Estimating the Relative Contribution of Environmental and Genetic Risk Factors to Different Aging Traits by Combining Correlated Variables into Weighted Risk Scores
Abstract
1. Introduction
2. Materials and Methods
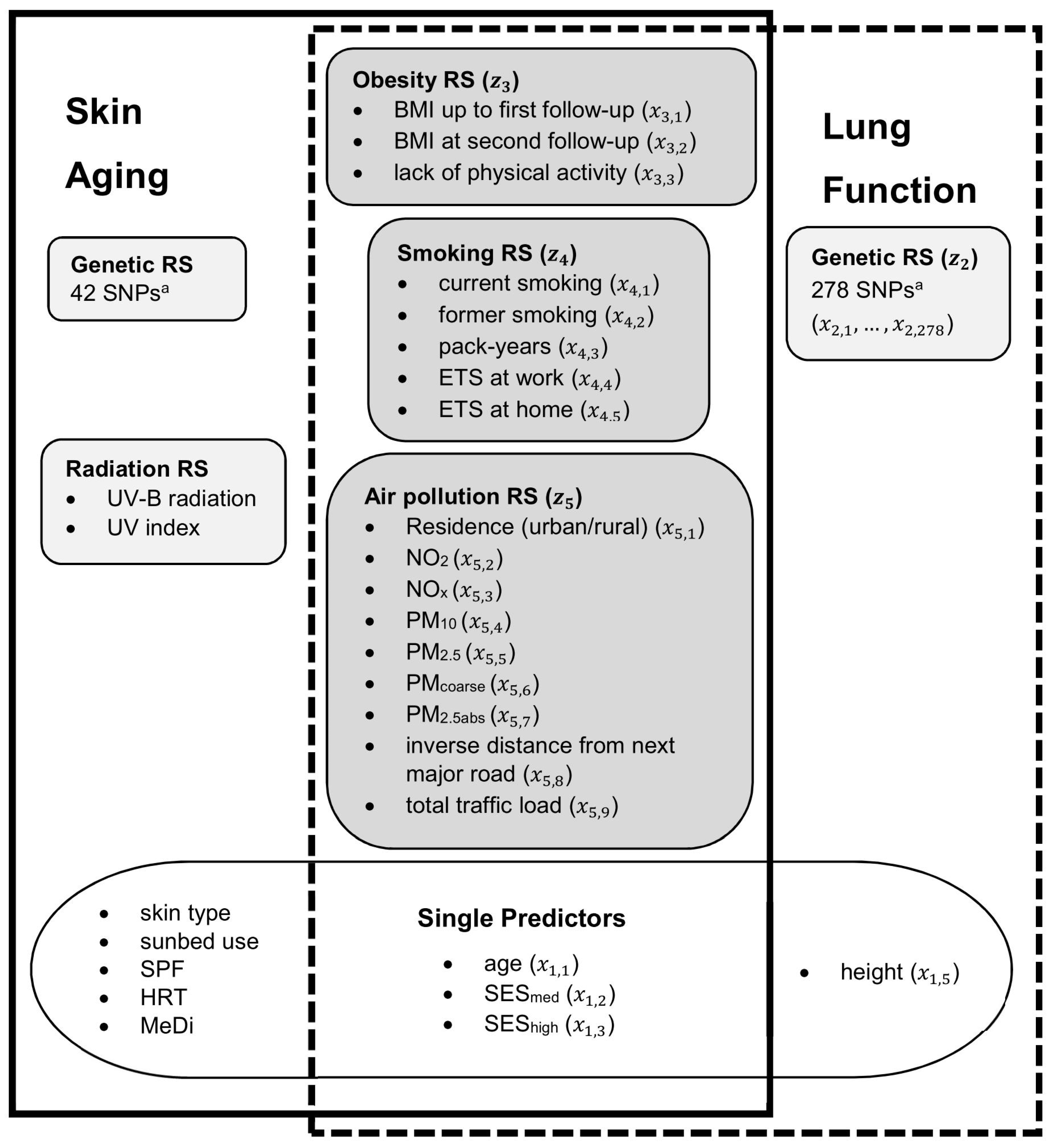
2.1. Choice of Predictors
2.2. Bootstrap Data Sets and Division into Training and Test Sample
2.3. Learning Risk Score Weights on Training Sample
2.4. Risk Scores, Linear Model and Relative Importance in the Test Sample
3. Results
3.1. Descriptive Analyses of the Outcomes and the Predictors
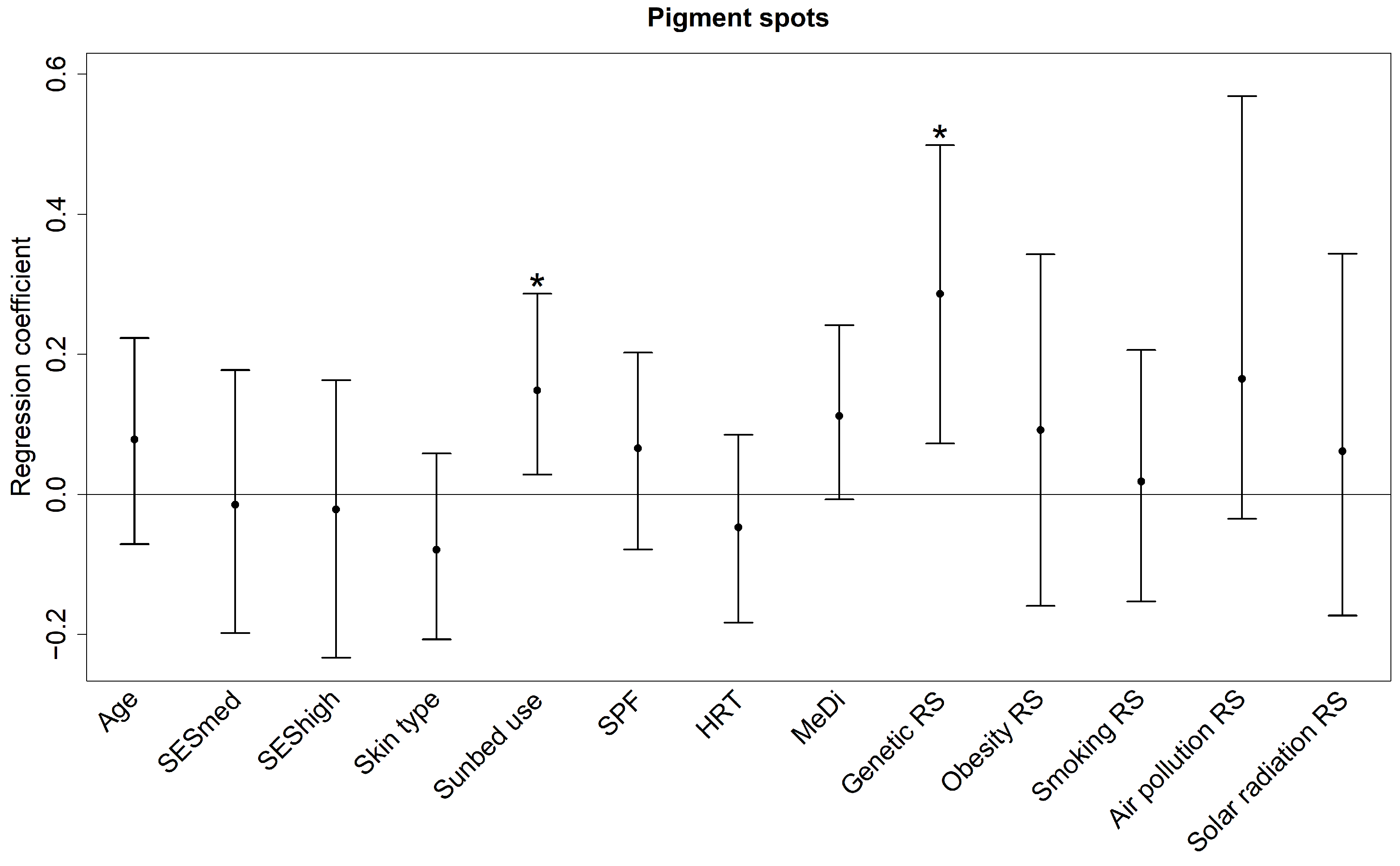
3.2. Skin Aging Outcome
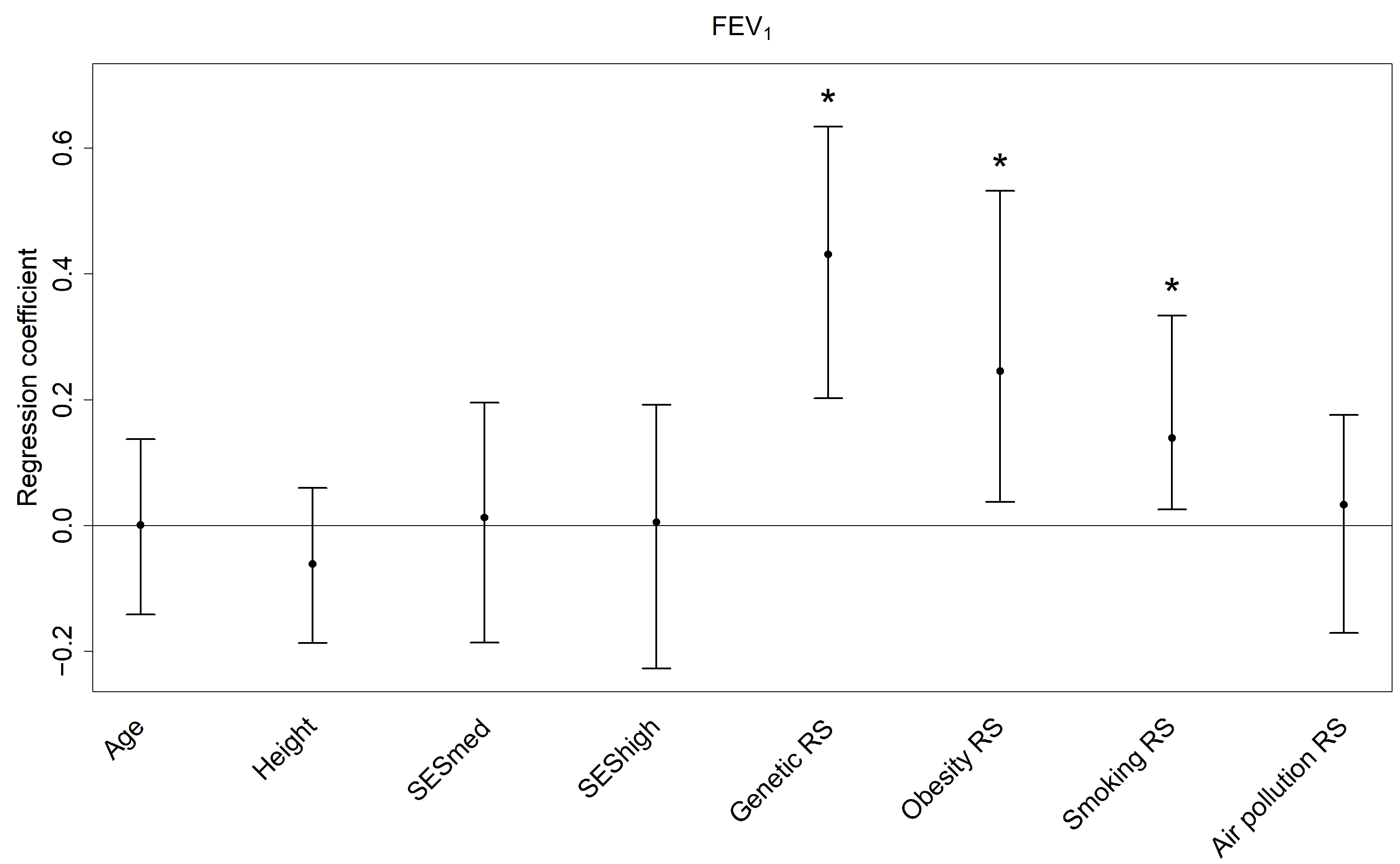
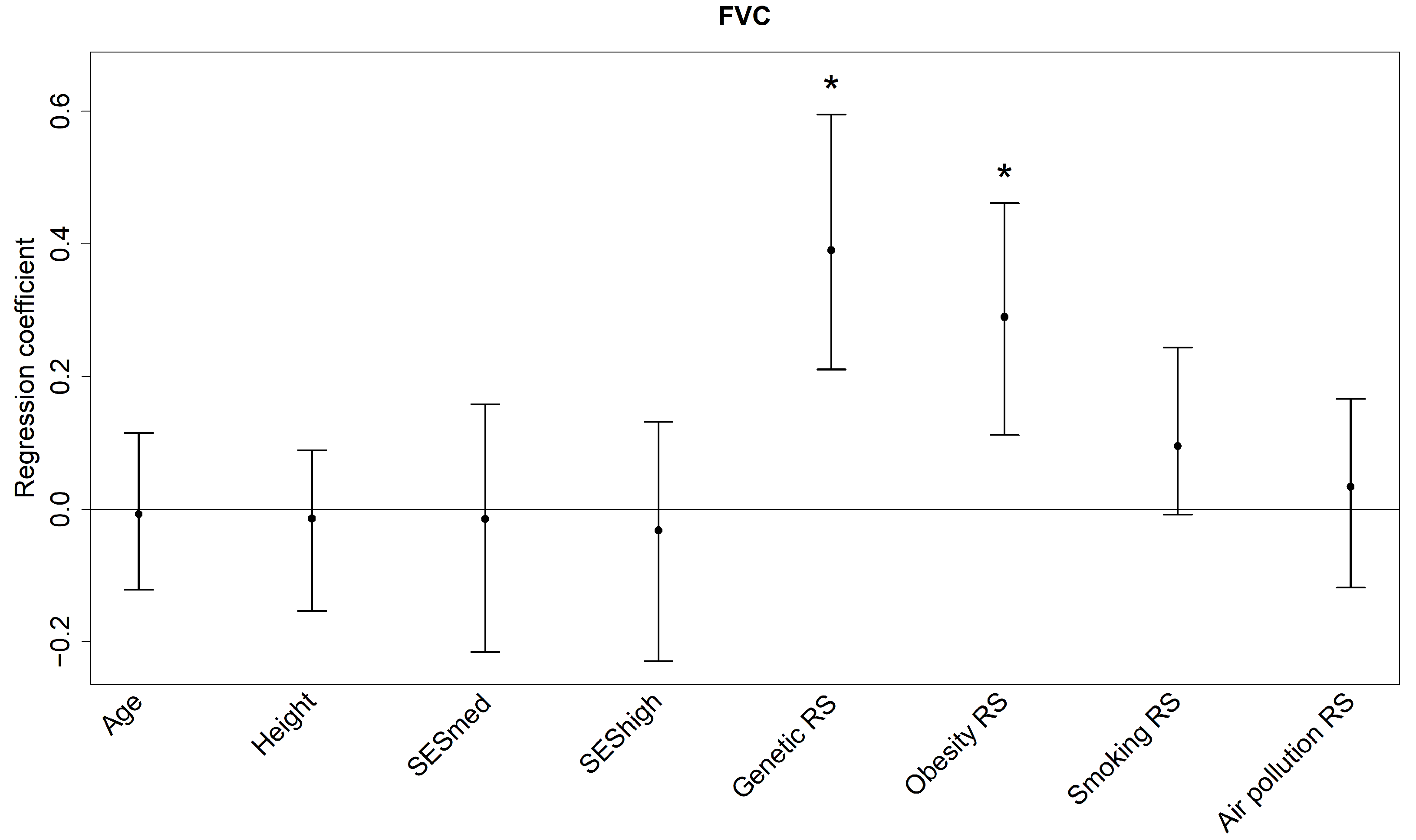
3.3. Lung Function Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wild, C.P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-N.; Hong, Y.-C. The exposome and the future of epidemiology: A vision and prospect. Environ. Health Toxicol. 2017, 32, e2017009. [Google Scholar] [CrossRef]
- Siroux, V.; Agier, L.; Slama, R. The exposome concept: A challenge and a potential driver for environmental health research. Eur. Respir. Rev. 2016, 25, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Guillien, A.; Cadiou, S.; Slama, R.; Siroux, V. The Exposome Approach to Decipher the Role of Multiple Environmental and Lifestyle Determinants in Asthma. Int. J. Environ. Res. Public Health 2021, 18, 1138. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.J.; Bhattacharya, J.; Butte, A.J. An Environment-Wide Association Study (EWAS) on Type 2 Diabetes Mellitus. PLoS ONE 2010, 5, e10746. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Sinisi, S.E.; van der Laan, M.J. Deletion/substitution/addition algorithm in learning with applications in genomics. Stat. Appl. Genet. Mol. Biol. 2004, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Carrico, C.; Gennings, C.; Wheeler, D.C.; Factor-Litvak, P. Characterization of Weighted Quantile Sum Regression for Highly Correlated Data in a Risk Analysis Setting. J. Agric. Biol. Environ. Stat. 2015, 20, 100–120. [Google Scholar] [CrossRef] [PubMed]
- Keil, A.P.; Buckley, J.P.; O’Brien, K.M.; Ferguson, K.K.; Zhao, S.; White, A.J. A Quantile-Based g-Computation Approach to Addressing the Effects of Exposure Mixtures. Environ. Health Perspect. 2020, 128, 047004. [Google Scholar] [CrossRef]
- Bobb, J.F.; Valeri, L.; Claus Henn, B.; Christiani, D.C.; Wright, R.O.; Mazumdar, M.; Godleski, J.J.; Coull, B.A. Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures. Biostatistics 2015, 16, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F. Power and Predictive Accuracy of Polygenic Risk Scores. PLoS Genet. 2013, 9, e1003348. [Google Scholar] [CrossRef]
- Forgetta, V.; Keller-Baruch, J.; Forest, M.; Durand, A.; Bhatnagar, S.; Kemp, J.P.; Nethander, M.; Evans, D.; Morris, J.A.; Kiel, D.P.; et al. Development of a polygenic risk score to improve screening for fracture risk: A genetic risk prediction study. PLoS Med. 2020, 17, e1003152. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.-H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Hüls, A.; Krämer, U.; Carlsten, C.; Schikowski, T.; Ickstadt, K.; Schwender, H. Comparison of weighting approaches for genetic risk scores in gene-environment interaction studies. BMC Genet. 2017, 18, 115. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-Y.; Lin, Y.-S.; Chan, C.-C.; Liu, Y.-L.; Tsai, S.-J.; Kuo, P.-H. Using Genetic Risk Score Approaches to Infer Whether an Environmental Factor Attenuates or Exacerbates the Adverse Influence of a Candidate Gene. Front. Genet. 2020, 11, 331. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Huang, C.-C.; Liu, Y.-L.; Tsai, S.-J.; Kuo, P.-H. Polygenic approaches to detect gene–environment interactions when external information is unavailable. Brief Bioinform. 2019, 20, 2236–2252. [Google Scholar] [CrossRef]
- Lindeman, R.H.; Merenda, P.F.; Gold, R.Z. Introduction to Bivariate and Multivariate Analysis; Scott Foresman & Co.: Glenview, IL, USA, 1980. [Google Scholar]
- Krutmann, J.; Schikowski, T.; Morita, A.; Berneburg, M. Environmentally-Induced (Extrinsic) Skin Aging: Exposomal Factors and Underlying Mechanisms. J. Invest. Dermatol. 2021, 141, 1096–1103. [Google Scholar] [CrossRef]
- Wheelock, C.E.; Rappaport, S.M. The role of gene–environment interactions in lung disease: The urgent need for the exposome. Eur. Respir. J. 2020, 55, 1902064. [Google Scholar] [CrossRef]
- Götschi, T.; Heinrich, J.; Sunyer, J.; Künzli, N. Long-term effects of ambient air pollution on lung function: A review. Epidemiology 2008, 19, 690–701. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. The Health Consequences of Smoking: A Report of the Surgeon General; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health: Atlanta, GA, USA, 2004.
- Schikowski, T.; Sugiri, D.; Ranft, U.; Gehring, U.; Heinrich, J.; Wichmann, H.E.; Krämer, U. Long-term air pollution exposure and living close to busy roads are associated with COPD in women. Respir. Res. 2005, 6, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Schikowski, T.; Sugiri, D.; Ranft, U.; Gehring, U.; Heinrich, J.; Wichmann, H.E.; Krämer, U. Does respiratory health contribute to the effects of long-term air pollution exposure on cardiovascular mortality? Respir. Res. 2007, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Schikowski, T.; Vossoughi, M.; Vierkötter, A.; Schulte, T.; Teichert, T.; Sugiri, D.; Fehsel, K.; Tzivian, L.; Bae, I.S.; Ranft, U.; et al. Association of air pollution with cognitive functions and its modification by APOE gene variants in elderly women. Environ. Res. 2015, 142, 10–16. [Google Scholar] [CrossRef]
- Miller, M.R.; Hankinson, J.; Brusasco, V.; Burgos, F.; Casaburi, R.; Coates, A.; Crapo, R.; Enright, P.; van der Grinten, C.P.; Gustafsson, P.; et al. Standardisation of spirometry. Eur. Respir. J. 2005, 26, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Quanjer, P.H.; Stanojevic, S.; Cole, T.J.; Baur, X.; Hall, G.L.; Culver, B.H.; Enright, P.L.; Hankinson, J.L.; Ip, M.S.M.; Zheng, J.; et al. Multi-ethnic reference values for spirometry for the 3–95-yr age range: The global lung function 2012 equations. Eur. Respir. J. 2012, 40, 1324–1343. [Google Scholar] [CrossRef] [PubMed]
- Tschachler, E.; Morizot, F. Ethnic Differences in Skin Aging. In Skin Aging; Krutmann, J., Gilchrest, B.A., Eds.; Springer GmbH: Berlin, Germany, 2006. [Google Scholar]
- Vierkötter, A.; Ranft, U.; Krämer, U.; Sugiri, D.; Reimann, V.; Krutmann, J. The SCINEXA: A novel, validated score to simultaneously assess and differentiate between intrinsic and extrinsic skin ageing. J. Dermatol. Sci. 2009, 53, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Forer, L.; Schönherr, S. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Endo, C.; Johnson, T.A.; Morino, R.; Nakazono, K.; Kamitsuji, S.; Akita, M.; Kawajiri, M.; Yamasaki, T.; Kami, A.; Hoshi, Y.; et al. Genome-wide association study in Japanese females identifies fifteen novel skin-related trait associations. Sci. Rep. 2018, 8, 8974. [Google Scholar] [CrossRef]
- Jacobs, L.C.; Hamer, M.A.; Gunn, D.A.; Deelen, J.; Lall, J.S.; van Heemst, D.; Uh, H.W.; Hofman, A.; Uitterlinden, A.G.; Griffiths, C.E.M.; et al. A Genome-Wide Association Study Identifies the Skin Color Genes IRF4, MC1R, ASIP, and BNC2 Influencing Facial Pigmented Spots. J. Invest. Dermatol. 2015, 135, 1735–1742. [Google Scholar] [CrossRef]
- Laville, V.; Clerc, S.L.; Ezzedine, K.; Jdid, R.; Taing, L.; Labib, T.; Coulonges, C.; Ulveling, D.; Carpentier, W.; Galan, P.; et al. A genome-wide association study in Caucasian women suggests the involvement of HLA genes in the severity of facial solar lentigines. Pigment Cell Melanoma Res. 2016, 29, 550–558. [Google Scholar] [CrossRef]
- Liu, F.; Hamer, M.A.; Deelen, J.; Lall, J.S.; Jacobs, L.; van Heemst, D.; Murray, P.G.; Wollstein, A.; de Craen, A.J.; Uh, H.W.; et al. The MC1R Gene and Youthful Looks. Curr. Biol. 2016, 26, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-G.; Leem, S.; Kim, B.; Kim, Y.; Lee, S.-G.; Song, H.J.; Seo, J.Y.; Park, S.G.; Won, H.-H.; Kang, N.G. GWAS Analysis of 17,019 Korean Women Identifies the Variants Associated with Facial Pigmented Spots. J. Invest. Dermatol. 2021, 141, 555–562. [Google Scholar] [CrossRef]
- Shrine, N.; Guyatt, A.L.; Erzurumluoglu, A.M.; Jackson, V.E.; Hobbs, B.D.; Melbourne, C.A.; Batini, C.; Fawcett, K.A.; Song, K.; Sakornsakolpat, P.; et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 2019, 51, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.; Schikowski, T.; Carsin, A.E.; Cai, Y.; Jacquemin, B.; Sanchez, M.; Vierkötter, A.; Marcon, A.; Keidel, D.; Sugiri, D.; et al. Adult lung function and long-term air pollution exposure. ESCAPE: A multicentre cohort study and meta-analysis. Eur. Respir. J. 2014, 45, 38–50. [Google Scholar] [CrossRef]
- Beelen, R.; Hoek, G.; Vienneau, D.; Eeftens, M.; Dimakopoulou, K.; Pedeli, X.; Tsai, M.-Y.; Künzli, N.; Schikowski, T.; Marcon, A.; et al. Development of NO2 and NOx land use regression models for estimating air pollution exposure in 36 study areas in Europe—The ESCAPE project. Atmos. Environ. 2013, 72, 10–23. [Google Scholar] [CrossRef]
- Eeftens, M.; Beelen, R.; de Hoogh, K.; Bellander, T.; Cesaroni, G.; Cirach, M.; Declercq, C.; Dedele, A.; Dons, E.; de Nazelle, A.; et al. Development of land use regression models for PM2.5, PM2.5 absorbance, PM10 and PMcoarse in 20 European study areas;results of the ESCAPE project. Environ. Sci. Technol. 2012, 46, 11195–11205. [Google Scholar] [CrossRef]
- Hüls, A.; Sugiri, D.; Fuks, K.; Krutmann, J.; Schikowski, T. Lentigine Formation in Caucasian Women—Interaction between Particulate Matter and Solar UVR. J. Invest. Dermatol. 2019, 139, 974–976. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Grömping, U. Relative Importance for Linear Regression in R: The Package relaimpo. J. Stat. Softw. 2006, 17, 1–27. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Vierkötter, A.; Schikowski, T.; Ranft, U.; Sugiri, D.; Matsui, M.; Krämer, U.; Krutmann, J. Airborne particle exposure and extrinsic skin aging. J. Invest. Dermatol. 2010, 130, 2719–2726. [Google Scholar] [CrossRef]
- Panagiotakos, D.B.; Pitsavos, C.; Arvaniti, F.; Stefanadis, C. Adherence to the Mediterranean food pattern predicts the prevalence of hypertension, hypercholesterolemia, diabetes and obesity, among healthy adults; the accuracy of the MedDietScore. Prev. Med. 2007, 44, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Schwender, H. “scrime”: Analysis of High-Dimensional Categorical Data Such as SNP Data. R Package Version 1.3.5. 2018. Available online: https://cran.r-project.org/web/packages/scrime/scrime.pdf (accessed on 8 November 2022).
- Wei, T.; Simko, V. R Package “corrplot”: Visualization of a Correlation Matrix. R Package Version 0.84. 2017. Available online: https://cran.r-project.org/web/packages/corrplot/corrplot.pdf (accessed on 8 November 2022).
| Variable | Categories | Mean (SD) or n (%) | |
|---|---|---|---|
| Skin aging analysis (n = 547) | Lung function analysis (n = 510) | ||
| Outcomes | |||
| Pigment spot z-score | 0.00 (1.00) | ||
| FEV1 z-score | 0.17 (1.02) | ||
| FVC z-score | 0.24 (0.90) | ||
| FEV1/FVC z-score | −0.22 (0.85) | ||
| Single Predictors | |||
| Age [years] | 73.58 (2.97) | 73.52 (2.99) | |
| Height [cm] | 162.83 (5.76) | ||
| SES: | 1 = “<10 years” | 94 (17.2%) | 89 (17.5%) |
| 2 = “10 years” | 275 (50.3%) | 249 (48.8%) | |
| 3 = “>10 years” | 178 (32.5%) | 172 (33.7%) | |
| Skin type: | dark (0; Fitzpatrick type 3 and 4) | 243 (44.4%) | |
| light (1; Fitzpatrick type 1 and 2) | 304 (55.6%) | ||
| Sunbed use: | never (0) | 459 (83.9%) | |
| ever (1) | 88 (16.1%) | ||
| SPF: | no (0) | 214 (39.1%) | |
| yes (1) | 333 (60.9%) | ||
| HRT: | no (0) | 319 (58.3%) | |
| yes (1) | 228 (41.7%) | ||
| MeDi score | 28.50 (2.79) | ||
| Obesity Risk Score | |||
| mean BMI up to FU 1 [kg/m2] | 26.70 (3.67) | 26.66 (3.66) | |
| BMI FU 2 [kg/m2] | 27.32 (4.34) | 27.29 (4.34) | |
| Lack of physical activity: | no (0) | 216 (39.5%) | 203 (39.8%) |
| yes (1) | 331 (60.5%) | 307 (60.2%) | |
| Smoking Risk Score | |||
| Current smoking: | no (0) | 535 (97.8%) | 498 (97.6%) |
| yes (1) | 12 (2.2%) | 12 (2.4%) | |
| Former smoking: | no (0) | 458 (83.7%) | 425 (83.3%) |
| yes (1) | 89 (16.3%) | 85 (16.7%) | |
| ETS at work: | never (0) | 314 (57.4%) | 294 (57.6%) |
| ever (1) | 233 (42.6%) | 216 (42.4%) | |
| ETS at home: | never (0) | 365 (66.7%) | 337 (66.1%) |
| ever (1) | 182 (33.3%) | 173 (33.9%) | |
| Packyears [packs/day × years] | 3.68 (12.46) | 3.83 (12.74) | |
| Air pollution Risk Score | |||
| Residence: | rural (0) | 260 (47.5%) | 249 (48.8%) |
| urban (1) | 287 (52.5%) | 261 (51.2%) | |
| NO2 [µg/m3] | 37.73 (11.46) | 37.21 (11.16) | |
| NOx [µg/m3] | 70.52 (32.66) | 69.17 (31.93) | |
| PM10 [µg/m3] | 49.27 (7.17) | 48.98 (7.38) | |
| PM2.5 [µg/m3] | 32.75 (4.65) | 32.56 (4.78) | |
| PMcoarse [µg/m3] | 17.57 (3.85) | 17.42 (3.91) | |
| PM2.5abs [10−5/m] | 2.74 (0.92) | 2.71 (0.92) | |
| Trafloadmajor [1000 vehicles × m/day] | 900.63 (2313.15) | 839.51 (2200.63) | |
| Invdistmajor [1/m] | 0.01 (0.02) | 0.01 (0.02) | |
| Radiation Risk Score | |||
| UV-B [J/m2] | 3140.38 (33.03) | ||
| UV index [40 W/m2] | 7.18 (0.09) | ||
| Predictor | Median | 95% Confidence Interval |
|---|---|---|
| Overall | 16.90% | 8.75%, 28.04% |
| Genetic RS | 4.15% | 0.25%, 11.88% |
| Sunbed use | 2.11% | 0.09%, 6.97% |
| Air pollution RS | 1.20% | 0.04%, 5.76% |
| MeDi | 1.14% | 0.04%, 5.11% |
| SES | 0.70% | 0.09%, 3.44% |
| Obesity RS | 0.66% | 0.04%, 4.59% |
| SPF | 0.59% | 0.04%, 3.33% |
| Skin type | 0.58% | 0.03%, 3.51% |
| Age | 0.52% | 0.04%, 3.37% |
| Solar radiation RS | 0.45% | 0.04%, 3.35% |
| HRT | 0.29% | 0.03%, 2.41% |
| Smoking RS | 0.29% | 0.02%, 2.74% |
| Predictor | Median | 95% Confidence Interval |
|---|---|---|
| Overall | 22.32% | 10.87%, 33.57% |
| Genetic RS | 10.99% | 2.92%, 23.29% |
| Smoking RS | 5.10% | 1.15%, 11.87% |
| Obesity RS | 2.41% | 0.17%, 7.62% |
| SES | 0.80% | 0.13%, 3.48% |
| Height | 0.45% | 0.02%, 2.92% |
| Air pollution RS | 0.36% | 0.02%, 2.69% |
| Age | 0.26% | 0.04%, 1.85% |
| Predictor | Median | 95% Confidence Interval |
|---|---|---|
| Overall | 23.36% | 10.86%, 33.67% |
| Genetic RS | 11.69% | 2.80%, 25.11% |
| Obesity RS | 5.62% | 1.21%, 12.50% |
| Smoking RS | 1.83% | 0.06%, 6.42% |
| SES | 0.90% | 0.13%, 3.71% |
| Air pollution RS | 0.42% | 0.03%, 3.40% |
| Age | 0.26% | 0.03%, 2.58% |
| Height | 0.24% | 0.02%, 1.96% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wigmann, C.; Hüls, A.; Krutmann, J.; Schikowski, T. Estimating the Relative Contribution of Environmental and Genetic Risk Factors to Different Aging Traits by Combining Correlated Variables into Weighted Risk Scores. Int. J. Environ. Res. Public Health 2022, 19, 16746. https://doi.org/10.3390/ijerph192416746
Wigmann C, Hüls A, Krutmann J, Schikowski T. Estimating the Relative Contribution of Environmental and Genetic Risk Factors to Different Aging Traits by Combining Correlated Variables into Weighted Risk Scores. International Journal of Environmental Research and Public Health. 2022; 19(24):16746. https://doi.org/10.3390/ijerph192416746
Chicago/Turabian StyleWigmann, Claudia, Anke Hüls, Jean Krutmann, and Tamara Schikowski. 2022. "Estimating the Relative Contribution of Environmental and Genetic Risk Factors to Different Aging Traits by Combining Correlated Variables into Weighted Risk Scores" International Journal of Environmental Research and Public Health 19, no. 24: 16746. https://doi.org/10.3390/ijerph192416746
APA StyleWigmann, C., Hüls, A., Krutmann, J., & Schikowski, T. (2022). Estimating the Relative Contribution of Environmental and Genetic Risk Factors to Different Aging Traits by Combining Correlated Variables into Weighted Risk Scores. International Journal of Environmental Research and Public Health, 19(24), 16746. https://doi.org/10.3390/ijerph192416746






