Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Pollen Collection
2.2. Pollen Detection by Morphological Optical Microscopy and DNA Metabarcoding
2.3. Statistical Analysis and Software for Brindisi and Lecce Samples
2.4. Back-Trajectory Cluster Analysis at the Brindisi and Lecce Monitoring Sites
3. Results and Discussion
3.1. Characterisation of Meteorological Parameters and Pollen Families in Brindisi
3.1.1. Pollen Grain Concentration Relationships with Meteorological Parameters in Brindisi
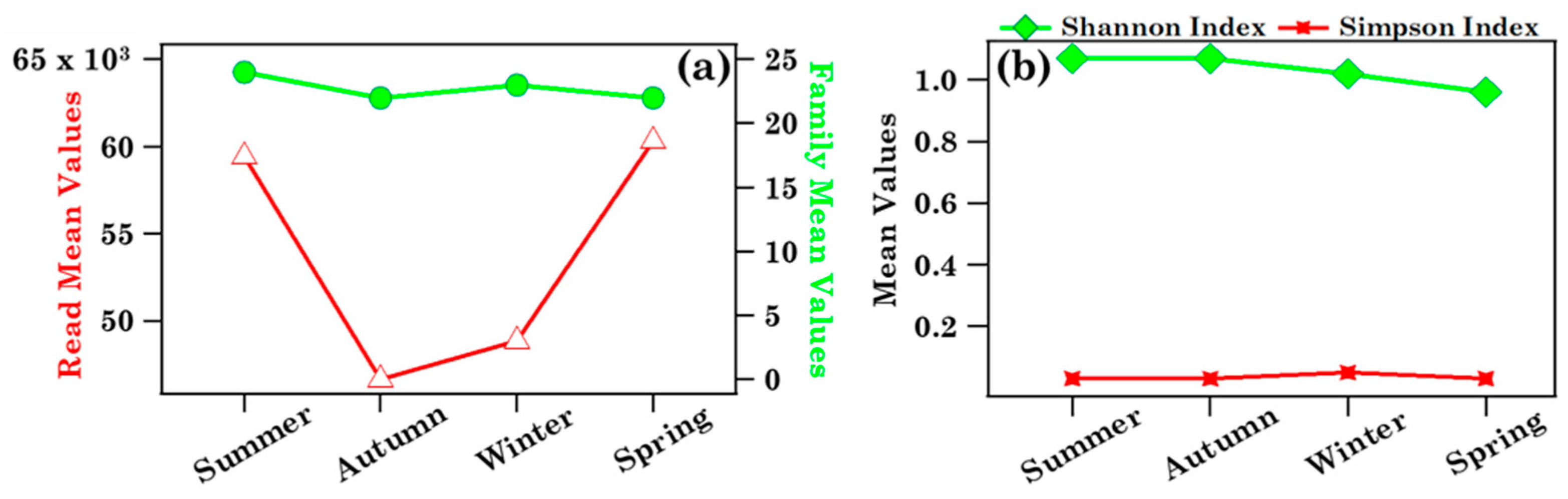
3.1.2. Biodiversity of Brindisi Samples and Analysis of Case Studies
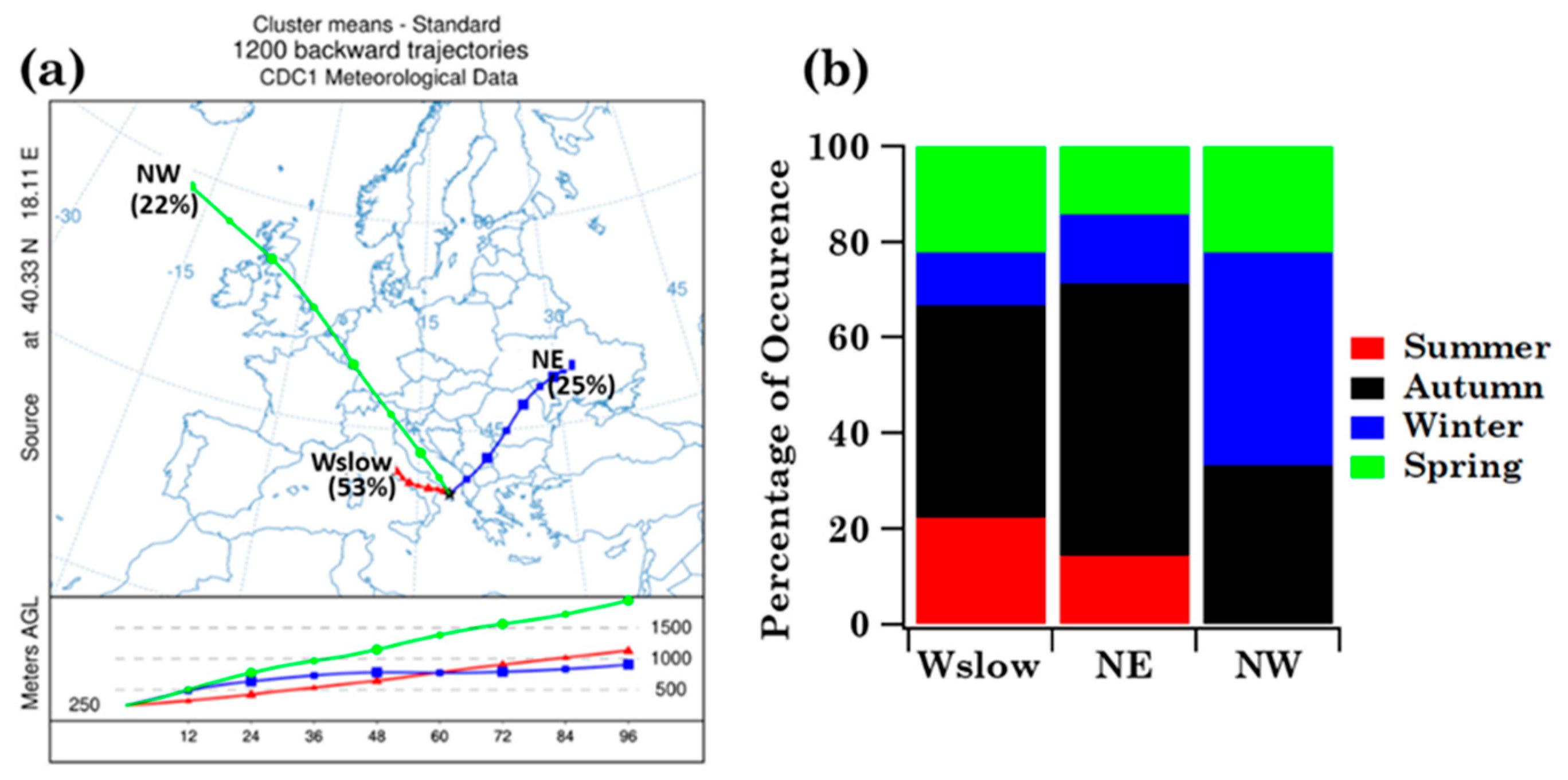
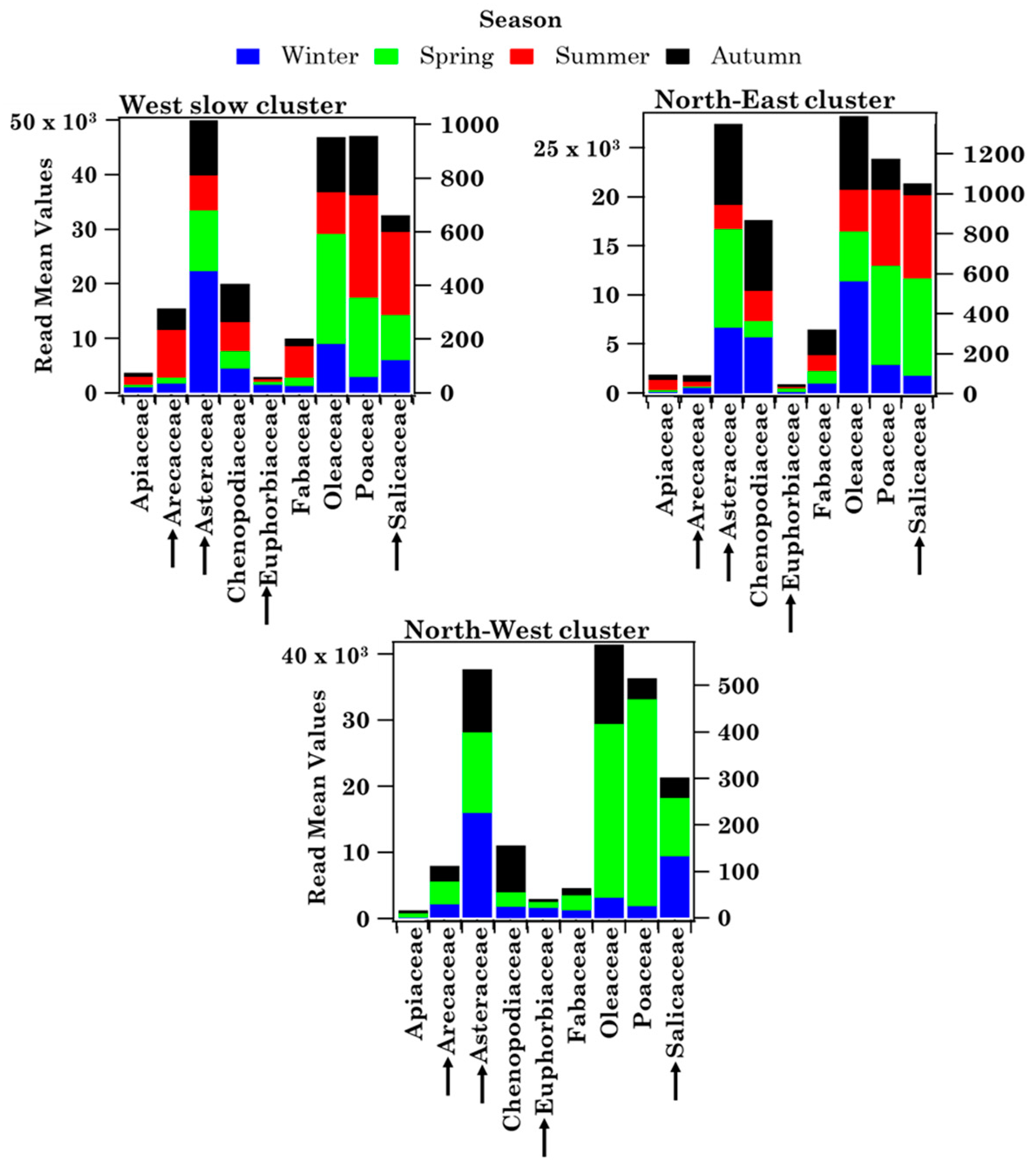
3.1.3. Impact of Advection Patterns on Pollen Grain Numbers and Families in Brindisi
3.2. Streptophyta Family Characterisation in Lecce by DNA-Metabarcoding Approach
3.2.1. Impact of Meteorological Parameters and Advection Patterns on the Pollen Families Detected in Lecce
3.2.2. Similarities and Dissimilarities between Lecce and Brindisi Samples
4. Summary and Conclusions
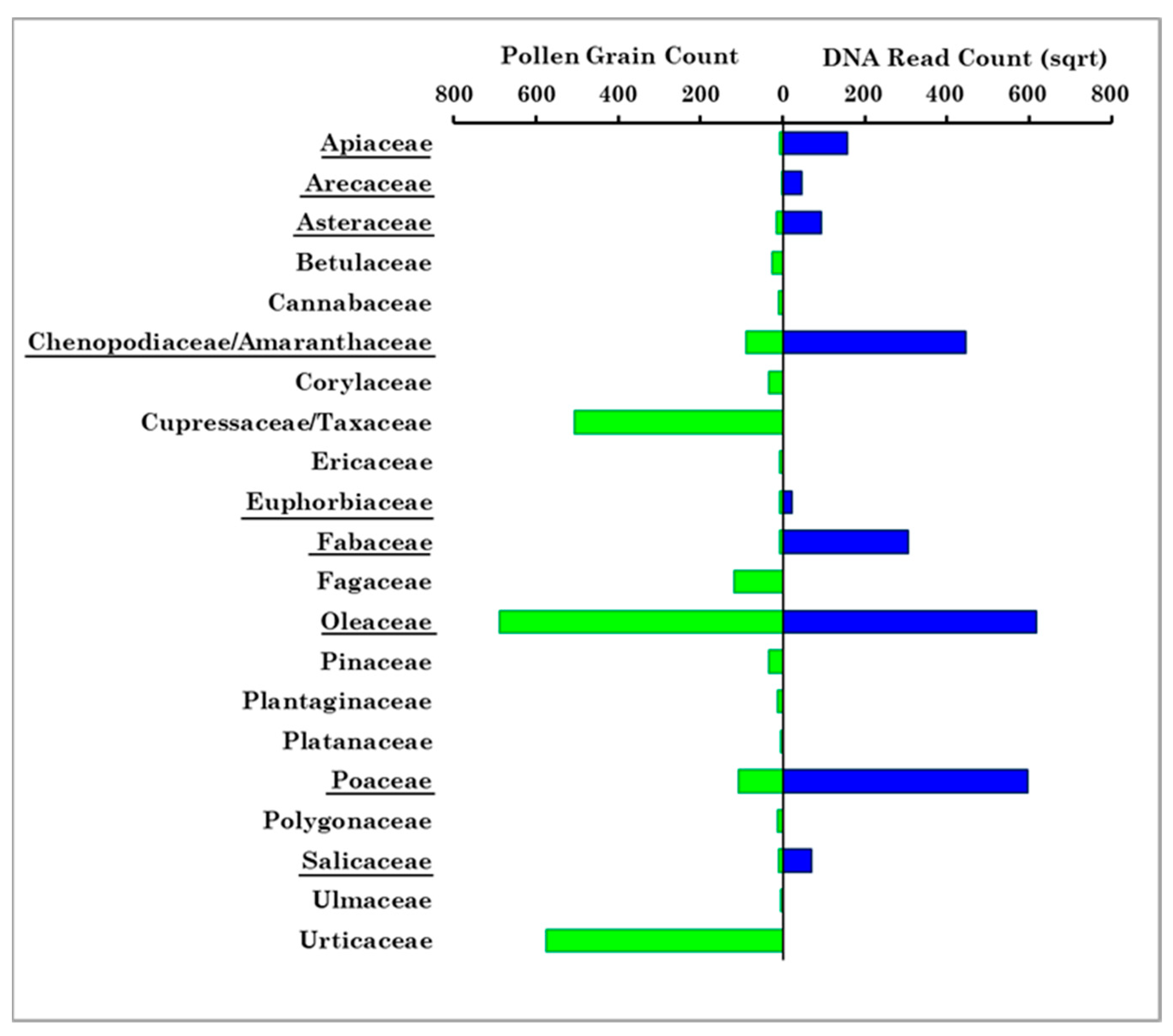
- Whole pollen grains, whose diameter ranges from about 10 to over 100 μm, are required for the pollen morphological detection by optical microscopy in Brindisi. Indeed, the used Hirst-type impactor allows collecting only pollen from a certain equivalent aerodynamic diameter, which cannot adapt to the induced air direction changes. Conversely, the PM10 sampler used in Lecce allows the collection of only airborne pollen grains and/or fragments with an aerodynamic diameter ≤10 μm. The deposition of whole pollen grains markedly decreases with increasing distance from the source because of their large size and, consequently, the long-range transport of pollen fragments ≤10 μm and/or cytoplasmic components is likely favoured over that of pollen grains. The residence time of pollen fragments and/or cytoplasmic contents in the atmosphere may also be longer than that of whole pollen grains. Hence, pollen families were detected in Brindisi mainly during their blooming period, while they were on average detected throughout the sampling time in Lecce. Note that the health impact of pollen fragments may be more dangerous than full-size pollen grains since the former can more easily penetrate the lower respiratory tract.
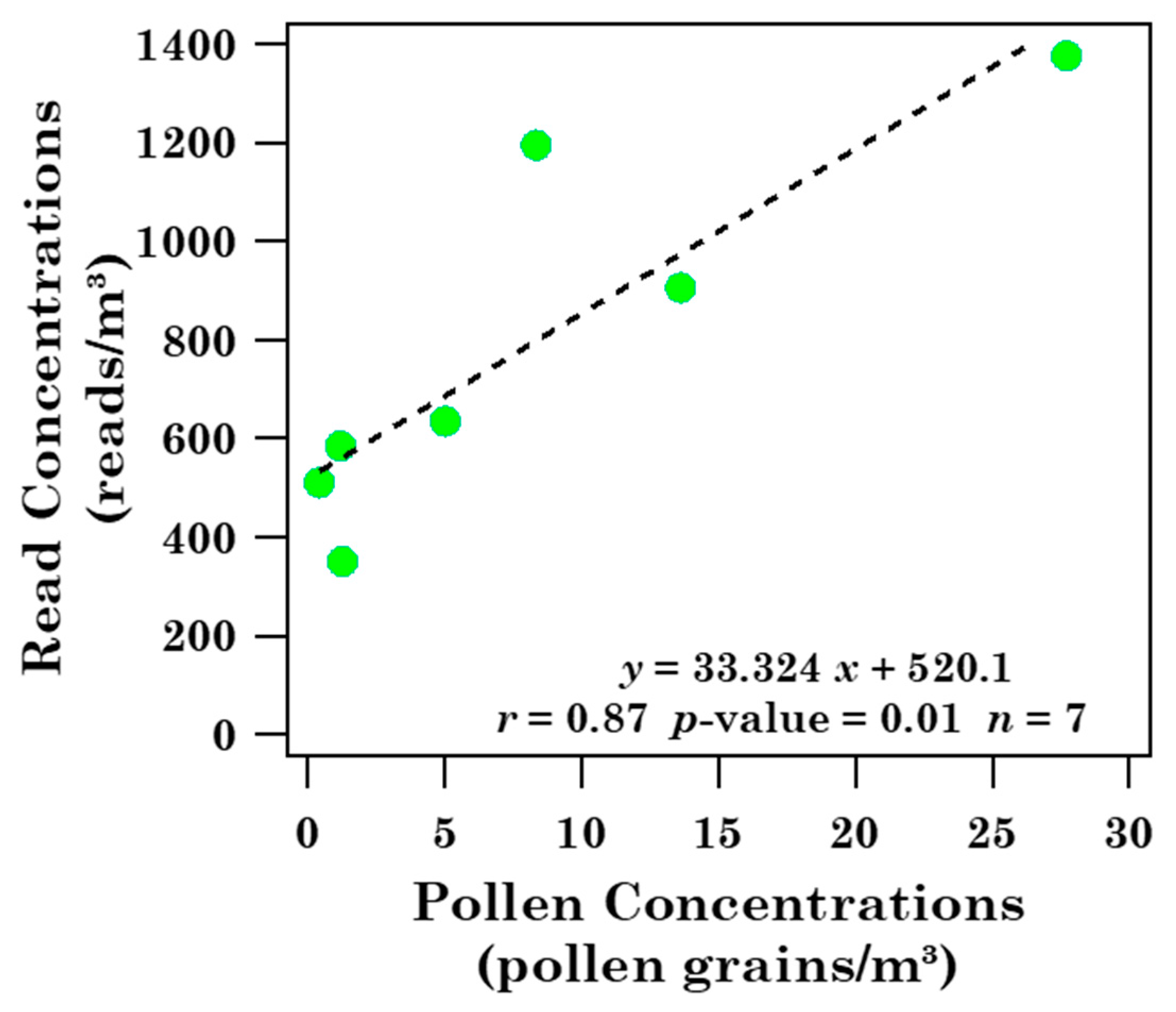
- Twenty-one pollen families were detected in Brindisi throughout the sampling period of this study and pollen grains were mainly correlated with temperature. In particular, no significant correlations with wind speed were observed, likely for the use of a Hirst-type trap to collect whole pollen grains and/or for the small number of samples collected during the one-year sampling. On the contrary, five out of the twenty-four pollen families detected in Lecce were positively correlated with WS.
- Few Urticaceae pollen grains were detected in Brindisi, only during heavy rainy days, also characterised by high RH values. Pollen grains may absorb moisture from the air and swell. Consequently, especially after heavy rain, swelling may cause the grains to rupture, preventing their identification by optical microscopy. No impact of heavy rain and/or high RH on the number of detected families and reads was observed in Lecce.
- Nine out of the twenty-four Strepthophyta pollen families detected in Lecce were in common with the twenty-one pollen families detected in Brindisi. The nine families were detected in all samples in Lecce, whereas they were detected mainly in spring in the samples collected in Brindisi.
- The four-day analytical back-trajectory analysis has shown that both sites were similarly affected by airflows, but their impact on the nine shared pollen family contributions varied differently with advection patterns at both sites.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martínez-Bracero, M.; Alcázar, P.; díaz de la Guardia, C.; González-Minero, F.J.; Ruiz, L.; Pérez, M.T.; Galán, C. Pollen calendars: A guide to common airborne pollen in Andalusia. Aerobiologia 2015, 31, 549–557. [Google Scholar] [CrossRef]
- Fernstrom, A.; Goldblatt, M. Aerobiology and Its Role in the Transmission of Infectious Diseases. J. Pathog. 2013, 2013, 493960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrmann, C.M.; Sugg, M.M.; Konrad, C.E. Airborne pollen characteristics and the influence of temperature and precipitation in Raleigh, North Carolina, USA (1999–2012). Aerobiologia 2016, 32, 683–696. [Google Scholar] [CrossRef]
- Tummon, F.; Arboledas, L.A.; Bonini, M.; Guinot, B.; Hicke, M.; Jacob, C.; Kendrovski, V.; McCairns, W.; Petermann, E.; Peuch, V.; et al. The need for Pan-European automatic pollen and fungal spore monitoring: A stakeholder workshop position paper. Clin. Transl. Allergy 2021, 11, e12015. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, G.; Lobefalo, G. Allergenic pollens in the southern Mediterranean area. J. Allergy Clin. Immunol. 1989, 83, 116–122. [Google Scholar] [CrossRef]
- D’Amato, G.; Cecchi, L.; Bonini, S.; Nunes, C.; Annesi-Maesano, I.; Behrendt, H.; Liccardi, G.; Popov, T.; Van Cauwenberge, P. Allergenic pollen and pollen allergy in Europe. Allergy 2007, 62, 976–990. [Google Scholar] [CrossRef]
- Gomes, C.; Ribeiro, H.; Abreu, I. Aerobiology of Cupressaceae in Porto city, Portugal. Aerobiologia 2018, 35, 97–103. [Google Scholar] [CrossRef]
- Clot, B.; Gilge, S.; Hajkova, L.; Magyar, D.; Scheifinger, H.; Sofiev, M.; Bütler, F.; Tummon, F. The EUMETNET AutoPollen programme: Establishing a prototype automatic pollen monitoring network in Europe. Aerobiologia 2020, 1–9. [Google Scholar] [CrossRef]
- Negral, L.; Moreno-Grau, S.; Galera, M.; Elvira-Rendueles, B.; Costa-Gómez, I.; Aznar, F.; Pérez-Badia, R.; Moreno, J. The effects of continentality, marine nature and the recirculation of air masses on pollen concentration: Olea in a Mediterranean coastal enclave. Sci. Total Environ. 2021, 790, 147999. [Google Scholar] [CrossRef]
- Pospiech, M.; Javůrková, Z.; Hrabec, P.; Štarha, P.; Ljasovská, S.; Bednář, J.; Tremlová, B. Identification of pollen taxa by different microscopy techniques. PLoS ONE 2021, 16, e0256808. [Google Scholar] [CrossRef]
- Calderon, C.; Ward, E.; Freeman, J.; McCartney, A. Detection of airborne fungal spores sampled by rotating-arm and Hirst-type spore traps using polymerase chain reaction assays. J. Aerosol Sci. 2002, 33, 283–296. [Google Scholar] [CrossRef]
- Swenson, S.J.; Gemeinholzer, B. Testing the effect of pollen exine rupture on metabarcoding with Illumina sequencing. PLoS ONE 2021, 16, e0245611. [Google Scholar] [CrossRef] [PubMed]
- Leontidou, K.; Vernesi, C.; De Groeve, J.; Cristofolini, F.; Vokou, D.; Cristofori, A. DNA metabarcoding of airborne pollen: New protocols for improved taxonomic identification of environmental samples. Aerobiologia 2017, 34, 63–74. [Google Scholar] [CrossRef]
- Gusareva, E.S.; Acerbi, E.; Lau, K.J.X.; Luhung, I.; Premkrishnan, B.N.V.; Kolundžija, S.; Purbojati, R.W.; Wong, A.; Houghton, J.N.I.; Miller, D.; et al. Microbial communities in the tropical air ecosystem follow a precise diel cycle. Proc. Natl. Acad. Sci. USA 2019, 116, 23299–23308. [Google Scholar] [CrossRef] [Green Version]
- Banchi, E.; Ametrano, C.G.; Tordoni, E.; Stanković, D.; Ongaro, S.; Tretiach, M.; Pallavicini, A.; Muggia, L.; Verardo, P.; Tassan, F.; et al. Environmental DNA assessment of airborne plant and fungal seasonal diversity. Sci. Total Environ. 2020, 738, 140249. [Google Scholar] [CrossRef]
- Fragola, M.; Perrone, M.; Alifano, P.; Talà, A.; Romano, S. Seasonal Variability of the Airborne Eukaryotic Community Structure at a Coastal Site of the Central Mediterranean. Toxins 2021, 13, 518. [Google Scholar] [CrossRef]
- Burton, N.C.; Grinshpun, S.A.; Reponen, T. Physical Collection Efficiency of Filter Materials for Bacteria and Viruses. Ann. Occup. Hyg. 2006, 51, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Mykytczuk, N.C.S.; Wilhelm, R.C.; Whyte, L.G. Planococcus halocryophilus sp. nov., an extreme sub-zero species from high Arctic permafrost. Int. J. Syst. Evol. Microbiol. 2012, 62, 1937–1944. [Google Scholar] [CrossRef] [Green Version]
- Romano, S.; Di Salvo, M.; Rispoli, G.; Alifano, P.; Perrone, M.R.; Talà, A. Airborne bacteria in the Central Mediterranean: Structure and role of meteorology and air mass transport. Sci. Total Environ. 2019, 697, 134020. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Haegeman, B.; Hamelin, J.; Moriarty, J.; Neal, P.; Dushoff, J.; Weitz, J.S. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013, 7, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Chernov, T.I.; Tkhakakhova, A.K.; Kutovaya, O.V. Assessment of diversity indices for the characterization of the soil prokaryotic community by metagenomic analysis. Eurasian Soil Sci. 2015, 48, 410–415. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; De León, A.V.-P.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [Green Version]
- Krebs, C.J. Species diversity measures. In Ecological Methodology; Krebs, C.J., Ed.; University of British Columbia: Vancouver, BC, Canada, 2014; pp. 532–593. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Bailey, D.T. Meteorological Monitoring Guidance for Regulatory Modelling Applications; Environmental Protection Agency Rep. EPA-454/R-99-005; DIANE Publishing: Collingdale, PA, USA, 2000; p. 168. Available online: http://www.epa.gov/scram001/guidance/met/mmgrma.pdf (accessed on 21 February 2022).
- D’Amato, G.; Liccardi, G.; Frenguelli, G. Thunderstorm-asthma and pollen allergy. Allergy 2007, 62, 11–16. [Google Scholar] [CrossRef]
- Rezanejad, F. Air pollution effects on structure, proteins and flavonoids in pollen grains of Thuja orientalis L. (Cupressaceae). Grana 2009, 48, 205–213. [Google Scholar] [CrossRef]
- Shahali, Y.; Pourpak, Z.; Moin, M.; Zare, A.; Majd, A. Impacts of air pollution exposure on the allergenic properties of Arizona cypress pollens. J. Phys. Conf. Ser. 2009, 151, 012027. [Google Scholar] [CrossRef] [Green Version]
- Magurran, A.E. An index of diversity. In Measuring Biological Diversity; Blackwell Science: Oxford, UK, 2004; Chapter 4. [Google Scholar]
- Pace, L.; Boccacci, L.; Casilli, M.; Di Carlo, P.; Fattorini, S. Correlations between weather conditions and airborne pollen concentration and diversity in a Mediterranean high-altitude site disclose unexpected temporal patterns. Aerobiologia 2017, 34, 75–87. [Google Scholar] [CrossRef]
- Perrone, M.; Becagli, S.; Orza, J.A.G.; Vecchi, R.; Dinoi, A.; Udisti, R.; Cabello, M. The impact of long-range-transport on PM1 and PM2.5 at a Central Mediterranean site. Atmos. Environ. 2013, 71, 176–186. [Google Scholar] [CrossRef]
- Kadam, K.; Karbhal, R.; Jayaraman, V.K.; Sawant, S.; Kulkarni-Kale, U. AllerBase: A comprehensive allergen knowledgebase. Database 2017, 2017, bax066. [Google Scholar] [CrossRef]
- Radauer, C.; Breiteneder, H. Allergen databases—A critical evaluation. Allergy 2019, 74, 2057–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chichiricco, G.; Pacini, E. Cupressus arizonica pollen wall zonation and in vitro hydration. Plant Syst. Evol. 2007, 270, 231–242. [Google Scholar] [CrossRef]
- Kraaijeveld, K.; De Weger, L.A.; García, M.V.; Buermans, H.; Frank, J.; Hiemstra, P.; Dunnen, J.T.D. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 2014, 15, 8–16. [Google Scholar] [CrossRef]
- Gottardini, E.; Cristofolini, F.; Cristofori, A.; Vannini, A.; Ferretti, M. Sampling bias and sampling errors in pollen counting in aerobiological monitoring in Italy. J. Environ. Monit. 2009, 11, 751–755. [Google Scholar] [CrossRef]
- Šikoparija, B.; Pejak-Šikoparija, T.; Radišić, P.; Smith, M.; Soldevilla, C.G. The effect of changes to the method of estimating the pollen count from aerobiological samples. J. Environ. Monit. 2010, 13, 384–390. [Google Scholar] [CrossRef]
- Adamov, S.; Lemonis, N.; Clot, B.; Crouzy, B.; Gehrig, R.; Graber, M.-J.; Sallin, C.; Tummon, F. On the measurement uncertainty of Hirst-type volumetric pollen and spore samplers. Aerobiologia 2021, 1–15. [Google Scholar] [CrossRef]
- Campbell, B.; Al-Kouba, J.; Timbrell, V.; Noor, M.; Massel, K.; Gilding, E.; Angel, N.; Kemish, B.; Hugenholtz, P.; Godwin, I.; et al. Tracking seasonal changes in diversity of pollen allergen exposure: Targeted metabarcoding of a subtropical aerobiome. Sci. Total Environ. 2020, 747, 141189. [Google Scholar] [CrossRef]
- Hofmann, F.; Otto, M.; Wosniok, W. Maize pollen deposition in relation to distance from the nearest pollen source under common cultivation—Results of 10 years of monitoring (2001 to 2010). Environ. Sci. Eur. 2014, 26, 24. [Google Scholar] [CrossRef] [Green Version]


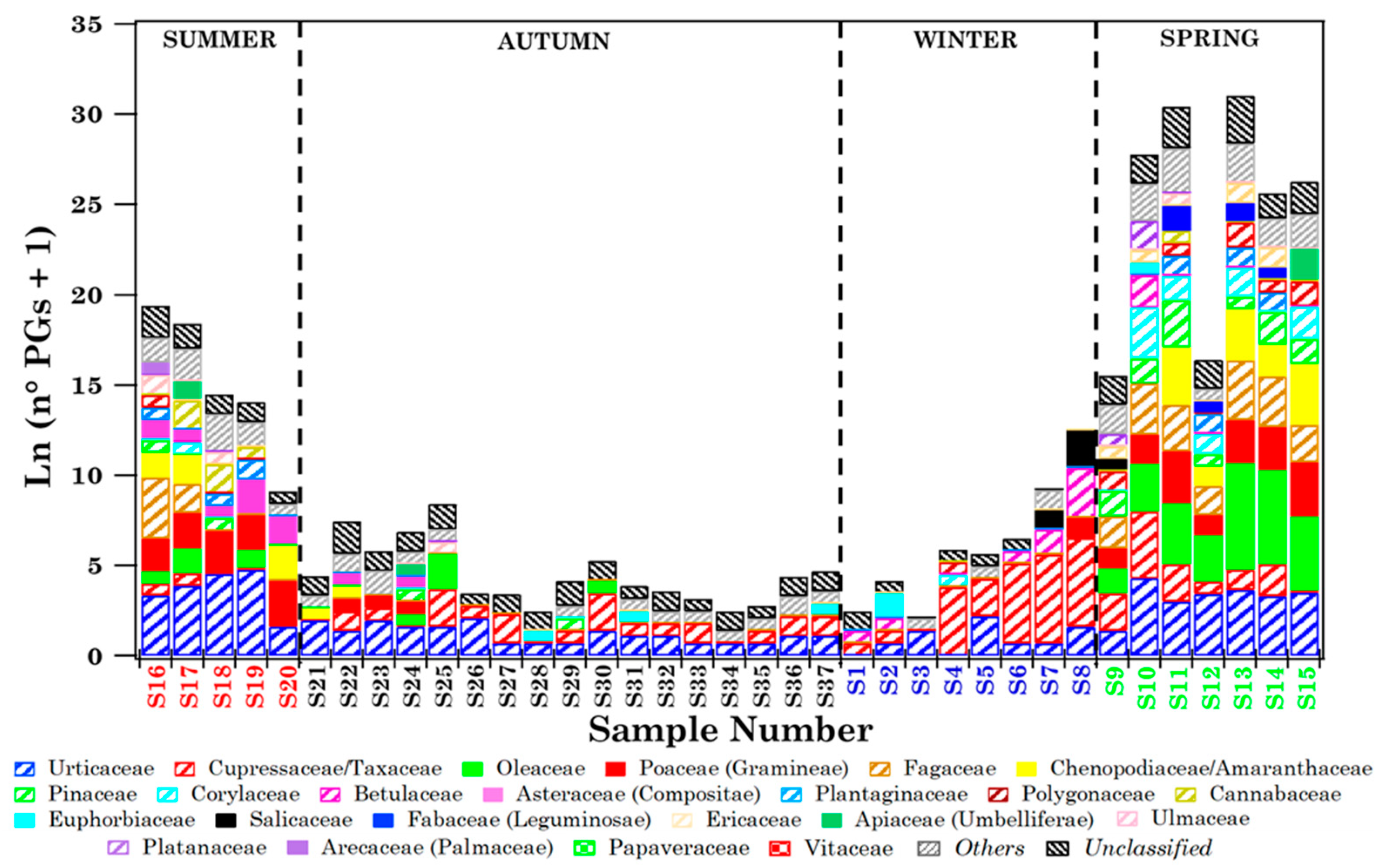
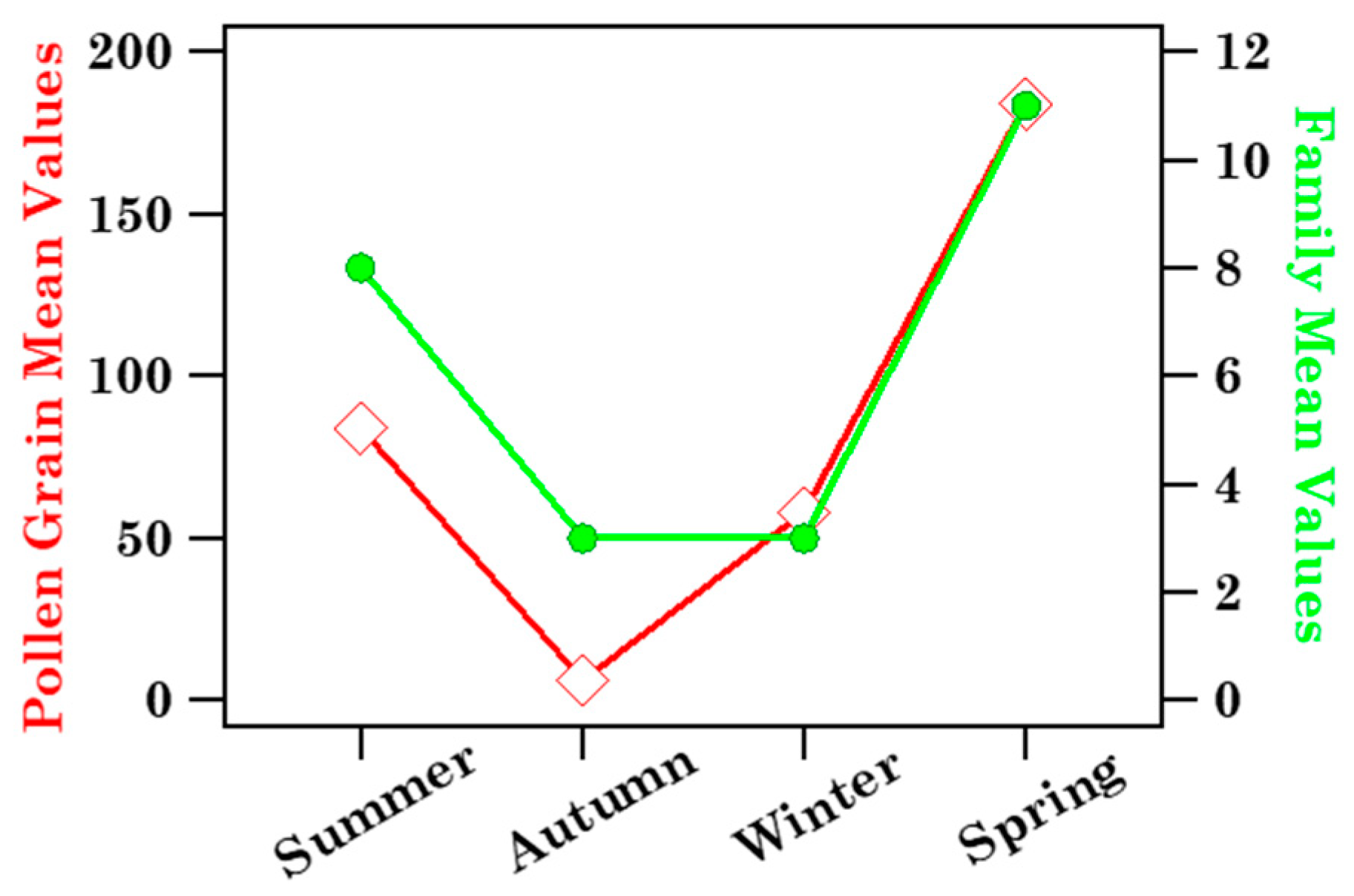
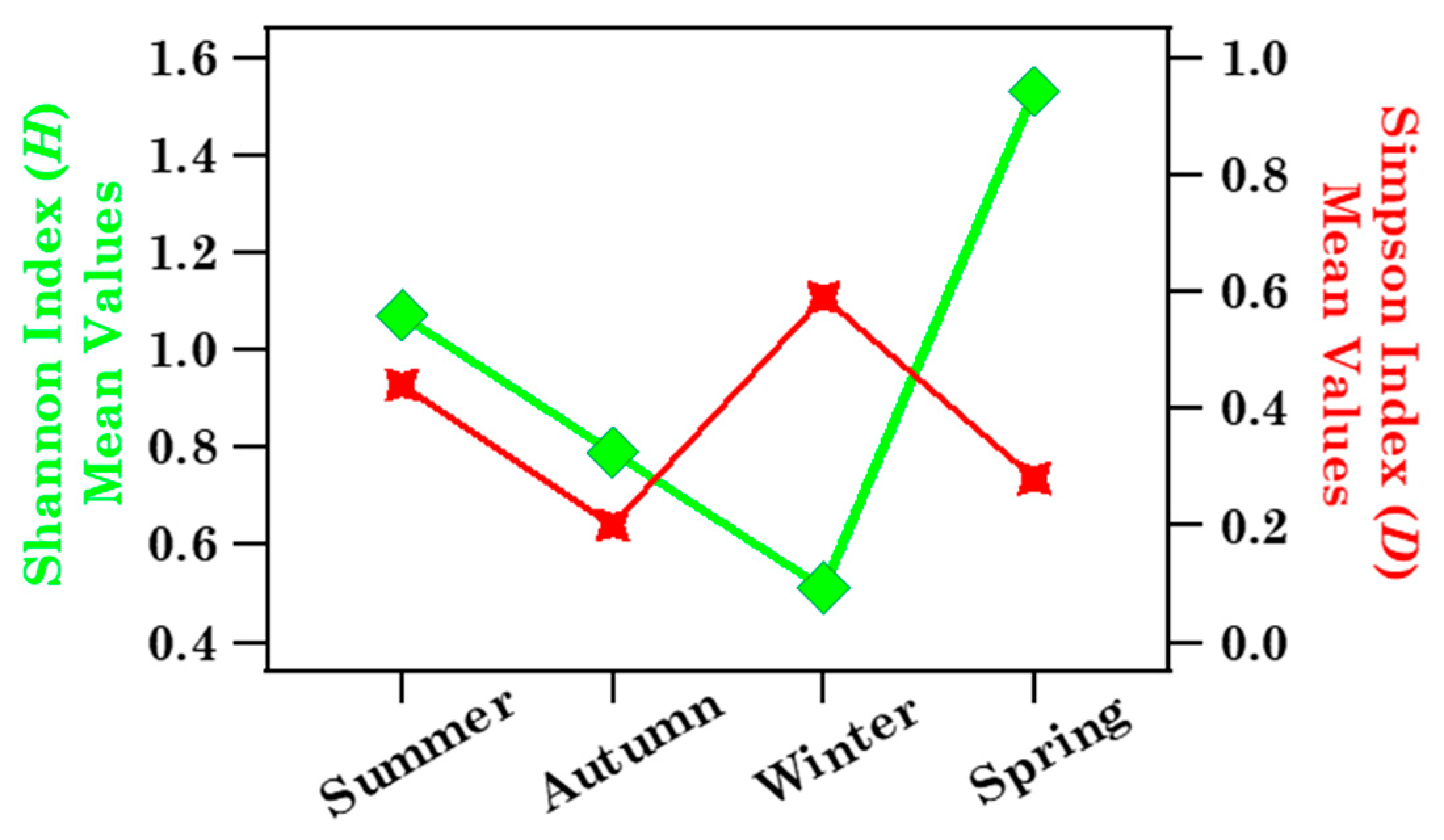
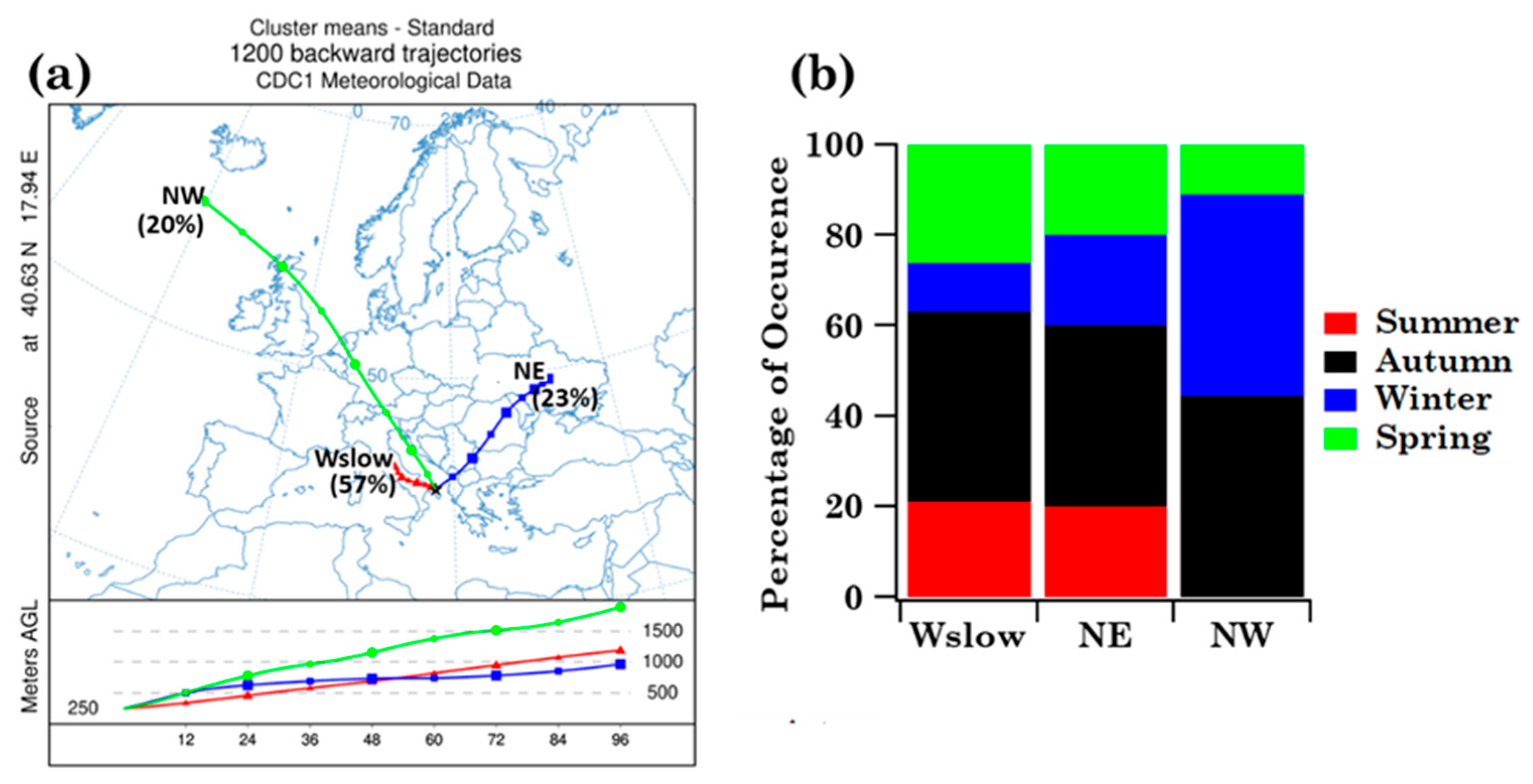


| Sample | Sampling Date (dd/mm/yy) | ST (h) | N° Tot. Pollen Grains | n° Families | At Family-Level | |
|---|---|---|---|---|---|---|
| Shannon Index (H) | Simpson Index (D) | |||||
| S1 | 07/01/2019 | 24 | 4 | 2 | 0.69 | 0.13 |
| S2 | 17/01/2019 | 24 | 7 | 4 | 1.20 | 0.24 |
| S3 | 24/01/2019 | 24 | 4 | 1 | 0.22 | 0.56 |
| S4 | 31/01/2019 | 24 | 47 | 3 | 0.23 | 0.88 |
| S5 | 07/02/2019 | 24 | 17 | 2 | 0.72 | 0.39 |
| S6 | 14/02/2019 | 24 | 85 | 3 | 0.14 | 0.93 |
| S7 | 21/02/2019 | 24 | 144 | 4 | 0.23 | 0.89 |
| S8 | 28/02/2019 | 24 | 166 | 5 | 0.65 | 0.70 |
| S9 | 04/04/2019 | 24 | 36 | 10 | 1.83 | 0.09 |
| S10 | 18/04/2019 | 24 | 186 | 11 | 1.69 | 0.22 |
| S11 | 09/05/2019 | 24 | 150 | 13 | 1.98 | 0.11 |
| S12 | 16/05/2019 | 24 | 62 | 10 | 1.49 | 0.28 |
| S13 | 23/05/2019 | 24 | 498 | 12 | 0.93 | 0.57 |
| S14 | 30/05/2019 | 24 | 259 | 11 | 1.12 | 0.50 |
| S15 | 06/06/2019 | 24 | 181 | 9 | 1.66 | 0.21 |
| S16 | 04/07/2018 | 48 | 81 | 12 | 1.54 | 0.24 |
| S17 | 11/07/2018 | 48 | 82 | 10 | 1.32 | 0.36 |
| S18 | 18/07/2018 | 48 | 119 | 7 | 0.68 | 0.61 |
| S19 | 25/07/2018 | 48 | 140 | 6 | 0.57 | 0.71 |
| S20 | 19/09/2018 | 48 | 28 | 4 | 1.25 | 0.27 |
| S21 | 03/10/2018 | 48 | 10 | 2 | 0.54 | 0.37 |
| S22 | 10/10/2018 | 48 | 15 | 5 | 1.13 | 0.07 |
| S23 | 17/10/2018 | 48 | 13 | 3 | 0.75 | 0.22 |
| S24 | 24/10/2018 | 48 | 12 | 6 | 1.40 | 0.15 |
| S25 | 07/11/2018 | 48 | 22 | 4 | 1.17 | 0.21 |
| S26 | 12/11/2018 | 24 | 9 | 2 | 0.44 | 0.62 |
| S27 | 14/11/2018 | 48 | 7 | 2 | 0.60 | 0.35 |
| S28 | 21/11/2018 | 48 | 4 | 2 | 0.69 | 0.13 |
| S29 | 28/11/2018 | 48 | 7 | 3 | 0.83 | 0.06 |
| S30 | 03/12/2018 | 24 | 13 | 3 | 0.87 | 0.35 |
| S31 | 10/12/2018 | 24 | 6 | 3 | 0.96 | 0.17 |
| S32 | 11/12/2018 | 24 | 6 | 2 | 0.66 | 0.14 |
| S33 | 12/12/2018 | 24 | 5 | 2 | 0.69 | 0.20 |
| S34 | 17/12/2018 | 24 | 4 | 1 | 0.35 | 0.06 |
| S35 | 18/12/2018 | 24 | 4 | 2 | 0.69 | 0.13 |
| S36 | 19/12/2018 | 24 | 8 | 2 | 0.69 | 0.13 |
| S37 | 20/12/2018 | 24 | 8 | 3 | 0.95 | 0.14 |
| Meteorological Parameters | Brindisi | Lecce |
|---|---|---|
| Significant Correlations | Significant Correlations | |
| Temperature (T) | Oleaceae (0.46 **), Pinaceae (0.39 *), Poaceae (0.69 **), Urticaceae (0.60 **), Others (0.40 *), n° PGs (0.38 *); Cupressaceae/Taxaceae (−0.36 *) | Poaceae (0.49 **); Asteraceae (−0.49 **), Euphorbiaceae (−0.40 *) |
| Relative Humidity (RH) | WS (−0.36 *) | |
| Pressure (P) | Cupressaceae/Taxaceae (0.38 *); Pinaceae (−0.33 *) | Oleaceae (−0.40 *) |
| Wind Speed (WS) | Fabaceae (0.44 **), Poaceae (0.36 *), Salicaceae (0.36 *); RH (−0.36 *) |
| Cluster | n° Families | n° Pollen Grains | n° Others and Unclassified Grains | At Family Level | ||
|---|---|---|---|---|---|---|
| Shannon Index (H) | Simpson Index (D) | |||||
| Wslow | mean ± SEM Min–Max | 6 ± 1 | 71 ± 26 | 6 ± 1 | 0.99 ± 0.13 | 0.34 ± 0.05 |
| 1–13 | 1–477 | 1–21 | 0.22–1.98 | 0.06–0.89 | ||
| NE | mean ± SEM Min–Max | 5 ± 2 | 68 ± 36 | 6 ± 1 | 0.97 ± 0.20 | 0.32 ± 0.11 |
| 2–11 | 8–175 | 2–11 | 0.56–1.69 | 0.07–0.71 | ||
| NW | mean ± SEM Min–Max | 4 ± 1 | 64 ± 30 | 2 ± 1 | 0.78 ± 0.14 | 0.43 ± 0.11 |
| 2–11 | 3–252 | 0–7 | 0.14–1.40 | 0.14–0.93 | ||
| Sample | n° Reads | n° Families | At Family Level | |
|---|---|---|---|---|
| Shannon Index (H) | Simpson Index (D) | |||
| S1 | 131,194 | 22 | 1.19 | 0.02 |
| S2 | 66,216 | 24 | 1.15 | 0.02 |
| S3 | 173,734 | 22 | 1.10 | 0.03 |
| S4 | 93,386 | 22 | 1.09 | 0.03 |
| S5 | 126,821 | 23 | 1.03 | 0.03 |
| S6 | 63,108 | 23 | 1.16 | 0.03 |
| S7 | 150,100 | 23 | 0.71 | 0.12 |
| S8 | 194,119 | 24 | 0.75 | 0.11 |
| S9 | 162,986 | 23 | 1.05 | 0.03 |
| S10 | 99,720 | 22 | 1.05 | 0.02 |
| S11 | 142,200 | 24 | 0.96 | 0.03 |
| S12 | 142,710 | 22 | 0.98 | 0.02 |
| S13 | 289,789 | 22 | 0.92 | 0.03 |
| S14 | 160,150 | 22 | 0.89 | 0.04 |
| S15 | 227,631 | 22 | 0.90 | 0.04 |
| S16 | 157,587 | 24 | 0.93 | 0.04 |
| S17 | 182,051 | 24 | 1.18 | 0.02 |
| S18 | 165,257 | 24 | 1.09 | 0.02 |
| S19 | 141,325 | 23 | 1.02 | 0.03 |
| S20 | 183,389 | 24 | 1.13 | 0.02 |
| S21 | 255,624 | 23 | 1.11 | 0.02 |
| S22 | 83,919 | 22 | 1.02 | 0.02 |
| S23 | 108,401 | 22 | 1.07 | 0.02 |
| S24 | 214,694 | 23 | 1.13 | 0.02 |
| S25 | 219,449 | 23 | 1.08 | 0.02 |
| S26 | 77,391 | 21 | 1.01 | 0.03 |
| S27 | 197,508 | 22 | 1.09 | 0.02 |
| S28 | 78,969 | 19 | 0.85 | 0.06 |
| S29 | 148,741 | 22 | 1.12 | 0.02 |
| S30 | 70,326 | 22 | 1.05 | 0.04 |
| S31 | 204,868 | 23 | 1.12 | 0.03 |
| S32 | 94,173 | 23 | 1.04 | 0.04 |
| S33 | 68,166 | 21 | 1.07 | 0.01 |
| S34 | 141,111 | 20 | 1.05 | 0.04 |
| S35 | 101,357 | 22 | 1.17 | 0.02 |
| S36 | 47,913 | 23 | 1.18 | 0.02 |
| S37 | 63,885 | 24 | 1.06 | 0.03 |
| Families | Winter | Spring | Summer | Autumn |
|---|---|---|---|---|
| Apiaceae | 4473 (0) | 3171 (5) | 6684 (2) | 10,500 (1) |
| Arecaceae | 240 (0) | 195 (0) | 729 (1) | 948 (0) |
| Asteraceae | 2306 (0) | 1749 (0) | 640 (14) | 4111 (2) |
| Chenopodiaceae | 31,845 (0) | 18,390 (75) | 24,312 (13) | 123,099 (2) |
| Euphorbiaceae | 177 (3) | 87 (1) | 33 (0) | 150 (3) |
| Fabaceae | 10,163 (0) | 11,974 (7) | 24,523 (0) | 46,467 (0) |
| Oleaceae | 48,879 (0) | 138,558 (675) | 34,392 (6) | 158,334 (8) |
| Poaceae | 23,849 (2) | 131,207 (64) | 82,341 (39) | 119,146 (3) |
| Salicaceae | 912 (9) | 1407 (1) | 1662 (0) | 894 (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fragola, M.; Arsieni, A.; Carelli, N.; Dattoli, S.; Maiellaro, S.; Perrone, M.R.; Romano, S. Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights. Int. J. Environ. Res. Public Health 2022, 19, 2624. https://doi.org/10.3390/ijerph19052624
Fragola M, Arsieni A, Carelli N, Dattoli S, Maiellaro S, Perrone MR, Romano S. Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights. International Journal of Environmental Research and Public Health. 2022; 19(5):2624. https://doi.org/10.3390/ijerph19052624
Chicago/Turabian StyleFragola, Mattia, Augusto Arsieni, Nicola Carelli, Sabrina Dattoli, Sante Maiellaro, Maria Rita Perrone, and Salvatore Romano. 2022. "Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights" International Journal of Environmental Research and Public Health 19, no. 5: 2624. https://doi.org/10.3390/ijerph19052624
APA StyleFragola, M., Arsieni, A., Carelli, N., Dattoli, S., Maiellaro, S., Perrone, M. R., & Romano, S. (2022). Pollen Monitoring by Optical Microscopy and DNA Metabarcoding: Comparative Study and New Insights. International Journal of Environmental Research and Public Health, 19(5), 2624. https://doi.org/10.3390/ijerph19052624







