Molecular Analysis of Pathogenicity, Adhesive Matrix Molecules (MSCRAMMs) and Biofilm Genes of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Food
Abstract
1. Introduction
2. Materials and Methods
2.1. Staphylococci Strains
2.2. Detection of the Ability of the Slime Production by the Congo Red Agar (CRA) Method
2.3. Biofilm Forming Ability Detection by the Microtiter Plate Method (MTP)
2.4. Detection of the Biofilm-Associated Genes
2.5. Detection of the Hemolysin Genes
2.6. Detection of IS256/IS257
2.7. Statistical Analysis
3. Results
3.1. Qualitative and Quantitative Analyses of the Biofilm Formation by the CoNS Isolates
3.2. Genetic Background Associated with the Biofilm and the Adherence in the CoNS Strains
3.3. Prevalence of the Hemolysin Genes and the Insertion Elements IS256/257 among the CoNS Strains
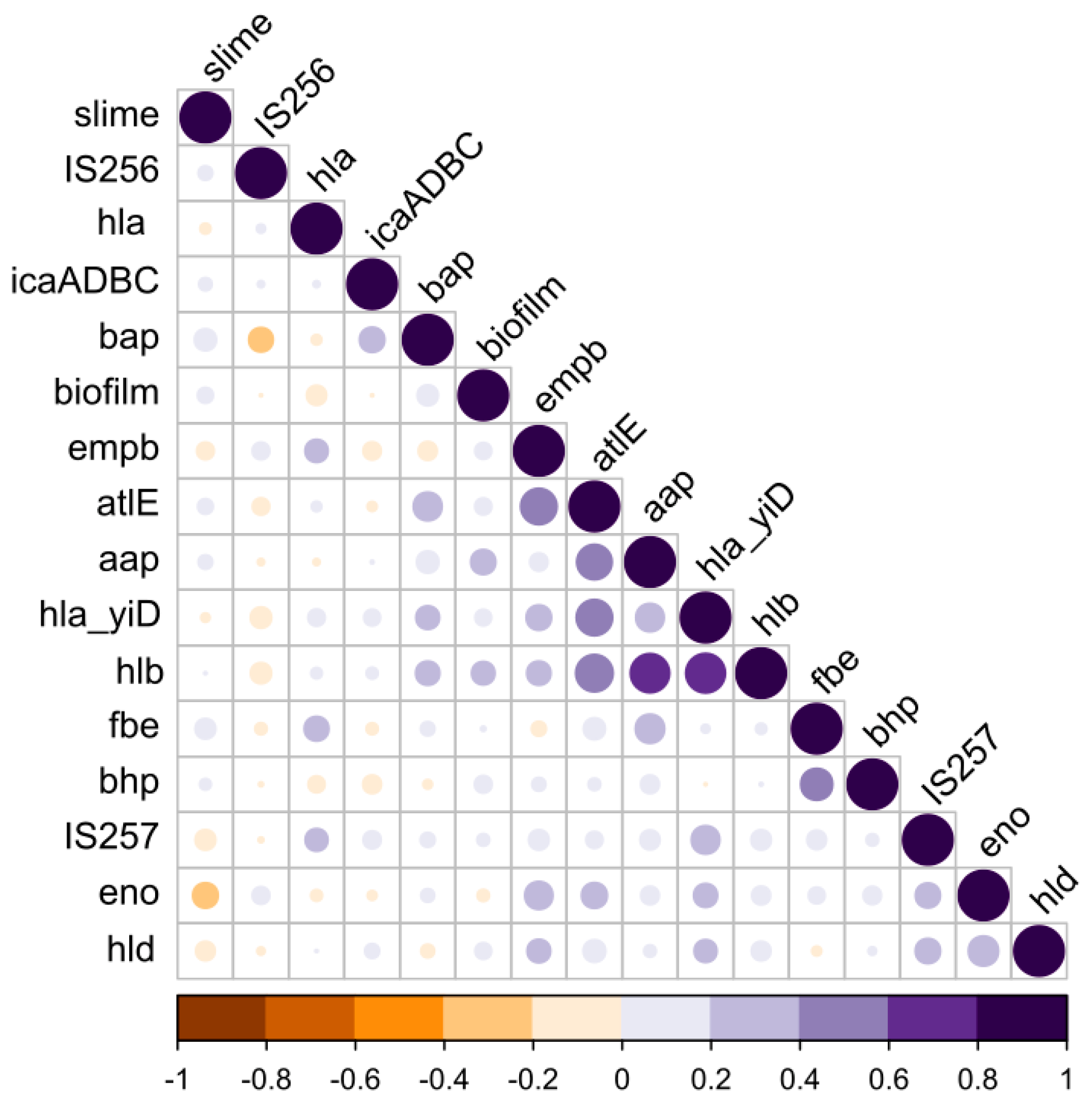
3.4. Phenotype vs. the Genotype Correlations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seo, K.S.; Bohach, G.A. Staphylococcus aureus. In Food Microbiology: Fundamentals and Frontiers, 4th ed.; Doyle, M.P., Buchanan, R.L., Eds.; ASM Press: Washington, DC, USA, 2013; pp. 547–573. [Google Scholar]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpińska, M.; Laniewska-Trokenheim, L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin-Phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015, 46, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Chajȩcka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpińska, M.; Łaniewska-Trokenheim, Ł. Retail ready-to-eat food as a potential vehicle for Staphylococcus spp. Harboring antibiotic resistance genes. J. Food Prot. 2014, 77, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Both, A.; Weißelberg, S.; Heilmann, C.; Rohde, H. Emergence of coagulase-negative staphylococci. Expert Rev. Anti-Infect. Ther. 2020, 18, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Favaro, M. Coagulase-Positive and Coagulase-Negative Staphylococci in Human Disease. In Pet-to-Man Travelling Staphylococci: A World in Progress; Academic Press: Cambridge, MA, USA, 2018; pp. 25–42. [Google Scholar]
- Kosecka-Strojek, M.; Buda, A.; Miȩdzobrodzki, J. Staphylococcal Ecology and Epidemiology. In Pet-to-Man Travelling Staphylococci: A World in Progress; Academic Press: Cambridge, MA, USA, 2018; pp. 11–24. [Google Scholar]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed]
- Chajęcka-Wierzchowska, W.; Gajewska, J.; Wiśniewski, P.; Zadernowska, A. Enterotoxigenic potential of coagulase-negative staphylococci from ready-to-eat food. Pathogens 2020, 9, 734. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 1–26. [Google Scholar] [CrossRef]
- Azara, E.; Longheu, C.; Sanna, G.; Tola, S. Biofilm formation and virulence factor analysis of Staphylococcus aureus isolates collected from ovine mastitis. J. Appl. Microbiol. 2017, 123, 372–379. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef]
- Vázquez-Sánchez, D.; Habimana, O.; Holck, A. Impact of food-related environmental factors on the adherence and biofilm formation of natural staphylococcus aureus isolates. Curr. Microbiol. 2013, 66, 110–121. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. Microbiologyopen 2020, 9, e00946. [Google Scholar] [CrossRef]
- Mack, D.; Becker, P.; Chatterjee, I.; Dobinsky, S.; Knobloch, J.K.M.; Peters, G.; Rohde, H.; Herrmann, M. Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: Functional molecules, regulatory circuits, and adaptive responses. Int. J. Med. Microbiol. 2004, 294, 203–212. [Google Scholar] [CrossRef]
- Heilmann, C.; Hussain, M.; Peters, G.; Götz, F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 1997, 24, 1013–1024. [Google Scholar] [CrossRef]
- Nilsson, M.; Frykberg, L.; Flock, J.I.; Pei, L.; Lindberg, M.; Guss, B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 1998, 66, 2666–2673. [Google Scholar] [CrossRef]
- Williams, R.J.; Henderson, B.; Sharp, L.J.; Nair, S.P. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 2002, 70, 6805–6810. [Google Scholar] [CrossRef]
- Heilmann, C.; Thumm, G.; Chhatwal, G.S.; Hartleib, J.; Uekötter, A.; Peters, G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 2003, 149, 2769–2778. [Google Scholar] [CrossRef]
- Löffler, B.; Tuchscherr, L.; Niemann, S.; Peters, G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014, 304, 170–176. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus Biofilm: A Complex Developmental Organism Graphical Abstract HHS Public Access. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef]
- Mack, D.; Riedewald, J.; Rohde, H.; Magnus, T.; Feucht, H.H.; Elsner, H.A.; Laufs, R.; Rupp, M.E. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect. Immun. 1999, 67, 1004–1008. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef]
- Agarwal, A.; Singh, K.P.; Jain, A. Medical significance and management of staphylococcal biofilm. FEMS Immunol. Med. Microbiol. 2010, 58, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Magro, G.; Biffani, S.; Minozzi, G.; Ehricht, R.; Monecke, S.; Luini, M.; Piccinini, R. Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins 2017, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Puah, S.M.; Tan, J.A.M.A.; Chew, C.H.; Chua, K.H. Diverse Profiles of Biofilm and Adhesion Genes in Staphylococcus Aureus Food Strains Isolated from Sushi and Sashimi. J. Food Sci. 2018, 83, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Moraveji, Z.; Tabatabaei, M.; Shirzad Aski, H.; Khoshbakht, R. Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, southern Iran. Iran. J. Vet. Res. 2014, 15, 326–330. [Google Scholar] [PubMed]
- Ebrahimi, A.; Taheri, M.A. Characteristics of staphylococci isolated from clinical and subclinical mastitis cows in Shahrekord, Iran. Iran. J. Vet. Res. 2009, 10, 273–277. [Google Scholar]
- Zhang, Y.Q.; Ren, S.X.; Li, H.L.; Wang, Y.X.; Fu, G.; Yang, J.; Qin, Z.Q.; Miao, Y.G.; Wang, W.Y.; Chen, R.S.; et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 2003, 49, 1577–1593. [Google Scholar] [CrossRef]
- Novick, R.P. Mobile genetic elements and bacterial toxinoses: The superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 2003, 49, 93–105. [Google Scholar] [CrossRef]
- Murugesan, S.; Mani, S.; Kuppusamy, I.; Krishnan, P. Role of insertion sequence element is256 as a virulence marker and its association with biofilm formation among methicillin-resistant Staphylococcus epidermidis from hospital and community settings in Chennai, South India. Indian J. Med. Microbiol. 2018, 36, 124–126. [Google Scholar] [CrossRef]
- Zakrzewski, A.J.; Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. A Comparison of Methods for Identifying Enterobacterales Isolates from Fish and Prawns. Pathogens 2022, 11, 410. [Google Scholar] [CrossRef]
- Li, X.; Xing, J.; Li, B.; Wang, P.; Liu, J. Use of tuf as a target for sequence-based identification of Gram-positive cocci of the genus Enterococcus, Streptococcus, coagulase-negative Staphylococcus, and Lactococcus. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 31. [Google Scholar] [CrossRef]
- Mathur, T.; Singhal, S.; Khan, S.; Upadhyay, D.J.; Fatma, T.; Rattan, A. Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods. Indian J. Med. Microbiol. 2006, 24, 25–29. [Google Scholar] [CrossRef]
- Gajewska, J.; Chajęcka-Wierzchowska, W. Biofilm formation ability and presence of adhesion genes among coagulase-negative and coagulase-positive staphylococci isolates from raw cow’s milk. Pathogens 2020, 9, 654. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćircović, I.; Ruzicka, F. Quantification of biofilm in microtiter plates. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence factors, antimicrobial resistance and biofilm formation in Enterococcus spp. isolated from retail shrimps. LWT Food Sci. Technol. 2016, 69, 117–122. [Google Scholar] [CrossRef]
- Nasaj, M.; Saeidi, Z.; Asghari, B.; Roshanaei, G.; Arabestani, M.R. Identification of hemolysin encoding genes and their association with antimicrobial resistance pattern among clinical isolates of coagulase-negative Staphylococci. BMC Res. Notes 2020, 13, 4–9. [Google Scholar] [CrossRef]
- Chessa, D.; Ganau, G.; Spiga, L.; Bulla, A.; Mazzarello, V.; Campus, G.V.; Rubino, S. Staphylococcus aureus and Staphylococcus epidermidis virulence strains as causative agents of persistent Infections in breast implants. PLoS ONE 2016, 11, e0146668. [Google Scholar] [CrossRef]
- Gonçalves, T.G.; Timm, C.D. Biofilm production by coagulase-negative Staphylococcus: A review. Arq. Inst. Biol. 2020, 87, 1–9. [Google Scholar] [CrossRef]
- Irlinger, F. Safety assessment of dairy microorganisms: Coagulase-negative staphylococci. Int. J. Food Microbiol. 2008, 126, 302–310. [Google Scholar] [CrossRef]
- Landeta, G.; Curiel, J.A.; Carrascosa, A.V.; Muñoz, R.; de las Rivas, B. Characterization of coagulase-negative staphylococci isolated from Spanish dry cured meat products. Meat Sci. 2013, 93, 387–396. [Google Scholar] [CrossRef]
- Petrelli, D.; Zampaloni, C.; D’ercole, S.; Prenna, M.; Ballarini, P.; Ripa, S.; Vitali, L.A. Analysis of different genetic traits and their association with biofilm formation in Staphylococcus epidermidis isolates from central venous catheter infections. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 773–781. [Google Scholar] [CrossRef]
- Atshan, S.S.; Nor Shamsudin, M.; Sekawi, Z.; Lung, L.T.T.; Hamat, R.A.; Karunanidhi, A.; Mateg Ali, A.; Ghaznavi-Rad, E.; Ghasemzadeh-Moghaddam, H.; Chong Seng, J.S.; et al. Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. J. Biomed. Biotechnol. 2012, 2012, e976972. [Google Scholar] [CrossRef] [PubMed]
- Serray, B.; Oufrid, S.; Hannaoui, I.; Bourjilate, F.; Soraa, N.; Mliji, M.; Sobh, M.; Hammoumi, A.; Timinouni, M.; Azhari, M. El Genes encoding adhesion factors and biofilm formation in methicillinresistant Staphylococcus aureus in Morocco. J. Infect. Dev. Ctries 2016, 10, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, J.; Li, H.; Zeng, P.; Li, J. Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among staphylococcus aureus strains isolated from different sources. Foodborne Pathog. Dis. 2013, 10, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Hook, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.; Pesce, M.; Franchelli, S.; Baldelli, I.; De Maria, A.; Marchese, A. Phenotypic and genotypic characterization of Staphylococci causing breast peri-implant infections in oncologic patients. BMC Microbiol. 2015, 15, 26. [Google Scholar] [CrossRef]
- França, A.; Gaio, V.; Lopes, N.; Melo, L.D.R. Virulence Factors in Coagulase-Negative Staphylococci. Pathogens 2021, 10, 170. [Google Scholar] [CrossRef]
- Heilbronner, S.; Foster, T.J. Staphylococcus lugdunensis: A skin commensal with invasive pathogenic potential. Clin. Microbiol. Rev. 2021, 34, e00205-20. [Google Scholar] [CrossRef]
- Argemi, X.; Matelska, D.; Ginalski, K.; Riegel, P.; Hansmann, Y.; Bloom, J.; Pestel-Caron, M.; Dahyot, S.; Lebeurre, J.; Prévost, G. Comparative genomic analysis of Staphylococcus lugdunensis shows a closed pan-genome and multiple barriers to horizontal gene transfer. BMC Genom. 2018, 19, 621. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Gajewska, J.S. epidermidis strains from artisanal cheese made from unpasteurized milk in Poland—Genetic characterization of antimicrobial resistance and virulence determinants. Int. J. Food Microbiol. 2019, 294, 55–59. [Google Scholar] [CrossRef]
- Mah, T. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Koskela, A.; Nilsdotter-Augustinsson, Å.; Persson, L.; Söderquist, B. Prevalence of the ica operon and insertion sequence IS256 among Staphylococcus epidermidis prosthetic joint infection isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 655–660. [Google Scholar] [CrossRef]
- Huseby, M.J.; Kruse, A.C.; Digre, J.; Kohler, P.L.; Vocke, J.A.; Mann, E.E.; Bayles, K.W.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 14407–14412. [Google Scholar] [CrossRef]
- Kmieciak, W.; Szewczyk, E.M. Cytolizyny-Czynniki zjadliwości Staphylococcus intermedius i Staphylococcus pseudintermedius. Postep. Mikrobiol. 2015, 54, 354–363. [Google Scholar]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef]
- Solati, S.M.; Tajbakhsh, E.; Khamesipour, F.; Gugnani, H.C. Prevalence of virulence genes of biofilm producing strains of Staphylococcus epidermidis isolated from clinical samples in Iran. AMB Express 2015, 5, 47. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penadés, J.R. Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation Staphylococcus aureus Surface Protein Involved in Biofilm Formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Tristan, A.; Ying, L.; Bes, M.; Etienne, J.; Vandenesch, F.; Lina, G. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 2003, 41, 4465–4467. [Google Scholar] [CrossRef]
- Rohde, H.; Burdelski, C.; Bartscht, K.; Hussain, M.; Buck, F.; Horstkotte, M.A.; Knobloch, J.K.M.; Heilmann, C.; Herrmann, M.; Mack, D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 2005, 55, 1883–1895. [Google Scholar] [CrossRef]
- Rohde, H.; Burandt, E.C.; Siemssen, N.; Frommelt, L.; Burdelski, C.; Wurster, S.; Scherpe, S.; Davies, A.P.; Harris, L.G.; Horstkotte, M.A.; et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 2007, 28, 1711–1720. [Google Scholar] [CrossRef]
| Species | Biofilm Formation (MTP) | Slime Production (CRA) | ||||
|---|---|---|---|---|---|---|
| Strong | Moderate | Weak | No Biofilm | Positive | Negative | |
| S. epidermidis (n = 21) | 10 (47.6%) | 1 (4.8%) | 0 | 10 (47.6%) | 6 (28.6%) | 15 (71.4%) |
| S. warneri (n = 14) | 8 (57.1%) | 1 (7.1%) | 2 (14.3%) | 3 (21.4%) | 2 (14.3%) | 12 (85.7%) |
| S. carnosus (n = 9) | 2 (22.2%) | 0 | 0 | 7 (77.8%) | 3 (33.3%) | 6 (66.7%) |
| S. simulans (n = 9) | 7 (77.8%) | 0 | 0 | 2 (22.2%) | 7 (77.8%) | 2 (22.2%) |
| S. xylosus (n = 8) | 8 (100%) | 0 | 0 | 0 | 6 (75%) | 2 (25.0%) |
| S. saprophyticus (n = 6) | 1 (16.7%) | 1 (16.7%) | 0 | 4 (66.7%) | 1 (16.7%) | 5 (83.3%) |
| S. pasteuri (n = 5) | 2 (40.0%) | 0 | 0 | 3 (60.0%) | 0 | 5 (100%) |
| S. heamolyticus (n = 4) | 0 | 1 (25.0%) | 1 (25.0%) | 2 (50.0%) | 1 (25%) | 3 (75.0%) |
| S. petrasii subsp. petrasii (n = 4) | 2 (50.0%) | 1 | 0 | 1 (25.0%) | 2 (50%) | 2 (50.0%) |
| S. lentus (n = 2) | 2 (100%) | 0 | 0 | 0 | 1 (50%) | 1 (50.0%) |
| S. piscifermentas (n = 2) | 2 (100%) | 0 | 0 | 0 | 0 | 2 (100%) |
| S. lugdenensis (n = 1) | 0 | 0 | 1 (100%) | 0 | 1 (100%) | 0 |
| Total (n = 85) | 44 (51.8%) | 5 (5.9%) | 4 (4.7%) | 32 (37.6%) | 30 (35.3%) | 55 (64.7%) |
| Biofilm Formation (MTP) | ica+ (n = 39) | ica− (n = 46) | Total (n = 85) | ||||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | ||
| strong | 20 | (51.3) | 24 | (52.1) | 44 | (51.8) | |
| moderate | 1 | (2.6) | 4 | (8.7) | 5 | (5.9) | |
| weak | 1 | (2.6) | 3 | (6.5) | 4 | (4.7) | |
| no biofilm | 13 | (33.3) | 19 | (4.1) | 32 | (37.6) | |
| Slime production CRA | n | (%) | n | (%) | n | (%) | |
| positive | 12 | (25.6) | 18 | (41.3) | 30 | (35.3) | |
| negative | 23 | (56.4) | 32 | (69.6) | 55 | (64.7) | |
| Species | No. of Strains | icaADBC | bap | eno | aap | fbe | bhp | embP | atlE | Biofilm Positive | Slime Production |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. epidermidis | 21 | 8 (38.1%) | 0 | 20 (95.2%) | 15 (71.4%) | 2 (9.5%) | 0 | 12 (57.1%) | 12 (57.1%) | 11 (52.4%) | 6 (28.6%) |
| S. warneri | 14 | 6 (42.9%) | 0 | 3 (21.4%) | 6 (42.9%) | 1 (7.1%) | 0 | 3 (21.4%) | 1 (7.1%) | 11 (78.6%) | 2 (14.3%) |
| S. carnosus | 9 | 3 (33.3%) | 0 | 6 (66.7%) | 2 (22.2%) | 0 | 0 | 0 | 0 | 2 (22.2%) | 3 (33.3%) |
| S. simulans | 9 | 7 (77.8%) | 4 (44.4%) | 4 (44.4%) | 6 (66.7%) | 2 (22.2%) | 0 | 0 | 6 (66.7%) | 7 (77.8%) | 7 (77.8%) |
| S. xylosus | 8 | 4 (50%) | 0 | 1 (12.5%) | 6 (75%) | 1 (12.5%) | 0 | 0 | 0 | 8 (100%) | 6 (75%) |
| S. saprophyticus | 6 | 4 (66.7%) | 0 | 6 (100%) | 3 (50%) | 2 (33.3%) | 1 (16.7%) | 0 | 0 | 2 (66.7%) | 1 (16.7%) |
| S. pasteuri | 5 | 2 (40%) | 0 | 1 (20%) | 2 (40%) | 0 | 0 | 2 (40%) | 2 (40%) | 2 (40%) | 0 |
| S. heamolyticus | 4 | 3 (75%) | 0 | 3 (75%) | 3 (75%) | 1 (25%) | 0 | 4 (100%) | 3 (75%) | 2 (50%) | 1 (25%) |
| S. petrasii subsp. petrasii | 4 | 1 (25%) | 0 | 1 (25%) | 0 | 0 | 0 | 1 (25%) | 0 | 3 (75%) | 2 (50%) |
| S. lentus | 2 | 0 | 0 | 2 (100%) | 2 (100%) | 1 (50%) | 0 | 2 (100%) | 2 (100%) | 2 (100%) | 1 (50%) |
| S. piscifermentas | 2 | 1 (50%) | 0 | 2 (100%) | 2 (100%) | 0 | 0 | 0 | 0 | 2 (100%) | 0 |
| S. lugdenensis | 1 | 0 | 0 | 0 | 1 (100%) | 0 | 0 | 0 | 1 (100%) | 1 (100%) | 1 (100%) |
| Total | 85 | 39 (45.9%) | 4 (4.7%) | 49 (57.6%) | 48 (56.5%) | 10 (11.8%) | 1 (1.2%) | 24 (28.2%) | 27 (31.8%) | 53 (62.4%) | 30 (35.3%) |
| Species | No. of Strains | hla_haem | hla_yiD | hlb | hld | IS256 | IS257 |
|---|---|---|---|---|---|---|---|
| S. epidermidis | 21 | 5 (23.8%) | 14 (66.7%) | 14 (66.7%) | 16 (76.2%) | 14 (66.7%) | 19 (90.5%) |
| S. warneri | 14 | 8 (57.1%) | 5 (35.7%) | 6 (42.9%) | 8 (57.1%) | 6 (42.9%) | 13 (92.9%) |
| S. carnosus | 9 | 0 | 5 (55.6%) | 3 (33.3%) | 1 (11.1%) | 6 (66.7%) | 8 (88.9%) |
| S. simulans | 9 | 3 (33.3%) | 7 (77.8%) | 6 (66.7%) | 3 (33.3%) | 1 (11.1%) | 9 (100%) |
| S. xylosus | 8 | 0 | 0 | 0 | 1 (12.5%) | 7 (87.5%) | 5 (62.5%) |
| S. saprophyticus | 6 | 2 (33.3%) | 0 | 0 | 3 (50.0%) | 2 (33.3%) | 5 (83.5%) |
| S. pasteuri | 5 | 3 (60.0%) | 4 (80.0%) | 2 (40.0%) | 1 (20.0%) | 2 (40.0%) | 5 (100%) |
| S. heamolyticus | 4 | 4 (100%) | 3 (75.0%) | 3 (75.0%) | 1 (25.0%) | 3 (75.0%) | 4 (100%) |
| S. petrasii subsp. petrasii | 4 | 3 (75.0%) | 1 (25.0%) | 2 (50.0%) | 1 (25.0%) | 1 (25.0%) | 2 (50.0%) |
| S. lentus | 2 | 0 | 2 (100%) | 2 (100%) | 0 | 2 (100%) | 2 (100%) |
| S. piscifermentas | 2 | 1 (50.0%) | 1 (50.0%) | 2 (100%) | 0 | 0 | 2 (100%) |
| S. lugdenensis | 1 | 0 | 1 (100%) | 1 (100%) | 0 | 1 (100%) | 0 |
| Total | 85 | 29 (34.1%) | 43 (50.6%) | 41 (48.2%) | 35 (41.2%) | 45 (52.9%) | 74 (87.1%) |
| Hemolysin Genes | ica+ | ica− | Total (n = 85) | |||
|---|---|---|---|---|---|---|
| No | (%) | No | (%) | No | (%) | |
| hla+ | 14 | (48.3) | 15 | (51.7) | 29 | (34.1) |
| hla_yiD+ | 22 | (51.1) | 21 | (48.8) | 43 | (50.6) |
| hlb+ | 19 | (46.3) | 22 | (53.7) | 41 | (48.2) |
| hld+ | 19 | (54.3) | 16 | (45.7) | 35 | (41.2) |
| Insertion sequences | No | (%) | No | (%) | No | (%) |
| IS256+ | 21 | (46.7) | 24 | (53.3) | 45 | (52.9) |
| IS257+ | 35 | (47.3) | 39 | (52.7) | 74 | (87.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chajęcka-Wierzchowska, W.; Gajewska, J.; Zakrzewski, A.J.; Caggia, C.; Zadernowska, A. Molecular Analysis of Pathogenicity, Adhesive Matrix Molecules (MSCRAMMs) and Biofilm Genes of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Food. Int. J. Environ. Res. Public Health 2023, 20, 1375. https://doi.org/10.3390/ijerph20021375
Chajęcka-Wierzchowska W, Gajewska J, Zakrzewski AJ, Caggia C, Zadernowska A. Molecular Analysis of Pathogenicity, Adhesive Matrix Molecules (MSCRAMMs) and Biofilm Genes of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Food. International Journal of Environmental Research and Public Health. 2023; 20(2):1375. https://doi.org/10.3390/ijerph20021375
Chicago/Turabian StyleChajęcka-Wierzchowska, Wioleta, Joanna Gajewska, Arkadiusz Józef Zakrzewski, Cinzia Caggia, and Anna Zadernowska. 2023. "Molecular Analysis of Pathogenicity, Adhesive Matrix Molecules (MSCRAMMs) and Biofilm Genes of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Food" International Journal of Environmental Research and Public Health 20, no. 2: 1375. https://doi.org/10.3390/ijerph20021375
APA StyleChajęcka-Wierzchowska, W., Gajewska, J., Zakrzewski, A. J., Caggia, C., & Zadernowska, A. (2023). Molecular Analysis of Pathogenicity, Adhesive Matrix Molecules (MSCRAMMs) and Biofilm Genes of Coagulase-Negative Staphylococci Isolated from Ready-to-Eat Food. International Journal of Environmental Research and Public Health, 20(2), 1375. https://doi.org/10.3390/ijerph20021375







