | Head and neck cancer | 333 | 3.34 × 10−19 |
| WNT8B | 240 | Head and neck cancer | 333 | 3.34 × 10−19 |
| WNT3A | 240 | Abdominal carcinoma | 362 | 3.64 × 10−19 |
| WNT8B | 240 | Abdominal carcinoma | 362 | 3.64 × 10−19 |
| WNT3A | 241 | Solid cancer | 381 | 6.93 × 10−18 |
| WNT8B | 241 | Solid cancer | 381 | 6.93 × 10−18 |
| WNT3A | 241 | Thyroid cancer | 318 | 1.21 × 10−17 |
| WNT8B | 241 | Thyroid cancer | 318 | 1.21 × 10−17 |
| WNT3A | 242 | Thyroid cancer | 319 | 1.26 × 10−17 |
| WNT8B | 242 | Thyroid cancer | 319 | 1.26 × 10−17 |
| WNT3A | 242 | Endocrine tumour | 321 | 1.44 × 10−17 |
| WNT8B | 242 | Endocrine tumour | 321 | 1.44 × 10−17 |
| Nearest Gene Name | Page | Functional Annotation | Number Genes Identified | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|
| ROCK1 | 236 | Focal Adhesion | 39 | 0.000934 | 0.226633 |
| ROCK1 | 236 | Vascular Smooth Muscle | 25 | 0.001862 | 0.400987 |
| ROCK1 | 236 | cGMP-PKG signalling | 30 | 0.003895 | 0.658065 |
| ROCK1 | 237 | Actin Cytoskeleton | 37 | 0.004587 | 0.717564 |
| ROCK1 | 237 | Sphingolipid signalling | 24 | 0.005596 | 0.786305 |
| ROCK1 | 237 | Axon guidance | 24 | 0.011178 | 0.954555 |
| ROCK1 | 237 | Oxytocin signalling | 25 | 0.039031 | 1.000000 |
| ROCK1 | 237 | TGFβ signalling | 16 | 0.040247 | 1.000000 |
| ROCK1 | 237 | cAMP signalling | 31 | 0.044117 | 1.000000 |
| ROCK1 | 237 | Leukocyte transendothelial migration | 20 | 0.046734 | 1.000000 |
| ROCK1 | 238 | Cancer pathways | 54 | 0.067770 | 1.000000 |
| ROCK1 | 238 | Platelet activation | 21 | 0.077866 | 1.000000 |
| Nearest Gene Name | Page | Status | Functional Annotation | Number Genes Identified | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|
| TGFβ | ||||||
| TGFB1 | 50 | Dependence | Fibrosis | 2.53 × 10−6 | 0.011012 | |
| TGFB1 | 237 | Chronic myeloid leukaemia | 16 | 7.73 × 10−20 | 0.095040 | |
| TGFB1 | 237 | Colorectal cancer | 14 | 0.004626 | 0.988843 | |
| TGFB1 | 237 | Hippo signalling | 26 | 0.004047 | 0.998936 | |
| TGFB1 | 237 | MAPK pathway | 39 | 0.007516 | 1.000000 | |
| TGFB1 | 237 | TGFβ signalling | 16 | 0.040246 | 1.000000 | |
| TGFB1 | 238 | Cancer pathways | 54 | 0.067770 | 1.000000 | |
| TGFB1 | 238 | Amoebiasis | 18 | 0.072579 | 1.000000 | |
| Nearest Gene Name | Page | Status | Gene Number | Chromosome Number | p-Value | Bonferroni Adjusted p-Value |
| SMADs | ||||||
| SMAD2 | 6 | Dependence | ENSG00000175387 | 18 | 9.32 × 10−9 | 0.000632 |
| SMAD7 | 26 | Dependence | ENSG00000101665 | 18 | 6.18 × 10−7 | 0.005518 |
| SMAD2 | 53 | Dependence | ENSG00000175387 | 18 | 2.90 × 10−6 | 0.011688 |
| SMAD9 | 85 | Dependence | ENSG00000120693 | 13 | 7.47 × 10−6 | 0.018446 |
| SMAD6 | 125 | Withdrawal | ENSG00000137834 | 15 | 2.26 × 10−8 | 0.001125 |
| SMAD3 | 166 | Withdrawal | ENSG00000166949 | 15 | 2.96 × 10−6 | 0.012904 |
| SMAD7 | 169 | Withdrawal | ENSG00000101665 | 18 | 3.40 × 10−6 | 0.013798 |
| SMAD2 | 184 | Withdrawal | ENSG00000175387 | 18 | 5.65 × 10−6 | 0.017449 |
| SMAD3 | 209 | Withdrawal | ENSG00000166949 | 15 | 1.07 × 10−5 | 0.023594 |
| SMAD2 | 236 | ENSG00000175387 | 18 | 0.003911 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reece, A.S.; Hulse, G.K. Epidemiological Patterns of Cannabis- and Substance- Related Congenital Uronephrological Anomalies in Europe: Geospatiotemporal and Causal Inferential Study. Int. J. Environ. Res. Public Health 2022, 19, 13769. https://doi.org/10.3390/ijerph192113769
Reece AS, Hulse GK. Epidemiological Patterns of Cannabis- and Substance- Related Congenital Uronephrological Anomalies in Europe: Geospatiotemporal and Causal Inferential Study. International Journal of Environmental Research and Public Health. 2022; 19(21):13769. https://doi.org/10.3390/ijerph192113769
Chicago/Turabian StyleReece, Albert Stuart, and Gary Kenneth Hulse. 2022. "Epidemiological Patterns of Cannabis- and Substance- Related Congenital Uronephrological Anomalies in Europe: Geospatiotemporal and Causal Inferential Study" International Journal of Environmental Research and Public Health 19, no. 21: 13769. https://doi.org/10.3390/ijerph192113769
APA StyleReece, A. S., & Hulse, G. K. (2022). Epidemiological Patterns of Cannabis- and Substance- Related Congenital Uronephrological Anomalies in Europe: Geospatiotemporal and Causal Inferential Study. International Journal of Environmental Research and Public Health, 19(21), 13769. https://doi.org/10.3390/ijerph192113769







