MALDI-TOF Mass Spectrometry Analysis and Human Post-Mortem Microbial Community: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Autopsy and Sampling
2.2. Microbiological Analysis
2.3. Bacterial Identification
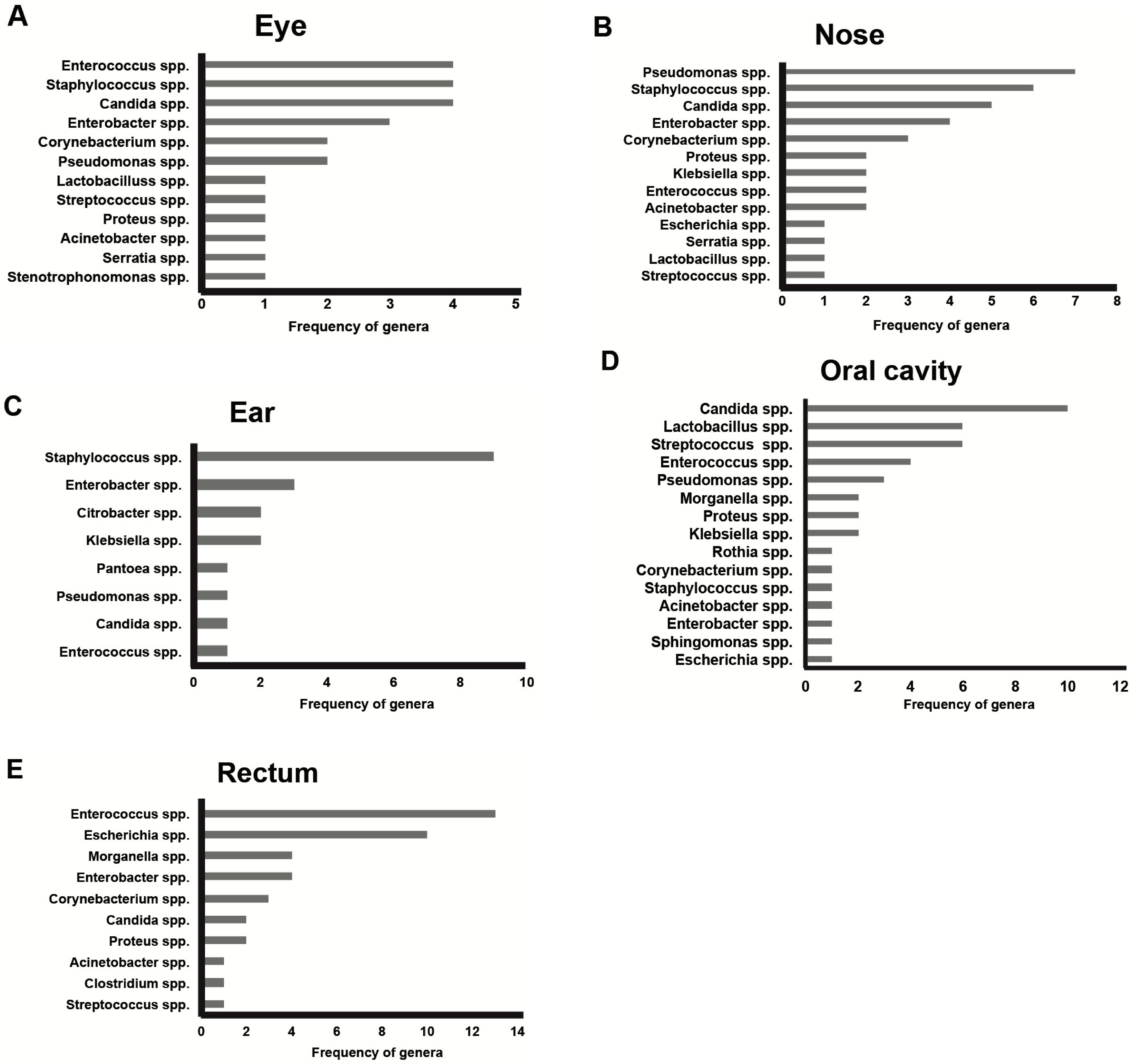
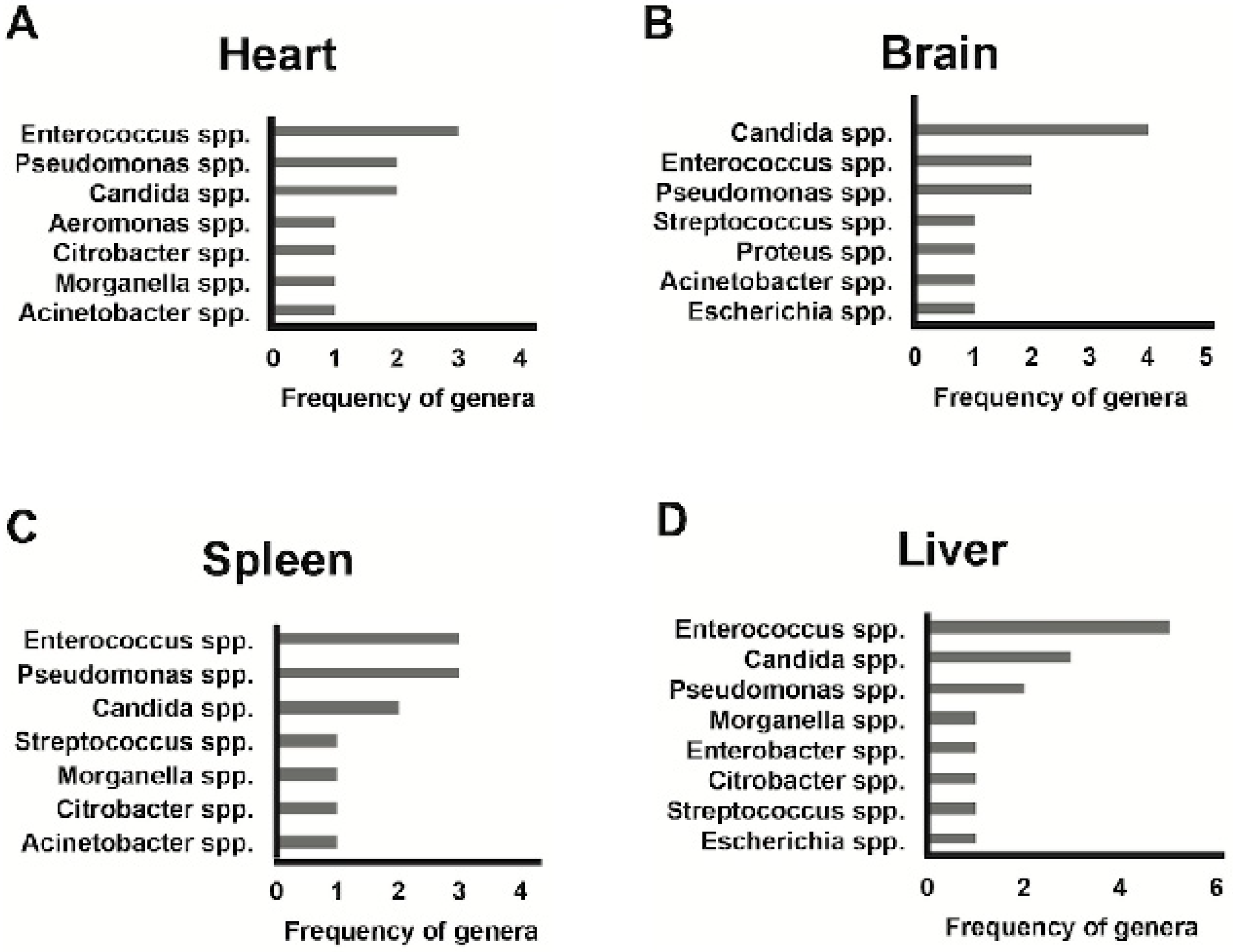
3. Results
4. Discussion
5. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knight, R.; Callewaert, C.; Marotz, C.; Hyde, E.R.; Debelius, J.W.; McDonald, D.; Sogin, M.L. The Microbiome and Human Biology. Annu. Rev. Genom. Hum. Genet. 2017, 18, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Hyde, E.R.; Haarmann, D.P.; Lynne, A.M.; Bucheli, S.R.; Petrosino, J.F. The living dead: Bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS ONE 2013, 8, e77733. [Google Scholar] [CrossRef] [PubMed]
- Benbow, M.E.; Tomberlin, J.K.; Tarone, A.M. Carrion Ecology, Evolution, and Their Applications; CRC Press: Boca Raton, FL, USA, 2015; pp. 28–45. [Google Scholar]
- Marchitelli, B. The Pathophysiology of Dying. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Javan, G.T.; Finley, S.J.; Abidin, Z.; Mulle, J.G. The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front. Microbiol. 2016, 7, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Can, I.; Javan, G.T.; Pozhitkov, A.E.; Noble, P.A. Distinctive thanatomicrobiome signatures found in the blood and internal organs of humans. J. Microbiol. Methods 2014, 106, 1–7. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2018, 9, 2868. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Wegener Parfrey, L.; Gonzalez, A.; Lauber, C.L.; Knights, D.; Ackermann, G.; Humphrey, G.C.; Gebert, M.J.; Van Treuren, W.; Berg-Lyons, D.; et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. ELife 2013, 2, e01104. [Google Scholar] [CrossRef]
- Pechal, J.L.; Crippen, T.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Tomberlin, J.K. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Legal Med. 2014, 128, 193–205. [Google Scholar] [CrossRef]
- Javan, G.T.; Finley, S.J.; Smith, T.; Miller, J.; Wilkinson, J.E. Cadaver Thanatomicrobiome Signatures: The Ubiquitous Nature of Clostridium Species in Human Decomposition. Front. Microbiol. 2017, 8, 2096. [Google Scholar] [CrossRef] [Green Version]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pechal, J.L.; Schmidt, C.J.; Jordan, H.R.; Benbow, M.E. A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition. Sci. Rep. 2018, 8, 5724. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Martora, F.; Della Pepa, M.E.; Folliero, V.; Luongo, L.; Bocelli, S.; Guida, F.; Mascolo, P.; Campobasso, C.P.; Maione, S.; et al. Post-mortem interval assessment by MALDI-TOF mass spectrometry analysis in murine cadavers. J. Appl. Microbiol. 2022, 132, 707–714. [Google Scholar] [CrossRef]
- Li, C.; Ma, D.; Deng, K.; Chen, Y.; Huang, P.; Wang, Z. Application of MALDI-TOF MS for Estimating the Postmortem Interval in Rat Muscle Samples. J. Forensic Sci. 2017, 62, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Seeley, E.H.; Caprioli, R.M. MALDI imaging mass spectrometry of human tissue: Method challenges and clinical perspectives. Trends Biotechnol. 2011, 29, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meding, S.; Nitsche, U.; Balluff, B.; Elsner, M.; Rauser, S.; Schöne, C.; Nipp, M.; Maak, M.; Feith, M.; Ebert, M.P.; et al. Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J. Proteome Res. 2012, 11, 1996–2003. [Google Scholar] [CrossRef]
- Mao, X.; He, J.; Li, T.; Lu, Z.; Sun, J.; Meng, Y.; Abliz, Z.; Chen, J. Application of imaging mass spectrometry for the molecular diagnosis of human breast tumors. Sci. Rep. 2016, 6, 21043. [Google Scholar] [CrossRef]
- Chun, L.P.; Miguel, M.J.; Junkins, E.N.; Forbes, S.L.; Carter, D.O. An initial investigation into the ecology of culturable aerobic postmortem bacteria. Sci. Justice 2015, 55, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerquist, C.K.; Garbus, B.R.; Miller, W.G.; Williams, K.E.; Yee, E.; Bates, A.H.; Boyle, S.; Harden, L.A.; Cooley, M.B.; Mandrell, R.E. Rapid Identification of Protein Biomarkers of Escherichia coli O157:H7 by Matrix-Assisted Laser Desorption Ionization-Time-of-Flight−Time-of-Flight Mass Spectrometry and Top-Down Proteomics. Anal. Chem. 2010, 82, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current Status of MALDI-TOF Mass Spectrometry in Clinical Microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Florio, W.; Tavanti, A.; Barnini, S.; Ghelardi, E.; Lupetti, A. Recent Advances and Ongoing Challenges in the Diagnosis of Microbial Infections by MALDI-TOF Mass Spectrometry. Front. Microbiol. 2018, 9, 1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholl, P.; Leonardo, M.A.; Rule, A.; Carlson, M.A.; Antoine, M.; Buckley, T. The Development of Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry for the Detection of Biological Warfare Agent Aerosols. Johns Hopkins APL Technol. Dig. 1999, 20, 343–350. [Google Scholar]
- Han, S.S.; Jeong, Y.S.; Choi, S.K. Current Scenario and Challenges in the Direct Identification of Microorganisms Using MALDI TOF MS. Microorganisms 2021, 9, 1917. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.S.; Kim, Y.H. Rapid and robust MALDI-TOF MS techniques for microbial identification: A brief overview of their diverse applications. J. Microbiol. 2018, 56, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Dell’Annunziata, F.; Ilisso, C.P.; Dell’Aversana, C.; Greco, G.; Coppola, A.; Martora, F.; Dal Piaz, F.; Donadio, G.; Falanga, A.; Galdiero, M.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the miRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020, 8, 1985. [Google Scholar] [CrossRef]
- Forger, L.V.; Woolf, M.S.; Simmons, T.L.; Swall, J.L.; Singh, B. A eukaryotic community-succession based method for postmortem interval (PMI) estimation of decomposing porcine remains. For. Sci. Int. 2019, 302, 109838. [Google Scholar] [CrossRef]
- Weatherbee, C.R.; Pechal, J.L.; Eric Benbow, M. The Dynamic Maggot Mass Microbiome. Ann. Ent. Soc. Am. 2017, 110, 45–53. [Google Scholar] [CrossRef]
- Pittner, S.; Bugelli, V.; Weitgasser, K.; Zissler, A.; Sanit, S.; Lutz, L.; Monticelli, F.; Campobasso, C.P.; Steinbacher, P.; Amendt, J. A field study to evaluate PMI estimation methods for advanced decomposition stages. Int. J. Legal Med. 2020, 134, 1361–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dell’Annunziata, F.; Folliero, V.; Giugliano, R.; De Filippis, A.; Santarcangelo, C.; Izzo, V.; Daglia, M.; Galdiero, M.; Arciola, C.R.; Franci, G. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 5985. [Google Scholar] [CrossRef] [PubMed]
- Hyde, E.R.; Haarmann, D.P.; Petrosino, J.F.; Lynne, A.M.; Bucheli, S.R. Initial insights into bacterial succession during human decomposition. Int. J. Legal Med. 2015, 129, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Pittner, S.; Bugelli, V.; Benbow, M.E.; Ehrenfellner, B.; Zissler, A.; Campobasso, C.P.; Oostra, R.J.; Aalders, M.; Zehner, R.; Lutz, L.; et al. The applicability of forensic time since death estimation methods for buried bodies in advanced decomposition stages. PLoS ONE 2020, 15, e0243395. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Bian, Y. Thanatomicrobiome composition profiling as a tool for forensic investigation. Forensic Sci. Res. 2018, 3, 105–110. [Google Scholar] [CrossRef]
- Buchan, B.W.; Ledeboer, N.A. Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev. 2014, 27, 783–822. [Google Scholar] [CrossRef] [Green Version]
- Kisand, V.; Wikner, J. Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl. Environ. Microbiol. 2003, 69, 3607–3616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, D.; Tomo, S.; Purohit, P.; Setia, P. Microbiome in Death and Beyond: Current Vistas and Future Trends. Front. Ecol. Evol. 2021, 9, 630397. [Google Scholar] [CrossRef]
- Dong, K.; Xin, Y.; Cao, F.; Huang, Z.; Sun, J.; Peng, M.; Liu, W.; Shi, P. Succession of oral microbiota community as a tool to estimate postmortem interval. Sci. Rep. 2019, 9, 13063. [Google Scholar] [CrossRef]
- Pechal, J.L.; Crippen, T.L.; Tarone, A.M.; Lewis, A.J.; Tomberlin, J.K.; Benbow, M.E. Microbial Community Functional Change during Vertebrate Carrion Decomposition. PLoS ONE 2013, 8, e79035. [Google Scholar] [CrossRef] [PubMed]
- Iancu, L.; Junkins, E.N.; Necula-Petrareanu, G.; Purcarea, C. Characterizing forensically important insect and microbial community colonization patterns in buried remains. Sci. Rep. 2015, 8, 15513. [Google Scholar] [CrossRef]
- Martora, F.; Pinto, F.; Folliero, V.; Cammarota, M.; Dell’Annunziata, F.; Squillaci, G.; Galdiero, M.; Franci, G. Isolation, characterization and analysis of pro-inflammatory potential of Klebsiella pneumoniae outer membrane vesicles. Microb. Pathog. 2019, 136, 103719. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Amorim, A. Microbial forensics: New breakthroughs and future prospects. Appl. Microbiol. Biotechnol. 2018, 102, 10377–10391. [Google Scholar] [CrossRef] [PubMed]
- Iancu, L.; Carter, D.O.; Junkins, E.N.; Purcarea, C. Using bacterial and necrophagous insect dynamics for post-mortem interval estimation during cold season: Novel case study in Romania. For. Sci. Int. 2015, 254, 106–117. [Google Scholar] [CrossRef]
- Javan, G.T.; Finley, S.J.; Can, I.; Wilkinson, J.E.; Hanson, J.D.; Tarone, A.M. Human Thanatomicrobiome Succession and Time Since Death. Sci. Rep. 2016, 6, 29598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucci, A.; Campobasso, C.P.; Cirnelli, A.; Lorenzini, G. A promising microbiological test for the diagnosis of drowning. Forensic Sci. Int. 2008, 182, 20–26. [Google Scholar] [CrossRef]
- Morris, J.A.; Harrison, L.M.; Partridge, S.M. Postmortem bacteriology: A re-evaluation. J. Clin. Pathol. 2006, 59, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Casadonte, R.; Caprioli, R.M. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 2011, 6, 1695–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
 Firmicutes,
Firmicutes,  Proteobacteria,
Proteobacteria,  Ascomycota,
Ascomycota,  Actinobacteria].
Actinobacteria].
 Firmicutes,
Firmicutes,  Proteobacteria,
Proteobacteria,  Ascomycota,
Ascomycota,  Actinobacteria].
Actinobacteria].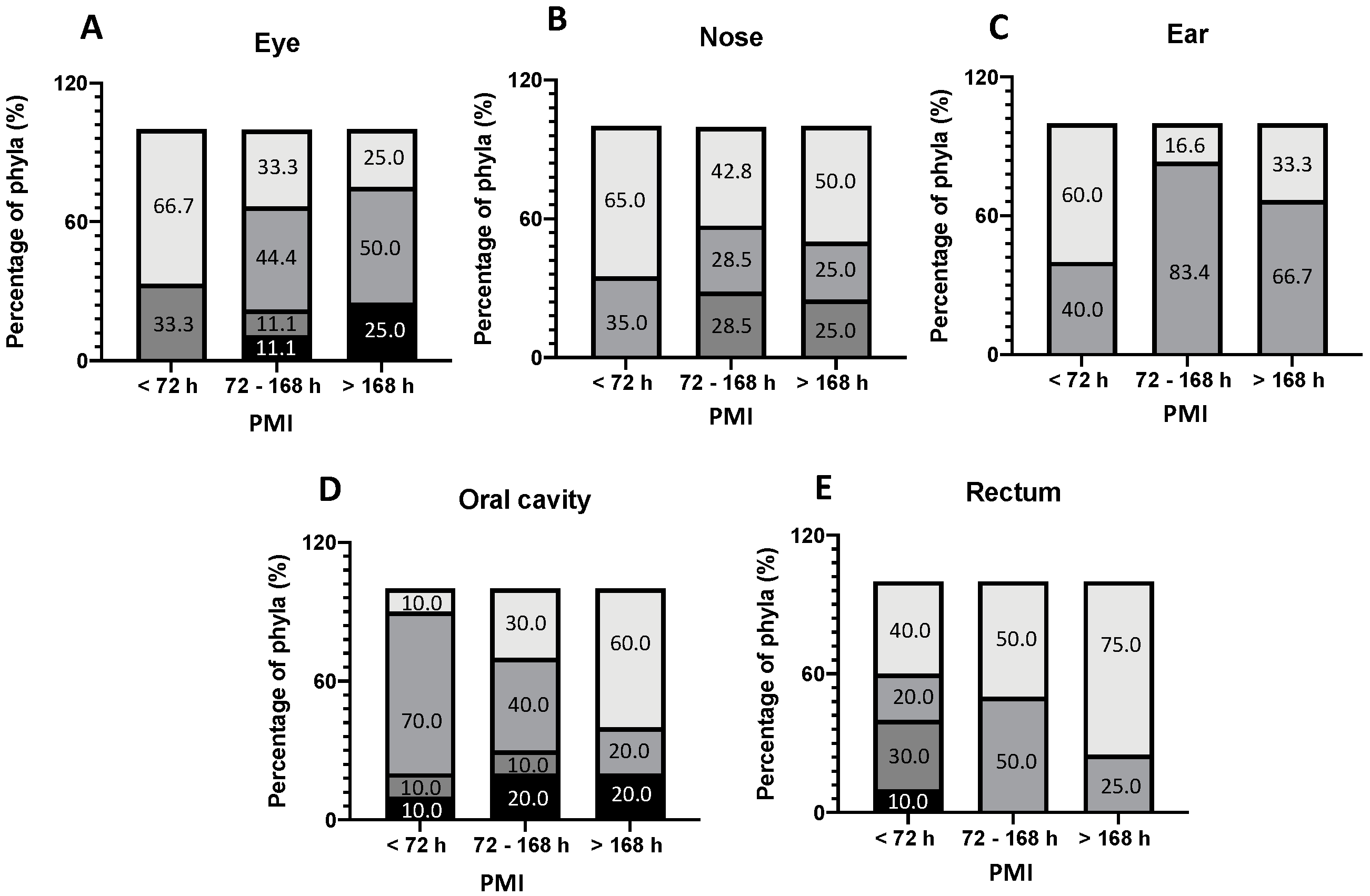
 Firmicutes,
Firmicutes,  Proteobacteria,
Proteobacteria,  Ascomycota,
Ascomycota,  Actinobacteria].
Actinobacteria].
 Firmicutes,
Firmicutes,  Proteobacteria,
Proteobacteria,  Ascomycota,
Ascomycota,  Actinobacteria].
Actinobacteria].
| Case # | Age/Sex | Death Scene | PMI (h) | Season | Stage of Decay | Cause of Death | Manner of Death |
|---|---|---|---|---|---|---|---|
| 1 | 53 y/M | Indoor (Home) | 24–36 | Winter | Fresh | Hanging | Suicide |
| 2 | 61 y/M | Indoor (Home) | 96–120 | Winter | Fresh with early signs of decomposition | Blunt injuries to the head | Homicide |
| 3 | 38 y/M | Outdoor | 36–48 | Winter | Fresh | Fatal fall from great height | Suicide |
| 4 | 57 y/M | Indoor (Hospital) | 72–96 | Spring | Fresh with early signs of decomposition | Respiratory failure due to pneumonia | Natural |
| 5 | 40 y/M | Indoor (Hospital) | 288–312 | Winter | Fresh/Bloated | Multi Organ Failure | Natural |
| 6 | 45 y/F | Indoor (Hospital) | 216–240 | Winter | Fresh/Bloated | Septic shock | Natural |
| 7 | 45 y/M | Indoor (Hospital) | 72–96 | Spring | Fresh | Gunshot injuries to the head | Homicide |
| 8 | 20 y/M | Indoor (Hospital) | 72–96 | Spring | Fresh | Blunt injuries to the head | Homicide |
| 9 | 80 y/M | Indoor (Hospital) | 36–48 | Spring | Fresh | Blunt Trauma | Road Accident |
| 10 | 6 m/M | Indoor (Home) | 24–36 | Spring | Fresh | Respiratory failure due to pneumonia | Natural |
| 11 | 51 y/M | Indoor (Prison) | 96–120 | Spring | Fresh with early signs of decomposition | Subarachnoid hemorrhage | Natural |
| 12 | 82 y/M | Indoor (Hospital) | 336–360 | Summer | Fresh/Bloated | Fatal fall from great height | Suicide |
| 13 | 4 m/M | Indoor (Home) | 24–36 | Summer | Fresh | Respiratory failure due to pneumonia | Natural |
| 14 | 41 y/M | Indoor (Hospital) | 168–192 | Summer | Fresh/Bloated | Liver failure in cirrhosis | Natural |
| 15 | 20 y/M | Indoor (Hospital) | 96–120 | Winter | Fresh with early signs of decomposition | Blunt Trauma | Road accident |
| 16 | 53 y/M | Outdoor | 240–264 | Spring | Bloated | Myocardial infarction due to coronary artery disease | Natural |
| 17 | 22 y/M | Indoor (Hospital) | 36–48 | Spring | Fresh | Blunt Trauma | Road accident |
| 18 | 70 y/M | Outdoor | 36–48 | Summer | Fresh | Arrhythmogenic cardiomyopathy | Natural |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campobasso, C.P.; Mastroianni, G.; Feola, A.; Mascolo, P.; Carfora, A.; Liguori, B.; Zangani, P.; Dell’Annunziata, F.; Folliero, V.; Petrillo, A.; et al. MALDI-TOF Mass Spectrometry Analysis and Human Post-Mortem Microbial Community: A Pilot Study. Int. J. Environ. Res. Public Health 2022, 19, 4354. https://doi.org/10.3390/ijerph19074354
Campobasso CP, Mastroianni G, Feola A, Mascolo P, Carfora A, Liguori B, Zangani P, Dell’Annunziata F, Folliero V, Petrillo A, et al. MALDI-TOF Mass Spectrometry Analysis and Human Post-Mortem Microbial Community: A Pilot Study. International Journal of Environmental Research and Public Health. 2022; 19(7):4354. https://doi.org/10.3390/ijerph19074354
Chicago/Turabian StyleCampobasso, Carlo Pietro, Gennaro Mastroianni, Alessandro Feola, Pasquale Mascolo, Anna Carfora, Bruno Liguori, Pierluca Zangani, Federica Dell’Annunziata, Veronica Folliero, Arianna Petrillo, and et al. 2022. "MALDI-TOF Mass Spectrometry Analysis and Human Post-Mortem Microbial Community: A Pilot Study" International Journal of Environmental Research and Public Health 19, no. 7: 4354. https://doi.org/10.3390/ijerph19074354








