DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Baseline Anthropometric, Clinical and Biochemical Variables
2.3. Tobacco Smoking and Adherence to the Mediterranean Diet
2.4. DNA Isolation and DNA-Methylation Analysis
2.5. Statistical Analysis and EWAS
3. Results
3.1. Participants Characteristics
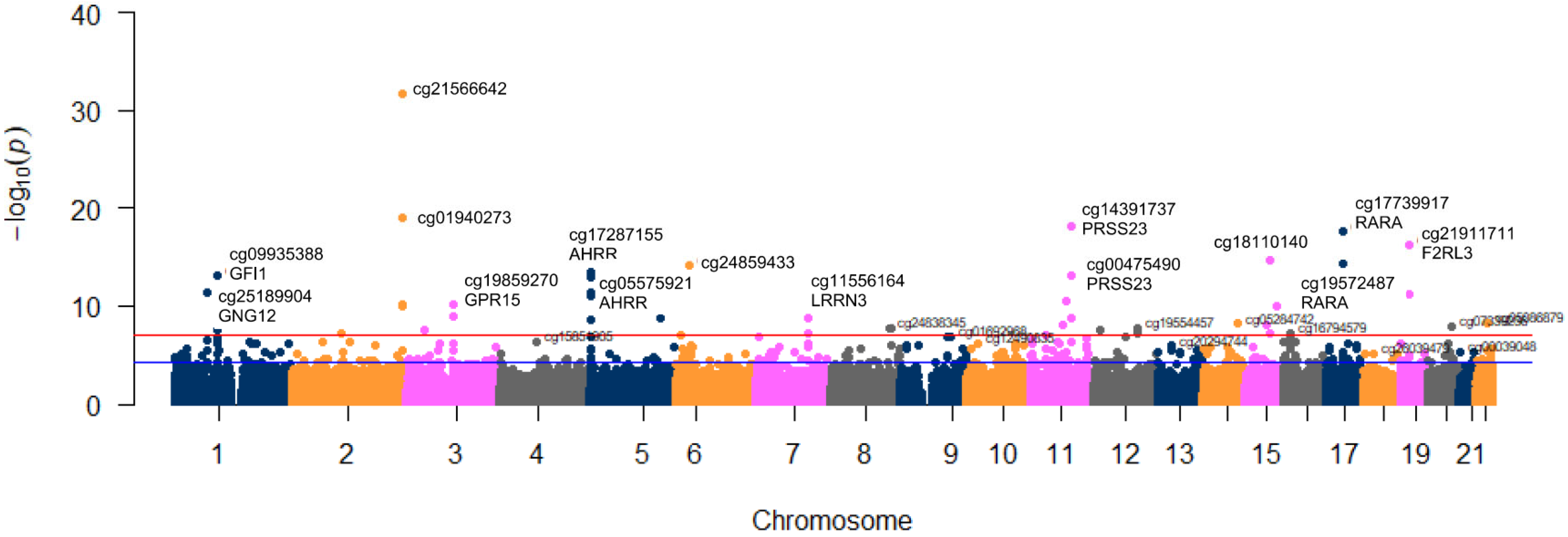
3.2. Association between Tobacco Smoking (5 Levels) and Its Epigenome-Wide Methylation Signatures
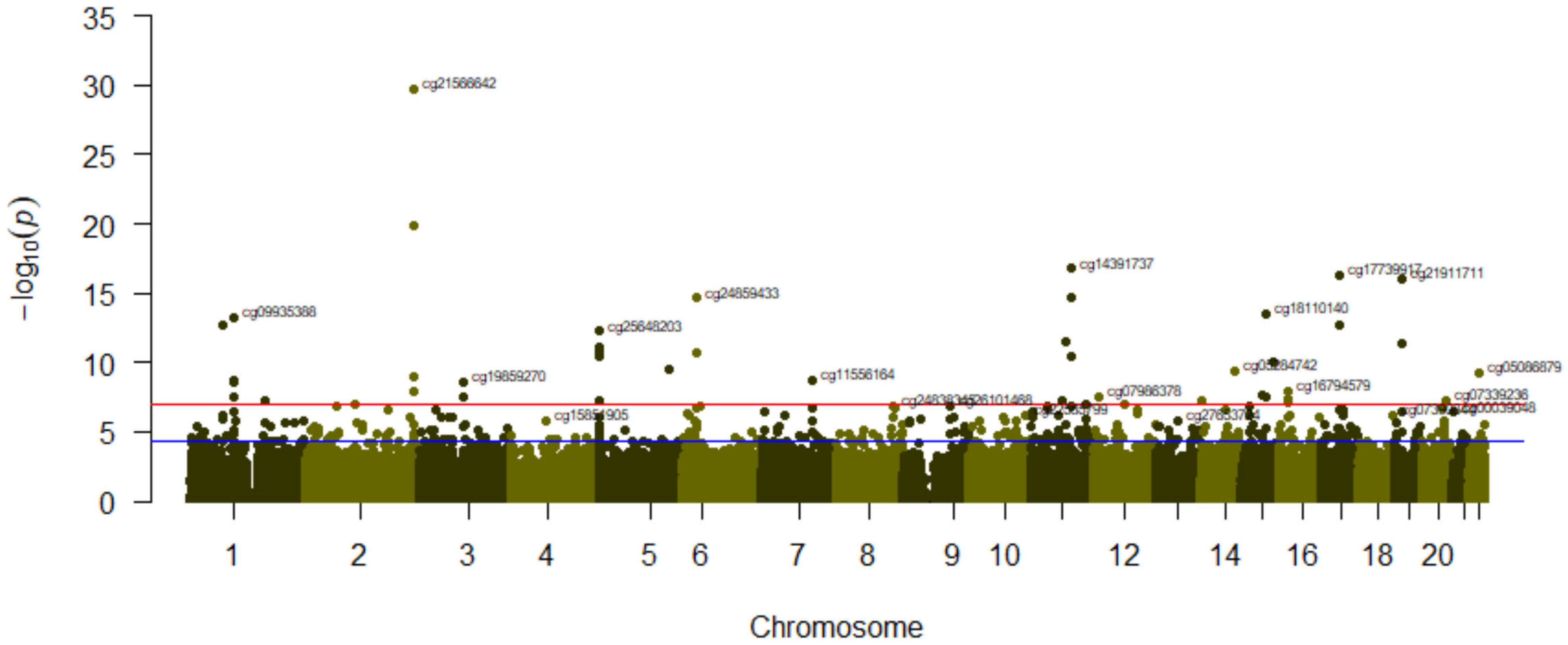
3.3. Association between Tobacco Smoking (Comparing Categories) and Its Epigenome-Wide Methylation Signatures
3.4. Gene Set Enrichment Analysis of the Differentially Methylated CpG Sites Using KEGG and GO
3.5. Dose-Response of DNA Methylation in Current Smokers
3.6. Methylation at Selected CpG as Predictors of Smoking Status
3.7. Association between Tobacco Smoking (5 Levels) and Its Epigenome-Wide Methylation Signatures by Sex
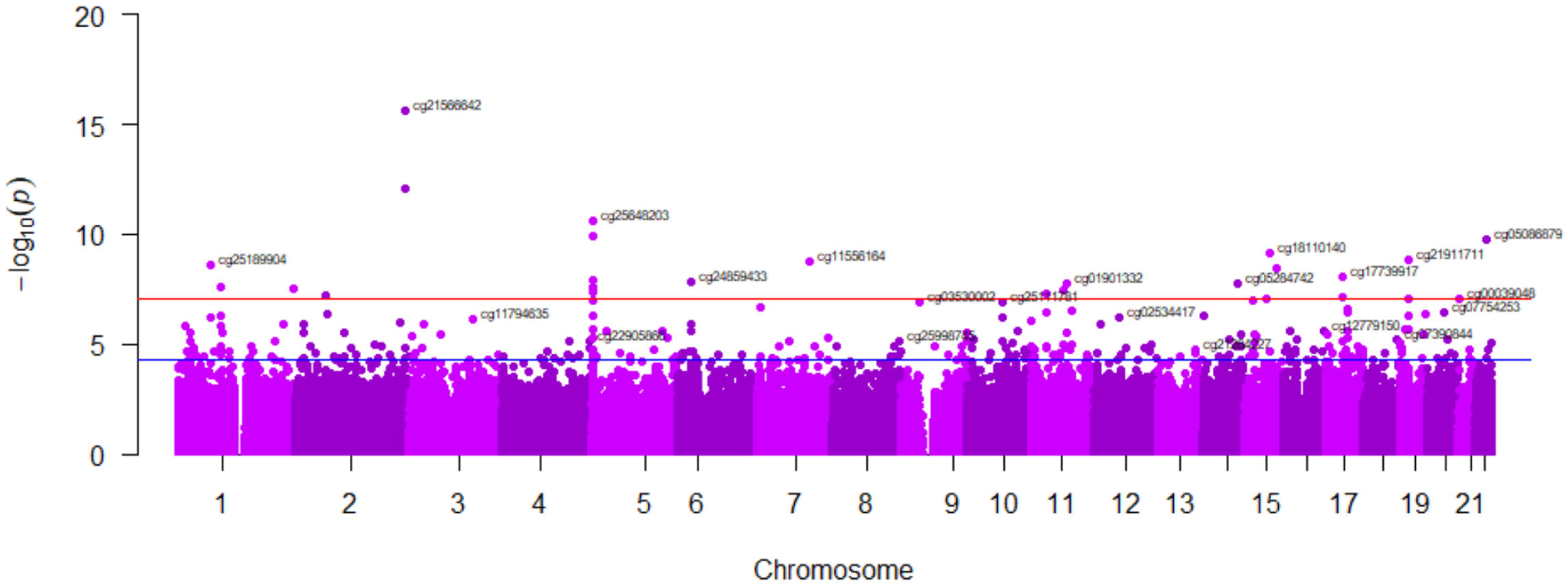
3.8. Modulation of Tobacco Smoking’s Epigenome-Wide Methylation Signature by Adherence to the Mediterranean Diet
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibbs, R.A. The Human Genome Project Changed Everything. Nat. Rev. Genet. 2020, 21, 575–576. [Google Scholar] [CrossRef]
- Lee, C.; Antonarakis, S.E.; Hamosh, A.; Burn, J. Three Decades of the Human Genome Organization. Am. J. Med. Genet. A 2021, 185, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Scott, A.J.; Brandt, M.; Hall, I.M. Genomic Analysis in the Age of Human Genome Sequencing. Cell 2019, 177, 70–84. [Google Scholar] [CrossRef] [Green Version]
- November, J. More than Moore’s Mores: Computers, Genomics, and the Embrace of Innovation. J. Hist. Biol. 2018, 51, 807–840. [Google Scholar] [CrossRef] [Green Version]
- Tanjo, T.; Kawai, Y.; Tokunaga, K.; Ogasawara, O.; Nagasaki, M. Practical Guide for Managing Large-Scale Human Genome Data in Research. J. Hum. Genet. 2021, 66, 39–52. [Google Scholar] [CrossRef]
- Tong, L.; Wu, H.; Wang, M.D.; Wang, G. Introduction of Medical Genomics and Clinical Informatics Integration for P-Health Care. Prog. Mol. Biol. Transl. Sci. 2022, 190, 1–37. [Google Scholar] [CrossRef]
- Bravo-Merodio, L.; Acharjee, A.; Russ, D.; Bisht, V.; Williams, J.A.; Tsaprouni, L.G.; Gkoutos, G.V. Translational Biomarkers in the Era of Precision Medicine. Adv. Clin. Chem. 2021, 102, 191–232. [Google Scholar] [CrossRef]
- Manolio, T.A. Implementing Genomics and Pharmacogenomics in the Clinic: The National Human Genome Research Institute’s Genomic Medicine Portfolio. Atherosclerosis 2016, 253, 225–236. [Google Scholar] [CrossRef] [Green Version]
- Goddard, K.A.B.; Lee, K.; Buchanan, A.H.; Powell, B.C.; Hunter, J.E. Establishing the Medical Actionability of Genomic Variants. Annu. Rev. Genom. Hum. Genet. 2022, 23, 173–192. [Google Scholar] [CrossRef]
- Ashley, E.A. Towards Precision Medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef]
- Cutler, D.M. Early Returns From the Era of Precision Medicine. JAMA 2020, 323, 109–110. [Google Scholar] [CrossRef]
- Schaffhausen, J. What Precisely Is Precision Medicine? Trends Pharmacol. Sci. 2017, 38, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feero, W.G. Introducing “Genomics and Precision Health”. JAMA 2017, 317, 1842–1843. [Google Scholar] [CrossRef] [Green Version]
- Meagher, K.M.; McGowan, M.L.; Settersten, R.A.; Fishman, J.R.; Juengst, E.T. Precisely Where Are We Going? Charting the New Terrain of Precision Prevention. Annu. Rev. Genom. Hum. Genet. 2017, 18, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Viana, J.N.; Edney, S.; Gondalia, S.; Mauch, C.; Sellak, H.; O’Callaghan, N.; Ryan, J.C. Trends and Gaps in Precision Health Research: A Scoping Review. BMJ Open 2021, 11, e056938. [Google Scholar] [CrossRef]
- Corella, D.; Ordovas, J.M. Integration of Environment and Disease into “omics” Analysis. Curr. Opin. Mol. Ther. 2005, 7, 569–576. [Google Scholar]
- Barouki, R.; Audouze, K.; Coumoul, X.; Demenais, F.; Gauguier, D. Integration of the Human Exposome with the Human Genomeo Advance Medicine. Biochimie 2018, 152, 155–158. [Google Scholar] [CrossRef]
- Vineis, P.; Robinson, O.; Chadeau-Hyam, M.; Dehghan, A.; Mudway, I.; Dagnino, S. What Is New in the Exposome? Environ. Int. 2020, 143, 105887. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Epigenomics: Technologies and Applications. Circ. Res. 2018, 122, 1191–1199. [Google Scholar] [CrossRef]
- Mehrmohamadi, M.; Sepehri, M.H.; Nazer, N.; Norouzi, M.R. A Comparative Overview of Epigenomic Profiling Methods. Front. Cell. Dev. Biol. 2021, 9, 714687. [Google Scholar] [CrossRef]
- Kronfol, M.M.; Dozmorov, M.G.; Huang, R.; Slattum, P.W.; McClay, J.L. The Role of Epigenomics in Personalized Medicine. Expert Rev. Precis. Med. Drug Dev. 2017, 2, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Corella, D.; Ordovas, J.M. Basic Concepts in Molecular Biology Related to Genetics and Epigenetics. Rev. Esp. Cardiol. Engl. Ed. 2017, 70, 744–753. [Google Scholar] [CrossRef]
- Zhang, W.; Song, M.; Qu, J.; Liu, G.-H. Epigenetic Modifications in Cardiovascular Aging and Diseases. Circ. Res. 2018, 123, 773–786. [Google Scholar] [CrossRef]
- Reichard, J.; Zimmer-Bensch, G. The Epigenome in Neurodevelopmental Disorders. Front. Neurosci. 2021, 15, 776809. [Google Scholar] [CrossRef]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA Methylation: A Historical Perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA Methylation: In the Right Place at the Right Time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.-H.; Wang, H.-T.; Kong, Q.-P. Dynamic DNA Methylation During Aging: A “Prophet” of Age-Related Outcomes. Front. Genet. 2019, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Liu, Y. DNA Methylation in Human Diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Zhong, J.; Agha, G.; Baccarelli, A.A. The Role of DNA Methylation in Cardiovascular Risk and Disease: Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies. Circ. Res. 2016, 118, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.A.H.; Ansari, S.A.; Mensah-Brown, E.P.K.; Emerald, B.S. The Role of DNA Methylation in the Pathogenesis of Type 2 Diabetes Mellitus. Clin. Epigenetics 2020, 12, 104. [Google Scholar] [CrossRef]
- Mahmoud, A.M. An Overview of Epigenetics in Obesity: The Role of Lifestyle and Therapeutic Interventions. Int. J. Mol. Sci. 2022, 23, 1341. [Google Scholar] [CrossRef]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA Methylation Landscape of Cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef]
- Sanchez-Mut, J.V.; Heyn, H.; Vidal, E.; Moran, S.; Sayols, S.; Delgado-Morales, R.; Schultz, M.D.; Ansoleaga, B.; Garcia-Esparcia, P.; Pons-Espinal, M.; et al. Human DNA Methylomes of Neurodegenerative Diseases Show Common Epigenomic Patterns. Transl. Psychiatry 2016, 6, e718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Morales, R.; Esteller, M. Opening up the DNA Methylome of Dementia. Mol. Psychiatry 2017, 22, 485–496. [Google Scholar] [CrossRef]
- Wielscher, M.; Mandaviya, P.R.; Kuehnel, B.; Joehanes, R.; Mustafa, R.; Robinson, O.; Zhang, Y.; Bodinier, B.; Walton, E.; Mishra, P.P.; et al. DNA Methylation Signature of Chronic Low-Grade Inflammation and Its Role in Cardio-Respiratory Diseases. Nat. Commun. 2022, 13, 2408. [Google Scholar] [CrossRef]
- Prunicki, M.; Cauwenberghs, N.; Lee, J.; Zhou, X.; Movassagh, H.; Noth, E.; Lurmann, F.; Hammond, S.K.; Balmes, J.R.; Desai, M.; et al. Air Pollution Exposure Is Linked with Methylation of Immunoregulatory Genes, Altered Immune Cell Profiles, and Increased Blood Pressure in Children. Sci. Rep. 2021, 11, 4067. [Google Scholar] [CrossRef]
- Schrott, R.; Song, A.; Ladd-Acosta, C. Epigenetics as a Biomarker for Early-Life Environmental Exposure. Curr. Environ. Health Rep. 2022, 9, 604–624. [Google Scholar] [CrossRef]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef] [Green Version]
- Smith, I.M.; Mydlarz, W.K.; Mithani, S.K.; Califano, J.A. DNA Global Hypomethylation in Squamous Cell Head and Neck Cancer Associated with Smoking, Alcohol Consumption and Stage. Int. J. Cancer 2007, 121, 1724–1728. [Google Scholar] [CrossRef]
- Breitling, L.P.; Yang, R.; Korn, B.; Burwinkel, B.; Brenner, H. Tobacco-Smoking-Related Differential DNA Methylation: 27K Discovery and Replication. Am. J. Hum. Genet. 2011, 88, 450–457. [Google Scholar] [CrossRef] [Green Version]
- Shenker, N.S.; Polidoro, S.; van Veldhoven, K.; Sacerdote, C.; Ricceri, F.; Birrell, M.A.; Belvisi, M.G.; Brown, R.; Vineis, P.; Flanagan, J.M. Epigenome-Wide Association Study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) Identifies Novel Genetic Loci Associated with Smoking. Hum. Mol. Genet. 2013, 22, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Philibert, R.A.; Beach, S.R.H.; Lei, M.-K.; Brody, G.H. Changes in DNA Methylation athe Aryl Hydrocarbon Receptor Repressor May Be a New Biomarker for Smoking. Clin. Epigenetics 2013, 5, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeilinger, S.; Kühnel, B.; Klopp, N.; Baurecht, H.; Kleinschmidt, A.; Gieger, C.; Weidinger, S.; Lattka, E.; Adamski, J.; Peters, A.; et al. Tobacco Smoking Leads to Extensive Genome-Wide Changes in DNA Methylation. PLoS ONE 2013, 8, e63812. [Google Scholar] [CrossRef]
- Tsaprouni, L.G.; Yang, T.-P.; Bell, J.; Dick, K.J.; Kanoni, S.; Nisbet, J.; Viñuela, A.; Grundberg, E.; Nelson, C.P.; Meduri, E.; et al. Cigarette Smoking Reduces DNA Methylation Levels at Multiple Genomic Loci but the Effect Is Partially Reversible upon Cessation. Epigenetics 2014, 9, 1382–1396. [Google Scholar] [CrossRef] [Green Version]
- Dogan, M.V.; Shields, B.; Cutrona, C.; Gao, L.; Gibbons, F.X.; Simons, R.; Monick, M.; Brody, G.H.; Tan, K.; Beach, S.R.H.; et al. The Effect of Smoking on DNA Methylation of Peripheral Blood Mononuclear Cells from African American Women. BMC Genom. 2014, 15, 151. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, L.M.; Wan, M.; Ding, J.; Taylor, J.R.; Lohman, K.; Su, D.; Bennett, B.D.; Porter, D.K.; Gimple, R.; Pittman, G.S.; et al. DNA Methylation of the Aryl Hydrocarbon Receptor Repressor Associations with Cigarette Smoking and Subclinical Atherosclerosis. Circ. Cardiovasc. Genet. 2015, 8, 707–716. [Google Scholar] [CrossRef]
- Harlid, S.; Xu, Z.; Panduri, V.; Sandler, D.P.; Taylor, J.A. CpG Sites Associated with Cigarette Smoking: Analysis of Epigenome-Wide Data from the Sister Study. Environ. Health Perspect. 2014, 122, 673–678. [Google Scholar] [CrossRef] [Green Version]
- Sayols-Baixeras, S.; Lluís-Ganella, C.; Subirana, I.; Salas, L.A.; Vilahur, N.; Corella, D.; Muñoz, D.; Segura, A.; Jimenez-Conde, J.; Moran, S.; et al. Identification of a New Locus and Validation of Previously Reported Loci Showing Differential Methylation Associated with Smoking. The REGICOR Study. Epigenetics 2015, 10, 1156–1165. [Google Scholar] [CrossRef]
- Li, S.; Wong, E.M.; Bui, M.; Nguyen, T.L.; Joo, J.-H.E.; Stone, J.; Dite, G.S.; Giles, G.G.; Saffery, R.; Southey, M.C.; et al. Causal Effect of Smoking on DNA Methylation in Peripheral Blood: A Twin and Family Study. Clin. Epigenetics 2018, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Hulls, P.M.; de Vocht, F.; Bao, Y.; Relton, C.L.; Martin, R.M.; Richmond, R.C. DNA Methylation Signature of Passive Smoke Exposure Is Less Pronounced than Active Smoking: The Understanding Society Study. Environ. Res. 2020, 190, 109971. [Google Scholar] [CrossRef]
- Sun, Y.-Q.; Richmond, R.C.; Suderman, M.; Min, J.L.; Battram, T.; Flatberg, A.; Beisvag, V.; Nøst, T.H.; Guida, F.; Jiang, L.; et al. Assessing the Role of Genome-Wide DNA Methylation between Smoking and Risk of Lung Cancer Using Repeated Measurements: The HUNT Study. Int. J. Epidemiol. 2021, 50, 1482–1497. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.P.; Hänninen, I.; Raitoharju, E.; Marttila, S.; Mishra, B.H.; Mononen, N.; Kähönen, M.; Hurme, M.; Raitakari, O.; Törönen, P.; et al. Epigenome-450K-Wide Methylation Signatures of Active Cigarette Smoking: The Young Finns Study. Biosci. Rep. 2020, 40, BSR20200596. [Google Scholar] [CrossRef] [PubMed]
- Barcelona, V.; Huang, Y.; Brown, K.; Liu, J.; Zhao, W.; Yu, M.; Kardia, S.L.R.; Smith, J.A.; Taylor, J.Y.; Sun, Y.V. Novel DNA Methylation Sites Associated with Cigarette Smoking among African Americans. Epigenetics 2019, 14, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, D.; Shi, J.; Liu, Y.; He, W.; Qu, W.; Wang, C.; Xing, H.; Cao, Y.; Li, J.; Zha, L. DNA Methylation Analysis for Smoking Status Prediction in the Chinese Population Based on the Methylation-Sensitive Single-Nucleotide Primer Extension Method. Forensic Sci. Int. 2022, 339, 111412. [Google Scholar] [CrossRef]
- Cardenas, A.; Ecker, S.; Fadadu, R.P.; Huen, K.; Orozco, A.; McEwen, L.M.; Engelbrecht, H.-R.; Gladish, N.; Kobor, M.S.; Rosero-Bixby, L.; et al. Epigenome-Wide Association Study and Epigenetic Age Acceleration Associated with Cigarette Smoking among Costa Rican Adults. Sci. Rep. 2022, 12, 4277. [Google Scholar] [CrossRef]
- Dugué, P.-A.; Jung, C.-H.; Joo, J.E.; Wang, X.; Wong, E.M.; Makalic, E.; Schmidt, D.F.; Baglietto, L.; Severi, G.; Southey, M.C.; et al. Smoking and Blood DNA Methylation: An Epigenome-Wide Association Study and Assessment of Reversibility. Epigenetics 2020, 15, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Langdon, R.J.; Yousefi, P.; Relton, C.L.; Suderman, M.J. Epigenetic Modelling of Former, Current and Never Smokers. Clin. Epigenetics 2021, 13, 206. [Google Scholar] [CrossRef]
- Wilhelm-Benartzi, C.S.; Koestler, D.C.; Karagas, M.R.; Flanagan, J.M.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J.; Houseman, E.A.; Brown, R. Review of Processing and Analysis Methods for DNA Methylation Array Data. Br. J. Cancer 2013, 109, 1394–1402. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Rodger, E.J.; Morison, I.M.; Eccles, M.R.; Stockwell, P.A. Tools and Strategies for Analysis of Genome-Wide and Gene-Specific DNA Methylation Patterns. Method. Mol. Biol. 2017, 1537, 249–277. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Drabløs, F.; Rye, M.B. DNA Methylation Data by Sequencing: Experimental Approaches and Recommendations for Tools and Pipelines for Data Analysis. Clin. Epigenetics 2019, 11, 193. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, C.; Castillo-Fernandez, J.E.; Domingo-Relloso, A.; Zhao, W.; El-Sayed Moustafa, J.S.; Tsai, P.-C.; Maddock, J.; Haack, K.; Cole, S.A.; Kardia, S.L.R.; et al. Novel DNA Methylation Signatures of Tobacco Smoking with Trans-Ethnic Effects. Clin. Epigenetics 2021, 13, 36. [Google Scholar] [CrossRef]
- Moran, S.; Arribas, C.; Esteller, M. Validation of a DNA Methylation Microarray for 850,000 CpG Sites of the Human Genome Enriched in Enhancer Sequences. Epigenomics 2016, 8, 389–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, H.R.; Burrows, K.; Min, J.L.; Tillin, T.; Mason, D.; Wright, J.; Santorelli, G.; Smith, G.D.; Lawlor, D.A.; Hughes, A.D.; et al. Characterisation of Ethnic Differences in DNA Methylation between UK-Resident South Asians and Europeans. Clin. Epigenetics 2022, 14, 130. [Google Scholar] [CrossRef] [PubMed]
- Fatumo, S.; Chikowore, T.; Choudhury, A.; Ayub, M.; Martin, A.R.; Kuchenbaecker, K. A Roadmap to Increase Diversity in Genomic Studies. Nat. Med. 2022, 28, 243–250. [Google Scholar] [CrossRef]
- Salas, L.A.; Peres, L.C.; Thayer, Z.M.; Smith, R.W.; Guo, Y.; Chung, W.; Si, J.; Liang, L. A Transdisciplinary Approach to Understand the Epigenetic Basis of Race/Ethnicity Health Disparities. Epigenomics 2021, 13, 1761–1770. [Google Scholar] [CrossRef]
- Maugeri, A. The Effects of Dietary Interventions on DNA Methylation: Implications for Obesity Management. Int. J. Mol. Sci. 2020, 21, 8670. [Google Scholar] [CrossRef]
- Fareed, M.M.; Ullah, S.; Qasmi, M.; Shityakov, S. The Role of Vitamins in DNA Methylation as Dietary Supplements or Neutraceuticals: A Systematic Review. Curr. Mol. Med. 2022. [Google Scholar] [CrossRef]
- Ungaro, P.; Nettore, I.C.; Franchini, F.; Palatucci, G.; Muscogiuri, G.; Colao, A.; Macchia, P.E. Epigenome Modulation Induced by Ketogenic Diets. Nutrients 2022, 14, 3245. [Google Scholar] [CrossRef]
- Li, X.; Qi, L. Epigenetics in Precision Nutrition. J. Pers. Med. 2022, 12, 533. [Google Scholar] [CrossRef]
- Salas-Salvadó, J.; Díaz-López, A.; Ruiz-Canela, M.; Basora, J.; Fitó, M.; Corella, D.; Serra-Majem, L.; Wärnberg, J.; Romaguera, D.; Estruch, R.; et al. Effect of a Lifestyle Intervention Program With Energy-Restricted Mediterranean Diet and Exercise on Weight Loss and Cardiovascular Risk Factors: One-Year Results of the PREDIMED-Plus Trial. Diabetes Care 2019, 42, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Coltell, O.; Asensio, E.M.; Sorlí, J.V.; Barragán, R.; Fernández-Carrión, R.; Portolés, O.; Ortega-Azorín, C.; Martínez-LaCruz, R.; González, J.I.; Zanón-Moreno, V.; et al. Genome-Wide Association Study (GWAS) on Bilirubin Concentrations in Subjects with Metabolic Syndrome: Sex-Specific GWAS Analysis and Gene-Diet Interactions in a Mediterranean Population. Nutrients 2019, 11, 90. [Google Scholar] [CrossRef] [Green Version]
- Bruegel, M.; Nagel, D.; Funk, M.; Fuhrmann, P.; Zander, J.; Teupser, D. Comparison of Five Automated Hematology Analyzers in a University Hospital Setting: Abbott Cell-Dyn Sapphire, Beckman Coulter DxH 800, Siemens Advia 2120i, Sysmex XE-5000, and Sysmex XN-2000. Clin. Chem. Lab. Med. 2015, 53, 1057–1071. [Google Scholar] [CrossRef]
- Malhotra, J.; Borron, C.; Freedman, N.D.; Abnet, C.C.; van den Brandt, P.A.; White, E.; Milne, R.L.; Giles, G.G.; Boffetta, P. Association between Cigar or Pipe Smoking and Cancer Risk in Men: A Pooled Analysis of Five Cohort Studies. Cancer Prev. Res. Phila. 2017, 10, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Teshima, A.; Laverty, A.A.; Filippidis, F.T. Burden of Current and Past Smoking across 28 European Countries in 2017: A Cross-Sectional Analysis. Tob. Induc. Dis. 2022, 20, 56. [Google Scholar] [CrossRef]
- Molina, L.; Sarmiento, M.; Peñafiel, J.; Donaire, D.; Garcia-Aymerich, J.; Gomez, M.; Ble, M.; Ruiz, S.; Frances, A.; Schröder, H.; et al. Validation of the Regicor Short Physical Activity Questionnaire for the Adult Population. PLoS ONE 2017, 12, e0168148. [Google Scholar] [CrossRef] [Green Version]
- Schröder, H.; Zomeño, M.D.; Martínez-González, M.A.; Salas-Salvadó, J.; Corella, D.; Vioque, J.; Romaguera, D.; Martínez, J.A.; Tinahones, F.J.; Miranda, J.L.; et al. Validity of the Energy-Restricted Mediterranean Diet Adherence Screener. Clin. Nutr. 2021, 40, 4971–4979. [Google Scholar] [CrossRef]
- Schröder, H.; Fitó, M.; Estruch, R.; Martínez-González, M.A.; Corella, D.; Salas-Salvadó, J.; Lamuela-Raventós, R.; Ros, E.; Salaverría, I.; Fiol, M.; et al. A Short Screener Is Valid for Assessing Mediterranean Diet Adherence among Older Spanish Men and Women. J. Nutr. 2011, 141, 1140–1145. [Google Scholar] [CrossRef] [Green Version]
- Coltell, O.; Sorlí, J.V.; Asensio, E.M.; Barragán, R.; González, J.I.; Giménez-Alba, I.M.; Zanón-Moreno, V.; Estruch, R.; Ramírez-Sabio, J.B.; Pascual, E.C.; et al. Genome-Wide Association Study for Serum Omega-3 and Omega-6 Polyunsaturated Fatty Acids: Exploratory Analysis of the Sex-Specific Effects and Dietary Modulation in Mediterranean Subjects with Metabolic Syndrome. Nutrients 2020, 12, 310. [Google Scholar] [CrossRef] [Green Version]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical Evaluation of the Illumina MethylationEPIC BeadChip Microarray for Whole-Genome DNA Methylation Profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [Green Version]
- Zindler, T.; Frieling, H.; Neyazi, A.; Bleich, S.; Friedel, E. Simulating ComBat: How Batch Correction Can Lead to the Systematic Introduction of False Positive Results in DNA Methylation Microarray Studies. BMC Bioinform. 2020, 21, 271. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, J.; Chen, W. Controlling Batch Effect in Epigenome-Wide Association Study. Method. Mol. Biol. 2022, 2432, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Min, J.L.; Hemani, G.; Smith, G.D.; Relton, C.; Suderman, M. Meffil: Efficient Normalization and Analysis of Very Large DNA Methylation Datasets. Bioinformatics 2018, 34, 3983–3989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murat, K.; Grüning, B.; Poterlowicz, P.W.; Westgate, G.; Tobin, D.J.; Poterlowicz, K. Ewastools: Infinium Human Methylation BeadChip Pipeline for Population Epigenetics Integrated into Galaxy. Gigascience 2020, 9, giaa049. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.; Jones, G.T. Data Analysis of DNA Methylation Epigenome-Wide Association Studies (EWAS): A Guide to the Principles of Best Practice. Methods Mol. Biol. 2022, 2458, 23–45. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.; Fernández, R.; Collet, S.; Kiyar, M.; Delgado-Zayas, E.; Gómez-Gil, E.; Van Den Eynde, T.; T’Sjoen, G.; Guillamon, A.; Mueller, S.C.; et al. Epigenetics Is Implicated in the Basis of Gender Incongruence: An Epigenome-Wide Association Analysis. Front. Neurosci. 2021, 15, 701017. [Google Scholar] [CrossRef]
- Fortin, J.-P.; Labbe, A.; Lemire, M.; Zanke, B.W.; Hudson, T.J.; Fertig, E.J.; Greenwood, C.M.; Hansen, K.D. Functional Normalization of 450k Methylation Array Data Improves Replication in Large Cancer Studies. Genome Biol. 2014, 15, 503. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.P.; van Dijk, S.; Phang, M.; Skilton, M.R.; Molloy, P.L.; Oytam, Y. Batch-Effect Detection, Correction and Characterisation in Illumina HumanMethylation450 and MethylationEPIC BeadChip Array Data. Clin. Epigenetics 2022, 14, 58. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-Value and M-Value Methods for Quantifying Methylation Levels by Microarray Analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Leung, Y.-K.; Chen, A.; Long, D.-X.; Hoyo, C.; Ho, S.-M. Differential Methylation Values in Differential Methylation Analysis. Bioinformatics 2019, 35, 1094–1097. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA Methylation Arrays as Surrogate Measures of Cell Mixture Distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [Green Version]
- Barton, S.J.; Melton, P.E.; Titcombe, P.; Murray, R.; Rauschert, S.; Lillycrop, K.A.; Huang, R.-C.; Holbrook, J.D.; Godfrey, K.M. In Epigenomic Studies, Including Cell-Type Adjustments in Regression Models Can Introduce Multicollinearity, Resulting in Apparent Reversal of Direction of Association. Front. Genet. 2019, 10, 816. [Google Scholar] [CrossRef]
- Kaushal, A.; Zhang, H.; Karmaus, W.J.J.; Ray, M.; Torres, M.A.; Smith, A.K.; Wang, S.-L. Comparison of Different Cell Type Correction Methods for Genome-Scale Epigenetics Studies. BMC Bioinform. 2017, 18, 216. [Google Scholar] [CrossRef] [Green Version]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA Methylation Studies: Statistical Insights from the Illumina EPIC Array. BMC Genom. 2019, 20, 366. [Google Scholar] [CrossRef] [Green Version]
- van Iterson, M.; van Zwet, E.W.; BIOS Consortium; Heijmans, B. T. Controlling Bias and Inflation in Epigenome- and Transcriptome-Wide Association Studies Using the Empirical Null Distribution. Genome Biol. 2017, 18, 19. [Google Scholar] [CrossRef] [Green Version]
- Maksimovic, J.; Oshlack, A.; Phipson, B. Gene Set Enrichment Analysis for Genome-Wide DNA Methylation Data. Genome Biol. 2021, 22, 173. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Wijesooriya, K.; Jadaan, S.A.; Perera, K.L.; Kaur, T.; Ziemann, M. Urgent Need for Consistent Standards in Functional Enrichment Analysis. PLoS Comput. Biol. 2022, 18, e1009935. [Google Scholar] [CrossRef]
- Tomczak, A.; Mortensen, J.M.; Winnenburg, R.; Liu, C.; Alessi, D.T.; Swamy, V.; Vallania, F.; Lofgren, S.; Haynes, W.; Shah, N.H.; et al. Interpretation of Biological Experiments Changes with Evolution of the Gene Ontology and Its Annotations. Sci. Rep. 2018, 8, 5115. [Google Scholar] [CrossRef]
- Winkler, T.W.; Kutalik, Z.; Gorski, M.; Lottaz, C.; Kronenberg, F.; Heid, I.M. EasyStrata: Evaluation and Visualization of Stratified Genome-Wide Association Meta-Analysis Data. Bioinformatics 2015, 31, 259–261. [Google Scholar] [CrossRef] [Green Version]
- Fraser, H.B.; Lam, L.L.; Neumann, S.M.; Kobor, M.S. Population-Specificity of Human DNA Methylation. Genome Biol. 2012, 13, R8. [Google Scholar] [CrossRef] [Green Version]
- Baglietto, L.; Ponzi, E.; Haycock, P.; Hodge, A.; Assumma, M.B.; Jung, C.-H.; Chung, J.; Fasanelli, F.; Guida, F.; Campanella, G.; et al. DNA Methylation Changes Measured in Pre-Diagnostic Peripheral Blood Samples Are Associated with Smoking and Lung Cancer Risk. Int. J. Cancer 2017, 140, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.E. Structure and Mechanism of Alkaline Phosphatase. Annu. Rev. Biophys. Biomol. Struct. 1992, 21, 441–483. [Google Scholar] [CrossRef] [PubMed]
- Nasab, N.H.; Raza, H.; Shim, R.S.; Hassan, M.; Kloczkowski, A.; Kim, S.J. Potent Alkaline Phosphatase Inhibitors, Pyrazolo-Oxothiazolidines: Synthesis, Biological Evaluation, Molecular Docking, and Kinetic Studies. Int. J. Mol. Sci. 2022, 23, 13262. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Kasela, S.; Carnero-Montoro, E.; van Iterson, M.; Štambuk, J.; Sharma, S.; van den Akker, E.; Klaric, L.; Benedetti, E.; Razdorov, G.; et al. IgG Glycosylation and DNA Methylation Are Interconnected with Smoking. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 637–648. [Google Scholar] [CrossRef]
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Babaei, M.S.; Sharp, G.; Hatcher, C.; Vega-Salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A Database of Epigenome-Wide Association Studies. Wellcome Open Res. 2022, 7, 41. [Google Scholar] [CrossRef]
- Joehanes, R.; Just, A.C.; Marioni, R.E.; Pilling, L.C.; Reynolds, L.M.; Mandaviya, P.R.; Guan, W.; Xu, T.; Elks, C.E.; Aslibekyan, S.; et al. Epigenetic Signatures of Cigarette Smoking. Circ. Cardiovasc. Genet. 2016, 9, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Domingo-Relloso, A.; Riffo-Campos, A.L.; Haack, K.; Rentero-Garrido, P.; Ladd-Acosta, C.; Fallin, D.M.; Tang, W.Y.; Herreros-Martinez, M.; Gonzalez, J.R.; Bozack, A.K.; et al. Cadmium, Smoking, and Human Blood DNA Methylation Profiles in Adults from the Strong Heart Study. Environ. Health Perspect. 2020, 128, 67005. [Google Scholar] [CrossRef]
- Brandstätter, O.; Schanz, O.; Vorac, J.; König, J.; Mori, T.; Maruyama, T.; Korkowski, M.; Haarmann-Stemmann, T.; von Smolinski, D.; Schultze, J.L.; et al. Balancing Intestinal and Systemic Inflammation through Cell Type-Specific Expression of the Aryl Hydrocarbon Receptor Repressor. Sci. Rep. 2016, 6, 26091. [Google Scholar] [CrossRef]
- Peach, C.J.; Edgington-Mitchell, L.E.; Bunnett, N.W.; Schmidt, B.L. Protease-Activated Receptors in Health and Disease. Physiol. Rev. 2023, 103, 717–785. [Google Scholar] [CrossRef]
- Tsuboi, Y.; Yamada, H.; Munetsuna, E.; Fujii, R.; Yamazaki, M.; Ando, Y.; Mizuno, G.; Hattori, Y.; Ishikawa, H.; Ohashi, K.; et al. Intake of Vegetables and Fruits Rich in Provitamin A Is Positively Associated with Aryl Hydrocarbon Receptor Repressor DNA Methylation in a Japanese Population. Nutr. Res. 2022, 107, 206–217. [Google Scholar] [CrossRef]
- Shorey-Kendrick, L.E.; McEvoy, C.T.; Ferguson, B.; Burchard, J.; Park, B.S.; Gao, L.; Vuylsteke, B.H.; Milner, K.F.; Morris, C.D.; Spindel, E.R. Vitamin C Prevents Offspring DNA Methylation Changes Associated with Maternal Smoking in Pregnancy. Am. J. Respir. Crit. Care Med. 2017, 196, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Lian, Y.; Wang, J.; Hu, L.; Luo, J.; Yu, J. KIF26B in the Prognosis and Immune Biomarking of Various Cancers: A Pan-Cancer Study. J. Oncol. 2022, 2022, 4829697. [Google Scholar] [CrossRef]
- Dizier, M.-H.; Margaritte-Jeannin, P.; Pain, L.; Sarnowski, C.; Brossard, M.; Mohamdi, H.; Lavielle, N.; Babron, M.-C.C.; Just, J.; Lathrop, M.; et al. Interactive Effect between ATPase-Related Genes and Early-Life Tobacco Smoke Exposure on Bronchial Hyper-Responsiveness Detected in Asthma-Ascertained Families. Thorax 2019, 74, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.-S.; Liu, Y.; Juan, H.; Yang, X. Overexpression of Human SPATA17 Protein Induces Germ Cell Apoptosis in Transgenic Male Mice. Mol. Biol. Rep. 2013, 40, 1905–1910. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Komaki, G.; Tajima, A.; Ando, T.; Ishikawa, M.; Nomoto, J.; Hata, K.; Oka, A.; Inoko, H.; Sasazuki, T.; et al. Identification of Novel Candidate Loci for Anorexia Nervosa at 1q41 and 11q22 in Japanese by a Genome-Wide Association Analysis with Microsatellite Markers. J. Hum. Genet. 2009, 54, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haenig, C.; Atias, N.; Taylor, A.K.; Mazza, A.; Schaefer, M.H.; Russ, J.; Riechers, S.-P.; Jain, S.; Coughlin, M.; Fontaine, J.-F.; et al. Interactome Mapping Provides a Network of Neurodegenerative Disease Proteins and Uncovers Widespread Protein Aggregation in Affected Brains. Cell Rep. 2020, 32, 108050. [Google Scholar] [CrossRef] [PubMed]
- Klebaner, D.; Huang, Y.; Hui, Q.; Taylor, J.Y.; Goldberg, J.; Vaccarino, V.; Sun, Y.V. X Chromosome-Wide Analysis Identifies DNA Methylation Sites Influenced by Cigarette Smoking. Clin. Epigenetics 2016, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Inkster, A.M.; Wong, M.T.; Matthews, A.M.; Brown, C.J.; Robinson, W.P. Who’s Afraid of the X? Incorporating the X and Y Chromosomes into the Analysis of DNA Methylation Array Data. Epigenetics Chromatin 2023, 16, 1. [Google Scholar] [CrossRef]
- Ambatipudi, S.; Cuenin, C.; Hernandez-Vargas, H.; Ghantous, A.; Le Calvez-Kelm, F.; Kaaks, R.; Barrdahl, M.; Boeing, H.; Aleksandrova, K.; Trichopoulou, A.; et al. Tobacco Smoking-Associated Genome-Wide DNA Methylation Changes in the EPIC Study. Epigenomics 2016, 8, 599–618. [Google Scholar] [CrossRef] [Green Version]
- Grieshober, L.; Graw, S.; Barnett, M.J.; Thornquist, M.D.; Goodman, G.E.; Chen, C.; Koestler, D.C.; Marsit, C.J.; Doherty, J.A. AHRR Methylation in Heavy Smokers: Associations with Smoking, Lung Cancer Risk, and Lung Cancer Mortality. BMC Cancer 2020, 20, 905. [Google Scholar] [CrossRef]
- Zhang, Y.; Elgizouli, M.; Schöttker, B.; Holleczek, B.; Nieters, A.; Brenner, H. Smoking-Associated DNA Methylation Markers Predict Lung Cancer Incidence. Clin. Epigenetics 2016, 8, 127. [Google Scholar] [CrossRef] [Green Version]
- Cappozzo, A.; McCrory, C.; Robinson, O.; Sterrantino, A.F.; Sacerdote, C.; Krogh, V.; Panico, S.; Tumino, R.; Iacoviello, L.; Ricceri, F.; et al. A Blood DNA Methylation Biomarker for Predicting Short-Term Risk of Cardiovascular Events. Clin. Epigenetics 2022, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, Y.; Yamada, H.; Munetsuna, E.; Fujii, R.; Yamazaki, M.; Ando, Y.; Mizuno, G.; Hattori, Y.; Ishikawa, H.; Ohashi, K.; et al. Increased Risk of Cancer Mortality by Smoking-Induced Aryl Hydrocarbon Receptor Repressor DNA Hypomethylation in Japanese Population: A Long-Term Cohort Study. Cancer Epidemiol. 2022, 78, 102162. [Google Scholar] [CrossRef]




| Total (n = 414) | Men (n = 186) | Women (n = 228) | p | |
|---|---|---|---|---|
| Age (years) | 65.1 ± 0.2 | 63.8 ± 0.4 | 66.1 ± 0.3 | <0.001 |
| BMI (kg/m2) | 32.3 ± 0.2 | 32.2 ± 0.3 | 32.4 ± 0.2 | 0.440 |
| SBP (mm Hg) | 141.9 ± 0.9 | 143.7 ± 1.4 | 140.5 ± 1.2 | 0.076 |
| DBP (mm Hg) | 81.0 ± 0.5 | 82.6 ± 0.7 | 79.6 ± 0.6 | 0.002 |
| Total cholesterol (mg/dL) | 195.7 ± 1.8 | 188.1 ± 2.8 | 202.0 ± 2.3 | <0.001 |
| LDL-C (mg/dL) | 124.3 ± 1.5 | 121.5 ± 2.4 | 126.6 ± 1.9 | 0.096 |
| HDL-C (mg/dL) | 51.6 ± 0.6 | 47.3 ± 0.8 | 55.1 ± 0.7 | <0.001 |
| Triglycerides 1 (mg/dL) | 141.3 ± 2.9 | 139.0 ± 4.0 | 143.1 ± 4.2 | 0.488 |
| Fasting glucose (mg/dL) | 113.4 ± 1.4 | 113.7 ± 2.2 | 113.2 ± 1.8 | 0.875 |
| Physical activity (MET·min/wk) | 1708 ± 78 | 1941 ± 133 | 1519 ± 89 | 0.007 |
| Adherence to MedDiet (17-I) 2 | 8.0 ± 2.8 | 7.9 ± 2.8 | 8.1 ± 2.7 | 0.210 |
| High Adherence MedDiet 3 (≥9) (n, %) | 172 (41.5) | 75 (40.3) | 97 (42.5) | 0.648 |
| Type 2 diabetes (n, %) | 164 (39.6) | 75 (40.3) | 89 (39.0) | 0.790 |
| Never smokers (n, %) | 188 (45.4) | 32 (17.2) | 156 (68.4) | <0.001 |
| Former smokers (>5 years) (n, %) | 139 (33.6) | 97 (52.2) | 42 (18.4) | <0.001 |
| Former smokers (1 to 5 years) (n, %) | 25 (6.0) | 16 (8.6) | 9 (3.9) | <0.001 |
| Former smokers (<1 year) (n, %) | 14 (3.4) | 11 (5.9) | 3 (1.3) | <0.001 |
| Current smokers (n, %) | 48 (11.6) | 30 (16.1) | 18 (7.9) | <0.001 |
| Number of cigarettes smoked per day 4 | 13.2 ± 1.1 | 14.4 ± 1.6 | 11.3 ± 1.4 | 0.186 |
| Number of years smoked 4 | 38.0 ± 1.6 | 39.1 ± 2.1 | 36.3 ± 2.6 | 0.428 |
| Number of pack-years 4,5 | 24.9 ± 2.4 | 27.9 ± 3.4 | 19.9 ± 2.4 | 0.105 |
| CpG Site | Gene Symbol | Chr | Position 3 | Included in 450 K 4 | p | r |
|---|---|---|---|---|---|---|
| cg21566642 | 2 | 233284661 | Y | 2.20 × 10−32 | −0.548 | |
| cg01940273 | 2 | 233284934 | Y | 1.02 × 10−19 | −0.435 | |
| cg14391737 | PRSS23 | 11 | 86513429 | N | 8.13 × 10−19 | −0.425 |
| cg17739917 | RARA | 17 | 38477572 | N | 2.30 × 10−18 | −0.420 |
| cg21911711 | F2RL3 | 19 | 16998668 | N | 6.15 × 10−17 | −0.403 |
| cg18110140 | 15 | 75350380 | N | 2.12 × 10−15 | −0.384 | |
| cg19572487 | RARA | 17 | 38476024 | Y | 4.26 × 10−15 | −0.381 |
| cg24859433 | 6 | 30720203 | Y | 6.33 × 10−15 | −0.378 | |
| cg17287155 | AHRR | 5 | 393347 | Y | 2.94 × 10−14 | −0.370 |
| cg00475490 | PRSS23 | 11 | 86517110 | N | 7.08 × 10−14 | −0.364 |
| cg09935388 | GFI1 | 1 | 92947588 | Y | 8.22 × 10−14 | −0.363 |
| cg05575921 | AHRR | 5 | 373378 | Y | 1.00 × 10−13 | −0.362 |
| cg15342087 | 6 | 30720209 | Y | 1.78 × 10−12 | −0.344 | |
| cg04551776 | AHRR | 5 | 393366 | Y | 3.50 × 10−12 | −0.340 |
| cg25189904 | GNG12 | 1 | 68299493 | Y | 4.27 × 10−12 | −0.339 |
| cg03636183 | F2RL3 | 19 | 17000585 | Y | 6.36 × 10−12 | −0.336 |
| cg26703534 | AHRR | 5 | 377358 | Y | 9.58 × 10−12 | −0.334 |
| cg25648203 | AHRR | 5 | 395444 | Y | 1.08 × 10−11 | −0.333 |
| cg01901332 | ARRB1 | 11 | 75031054 | Y | 3.21 × 10−11 | −0.325 |
| cg27241845 | 2 | 233250370 | Y | 5.77 × 10−11 | −0.321 | |
| cg19859270 | GPR15 | 3 | 98251294 | Y | 8.02 × 10−11 | −0.319 |
| cg16841366 | 2 | 233286192 | N | 1.04 × 10−10 | −0.317 | |
| cg23161492 | ANPEP | 15 | 90357202 | Y | 1.17 × 10−10 | −0.316 |
| cg02978227 | 3 | 98292027 | N | 1.04 × 10−9 | −0.300 | |
| cg11660018 | PRSS23 | 11 | 86510915 | Y | 1.44 × 10−9 | −0.298 |
| cg14580211 | C5orf62 | 5 | 150161299 | Y | 1.66 × 10−9 | −0.297 |
| cg11556164 | LRRN3 | 7 | 110738315 | Y | 2.00 × 10−9 | −0.296 |
| cg12806681 | AHRR | 5 | 368394 | Y | 2.96 × 10−9 | −0.293 |
| cg05086879 | MGAT3 | 22 | 39861490 | N | 4.79 × 10−9 | −0.289 |
| cg05284742 | ITPK1 | 14 | 93552128 | Y | 6.54 × 10−9 | −0.286 |
| cg17738628 | 15 | 67155520 | N | 7.50 × 10−9 | −0.285 | |
| cg21611682 | LRP5 | 11 | 68138269 | Y | 8.72 × 10−9 | −0.284 |
| cg07339236 | ATP9A | 20 | 50312490 | Y | 1.35 × 10−8 | −0.281 |
| cg24838345 | MTSS1 | 8 | 125737353 | Y | 1.54 × 10−8 | −0.280 |
| cg19554457 | NUDT4 | 12 | 93774772 | Y | 1.61 × 10−8 | −0.279 |
| cg25305703 | 8 | 128378218 | Y | 1.87 × 10−8 | −0.278 | |
| cg04535902 | GFI1 | 1 | 92947332 | Y | 2.30 × 10−8 | −0.276 |
| cg09945032 | 3 | 38871019 | N | 2.59 × 10−8 | −0.275 | |
| cg07986378 | ETV6 | 12 | 11898284 | Y | 2.79 × 10−8 | −0.275 |
| cg08714510 | UXS1 | 2 | 106755481 | N | 5.78 × 10−8 | −0.268 |
| cg15533935 | NUDT4P2 | 12 | 93774767 | N | 6.20 × 10−8 | −0.268 |
| cg16794579 | XYLT1 | 16 | 17562419 | Y | 6.28 × 10−8 | −0.268 |
| cg00310412 | SEMA7A | 15 | 74724918 | Y | 7.04 × 10−8 | −0.267 |
| cg05221370 | LRRN3 | 7 | 110738836 | Y | 7.13 × 10−8 | −0.267 |
| cg15394081 | LMO2 | 11 | 33893330 | N | 8.31 × 10−8 | −0.265 |
| cg06321596 | XYLT1 | 16 | 17562960 | Y | 8.61 × 10−8 | −0.265 |
| A-Smoking (3 Categories) | B-Never (N) Versus Current Smokers (C) | |||||||
|---|---|---|---|---|---|---|---|---|
| CpG Site | Gene Symbol | Chr | p4 (3 Categories) | CpG Site | Gene Symbol | Chr | p5 (N vs. C) | Beta Difference 6 |
| cg21566642 | 2 | 1.39 × 10−29 | cg21566642 | 2 | 1.68 × 10−30 | 0.143 | ||
| cg14391737 | PRSS23 | 11 | 6.95 × 10−20 | cg01940273 | 2 | 1.30 × 10−20 | 0.097 | |
| cg01940273 | 2 | 1.44 × 10−19 | cg14391737 | PRSS23 | 11 | 1.71 × 10−17 | 0.107 | |
| cg17739917 | RARA | 17 | 3.44 × 10−16 | cg17739917 | RARA | 17 | 5.13 × 10−17 | 0.112 |
| cg21911711 | F2RL3 | 19 | 8.14 × 10−16 | cg21911711 | F2RL3 | 19 | 8.76 × 10−17 | 0.085 |
| cg00475490 | PRSS23 | 11 | 9.66 × 10−16 | cg00475490 | PRSS23 | 11 | 1.73 × 10−15 | 0.046 |
| cg24859433 | 6 | 1.96 × 10−14 | cg24859433 | 6 | 2.29 × 10−15 | 0.048 | ||
| cg18110140 | 15 | 2.36 × 10−13 | cg18110140 | 15 | 3.04 × 10−14 | 0.094 | ||
| cg09935388 | GFI1 | 1 | 5.94 × 10−13 | cg09935388 | GFI1 | 1 | 6.54 × 10−14 | 0.105 |
| cg25648203 | AHRR | 5 | 9.21 × 10−13 | cg19572487 | RARA | 17 | 1.78 × 10−13 | 0.073 |
| cg25189904 | GNG12 | 1 | 1.46 × 10−12 | cg25189904 | GNG12 | 1 | 1.90 × 10−13 | 0.110 |
| cg19572487 | RARA | 17 | 1.55 × 10−12 | cg25648203 | AHRR | 5 | 5.67 × 10−13 | 0.065 |
| cg26703534 | AHRR | 5 | 2.44 × 10−11 | cg01901332 | ARRB1 | 11 | 3.61 × 10−12 | 0.078 |
| cg01901332 | ARRB1 | 11 | 2.63 × 10−11 | cg03636183 | F2RL3 | 19 | 4.54 × 10−12 | 0.132 |
| cg03636183 | F2RL3 | 19 | 3.84 × 10−11 | cg17287155 | AHRR | 5 | 7.31 × 10−12 | 0.029 |
| CpG Site | Gene Symbol | Chr | p2 (F vs. C) | Beta Difference 3 |
|---|---|---|---|---|
| cg21566642 | 2 | 2.51 × 10−16 | 0.0935 | |
| cg01940273 | 2 | 8.57 × 10−13 | 0.0706 | |
| cg25648203 | AHRR | 5 | 2.39 × 10−11 | 0.0563 |
| cg26703534 | AHRR | 5 | 1.20 × 10−10 | 0.0516 |
| cg05086879 | MGAT3 | 22 | 1.82 × 10−10 | 0.0452 |
| cg18110140 | 15 | 7.19 × 10−10 | 0.0719 | |
| cg21911711 | F2RL3 | 19 | 1.49 × 10−9 | 0.0588 |
| cg11556164 | LRRN3 | 7 | 1.66 × 10−9 | 0.0355 |
| cg25189904 | GNG12 | 1 | 2.65 × 10−9 | 0.0824 |
| cg23161492 | ANPEP | 15 | 3.85 × 10−9 | 0.0631 |
| cg17739917 | RARA | 17 | 8.78 × 10−9 | 0.0714 |
| cg04551776 | AHRR | 5 | 1.21 × 10−8 | 0.0440 |
| cg24859433 | 6 | 1.38 × 10−8 | 0.0332 | |
| cg01901332 | ARRB1 | 11 | 1.67 × 10−8 | 0.0607 |
| cg05284742 | ITPK1 | 14 | 1.71 × 10−8 | 0.0437 |
| Pathway Name | Enrichment Score | Enrichment p 1 | Bonferroni (Enrichment p) 2 |
|---|---|---|---|
| Parathyroid hormone synthesis, secretion and action | 26.912 | 2.05 × 10−12 | 3.90 × 10−10 |
| VEGF signaling pathway | 22.448 | 1.78 × 10−10 | 3.39 × 10−8 |
| Morphine addiction | 21.911 | 3.05 × 10−10 | 5.80 × 10−8 |
| Endocrine resistance | 20.622 | 1.11 × 10−9 | 2.10 × 10−7 |
| Chemokine signaling pathway | 18.663 | 7.85 × 10−9 | 1.49 × 10−6 |
| Phospholipase D signaling pathway | 18.406 | 1.01 × 10−8 | 1.93 × 10−6 |
| Non-small cell lung cancer | 18.024 | 1.49 × 10−8 | 2.83 × 10−6 |
| Glioma | 17.219 | 3.33 × 10−8 | 6.32 × 10−6 |
| Cholinergic synapse | 17.090 | 3.78 × 10−8 | 7.19 × 10−6 |
| Hepatocellular carcinoma | 15.770 | 1.42 × 10−7 | 2.69 × 10−5 |
| Human cytomegalovirus infection | 15.279 | 2.32 × 10−7 | 4.40 × 10−5 |
| ErbB signaling pathway | 15.148 | 2.64 × 10−7 | 5.02 × 10−5 |
| Relaxin signaling pathway | 14.613 | 4.51 × 10−7 | 8.56 × 10−5 |
| Transcriptional misregulation in cancer | 13.791 | 1.03 × 10−6 | 1.95 × 10−4 |
| Small cell lung cancer | 13.644 | 1.19 × 10−6 | 2.26 × 10−4 |
| Apelin signaling pathway | 13.353 | 1.59 × 10−6 | 3.02 × 10−4 |
| Pathways in cancer | 13.329 | 1.63 × 10−6 | 3.09 × 10−4 |
| Fc gamma R-mediated phagocytosis | 12.986 | 2.29 × 10−6 | 4.36 × 10−4 |
| Circadian entrainment | 12.829 | 2.68 × 10−6 | 5.09 × 10−4 |
| Adrenergic signaling in cardiomyocytes | 12.365 | 4.27 × 10−6 | 8.11 × 10−4 |
| GnRH secretion | 11.470 | 1.04 × 10−5 | 1.98 × 10−3 |
| Growth hormone synthesis, secretion and action | 10.053 | 4.31 × 10−5 | 8.18 × 10−3 |
| Melanoma | 10.035 | 4.38 × 10−5 | 8.33 × 10−3 |
| Calcium signaling pathway | 9.991 | 4.58 × 10−5 | 8.70 × 10−3 |
| Platelet activation | 9.558 | 7.06 × 10−5 | 1.34 × 10−2 |
| Axon guidance | 9.510 | 7.41 × 10−5 | 1.41 × 10−2 |
| Pancreatic cancer | 9.430 | 8.03 × 10−5 | 1.53 × 10−2 |
| Chronic myeloid leukemia | 9.430 | 8.03 × 10−5 | 1.53 × 10−2 |
| Dopaminergic synapse | 8.928 | 1.33 × 10−4 | 2.52 × 10−2 |
| Kaposi sarcoma-associated herpesvirus infection | 8.742 | 1.60 × 10−4 | 3.04 × 10−2 |
| Bladder cancer | 8.386 | 2.28 × 10−4 | 4.33 × 10−2 |
| Chemical carcinogenesis—receptor activation | 7.854 | 3.88 × 10−4 | 7.38 × 10−2 |
| GABAergic synapse | 7.833 | 3.96 × 10−4 | 7.53 × 10−2 |
| Human immunodeficiency virus 1 infection | 7.803 | 4.09 × 10−4 | 7.76 × 10−2 |
| Breast cancer | 7.713 | 4.47 × 10−4 | 8.49 × 10−2 |
| Gastric cancer | 7.580 | 5.11 × 10−4 | 9.70 × 10−2 |
| Men (n = 186) | Women (n = 228) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CpG Site | Gene Symbol | Chr | p | r | CpG Site | Gene Symbol | Chr | p | r |
| cg21566642 | 2 | 2.20 × 10−15 | −0.561 | cg21566642 | 2 | 4.07 × 10−16 | −0.522 | ||
| cg03636183 | F2RL3 | 19 | 1.92 × 10−11 | −0.487 | cg14391737 | PRSS23 | 11 | 2.51 × 10−12 | −0.458 |
| cg01940273 | 2 | 3.73 × 10−10 | −0.458 | cg05575921 | AHRR | 5 | 1.55 × 10−11 | −0.443 | |
| cg17287155 | AHRR | 5 | 8.37 × 10−9 | −0.425 | cg09935388 | GFI1 | 1 | 5.80 × 10−11 | −0.431 |
| cg17739917 | RARA | 17 | 1.28 × 10−8 | −0.420 | cg17739917 | RARA | 17 | 1.32 × 10−10 | −0.424 |
| cg14391737 | PRSS23 | 11 | 3.89 × 10−8 | −0.407 | cg00475490 | PRSS23 | 11 | 2.57 × 10−10 | −0.418 |
| cg19572487 | RARA | 17 | 9.96 × 10−8 | −0.396 | cg01940273 | 2 | 8.28 × 10−10 | −0.407 | |
| cg04551776 | AHRR | 5 | 1.37 × 10−7 | −0.392 | cg21911711 | F2RL3 | 19 | 9.46 × 10−10 | −0.405 |
| cg15342087 | 6 | 1.48 × 10−7 | −0.391 | cg24859433 | 6 | 1.94 × 10−9 | −0.398 | ||
| cg27241845 | 2 | 1.56 × 10−7 | −0.390 | cg25966498 | SPATA17 | 1 | 9.11 × 10−9 | −0.383 | |
| cg21911711 | F2RL3 | 19 | 2.96 × 10−7 | −0.382 | cg18110140 | 15 | 1.08 × 10−8 | −0.381 | |
| cg18110140 | 15 | 1.25 × 10−6 | −0.363 | cg11556164 | LRRN3 | 7 | 1.65 × 10−8 | −0.376 | |
| cg23161492 | ANPEP | 15 | 1.28 × 10−6 | −0.362 | cg19572487 | RARA | 17 | 2.51 × 10−8 | −0.372 |
| cg24090911 | AHRR | 5 | 1.32 × 10−6 | −0.362 | cg25648203 | AHRR | 5 | 6.15 × 10−8 | −0.362 |
| cg24859433 | 6 | 2.12 × 10−6 | −0.355 | cg25189904 | GNG12 | 1 | 7.29 × 10−8 | −0.360 | |
| cg01901332 | ARRB1 | 11 | 3.67 × 10−6 | −0.348 | cg21929649 | EFTUD2 | 17 | 2.90 × 10−7 | −0.344 |
| Low Adherence to Mediterranean Diet (n = 242) | High Adherence to Mediterranean Diet (n = 172) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CpG Site | Gene Symbol | Chr | p | r | CpG Site | Gene Symbol | Chr | p | r |
| cg21566642 | 2 | 1.34 × 10−22 | −0.592 | cg21566642 | 2 | 5.27 × 10−10 | −0.474 | ||
| cg03636183 | F2RL3 | 19 | 1.15 × 10−16 | −0.516 | cg14391737 | PRSS23 | 11 | 6.21 × 10−8 | −0.419 |
| cg01940273 | 2 | 3.72 × 10−15 | −0.494 | cg02330394 | C6orf52 | 6 | 3.96 × 10−7 | −0.395 | |
| cg05575921 | AHRR | 5 | 4.31 × 10−15 | −0.493 | cg04569608 | PLCB3 | 11 | 6.31 × 10−7 | −0.389 |
| cg24859433 | 6 | 5.52 × 10−15 | −0.491 | cg00475490 | PRSS23 | 11 | 8.17 × 10−7 | −0.385 | |
| cg15342087 | 6 | 7.71 × 10−15 | −0.489 | cg14175932 | 14 | 1.81 × 10−6 | −0.374 | ||
| cg17739917 | RARA | 17 | 1.41 × 10−14 | −0.484 | cg26742440 | RAB6A | 11 | 1.98 × 10−6 | −0.372 |
| cg09935388 | GFI1 | 1 | 4.39 × 10−13 | −0.459 | cg22024876 | KBTBD3 | 11 | 2.37 × 10−6 | −0.370 |
| cg19859270 | GPR15 | 3 | 4.79 × 10−13 | −0.459 | cg02286229 | 11 | 2.93 × 10−6 | −0.367 | |
| cg21911711 | F2RL3 | 19 | 7.56 × 10−13 | −0.455 | cg21619351 | SCAF1 | 19 | 4.83 × 10−6 | −0.359 |
| cg27241845 | 2 | 5.28 × 10−12 | −0.440 | cg13496340 | TIGD7 | 16 | 5.06 × 10−6 | −0.358 | |
| cg18110140 | 15 | 6.71 × 10−12 | −0.438 | cg26815336 | 6 | 7.03 × 10−6 | −0.353 | ||
| cg14391737 | PRSS23 | 11 | 9.53 × 10−12 | −0.435 | cg21911711 | F2RL3 | 19 | 8.06 × 10−6 | −0.351 |
| cg19572487 | RARA | 17 | 1.34 × 10−11 | −0.432 | cg01697541 | TMEM97 | 17 | 8.57 × 10−6 | −0.350 |
| cg25648203 | AHRR | 5 | 1.43 × 10−11 | −0.431 | cg08376211 | MAP7D1 | 1 | 9.11 × 10−6 | 0.349 |
| cg26703534 | AHRR | 5 | 1.80 × 10−11 | −0.430 | cg22078572 | KDSR | 18 | 9.73 × 10−6 | −0.348 |
| cg18316974 | GFI1 | 1 | 7.05 × 10−11 | −0.418 | cg22488975 | EDEM3 | 1 | 1.11 × 10−5 | −0.346 |
| cg12806681 | AHRR | 5 | 9.23 × 10−11 | −0.415 | cg22692169 | LINS1 | 15 | 1.11 × 10−5 | −0.346 |
| cg02978227 | 3 | 1.41 × 10−10 | −0.412 | cg01207684 | ADCY9 | 16 | 1.37 × 10−5 | −0.343 | |
| cg18146737 | GFI1 | 1 | 1.46 × 10−10 | −0.411 | cg17739917 | RARA | 17 | 1.64 × 10−5 | −0.340 |
| cg25189904 | GNG12 | 1 | 3.59 × 10−10 | −0.403 | cg16552945 | ARHGDIG | 16 | 1.78 × 10−5 | −0.338 |
| cg17287155 | AHRR | 5 | 3.74 × 10−10 | −0.403 | cg00502002 | ANAPC16 | 10 | 2.03 × 10−5 | 0.336 |
| cg23576855 | AHRR | 5 | 7.03 × 10−10 | −0.397 | cg18149653 | 12 | 2.23 × 10−5 | −0.335 | |
| cg04551776 | AHRR | 5 | 8.71 × 10−10 | −0.395 | cg08870961 | 12 | 2.37 × 10−5 | 0.334 | |
| cg24838345 | MTSS1 | 8 | 2.35 × 10−9 | −0.386 | cg19355087 | NKX6-2 | 10 | 2.38 × 10−5 | −0.333 |
| cg12876356 | GFI1 | 1 | 5.04 × 10−9 | −0.378 | cg01940273 | 2 | 2.42 × 10−5 | −0.333 | |
| cg07986378 | ETV6 | 12 | 5.25 × 10−9 | −0.378 | cg16915863 | LOC400043 | 12 | 2.43 × 10−5 | 0.333 |
| cg11660018 | PRSS23 | 11 | 5.69 × 10−9 | −0.377 | cg21897843 | GAB2 | 11 | 2.63 × 10−5 | 0.332 |
| cg09945032 | 3 | 1.36 × 10−8 | −0.368 | cg18866308 | G3BP1 | 5 | 2.82 × 10−5 | −0.331 | |
| cg04535902 | GFI1 | 1 | 1.44 × 10−8 | −0.367 | cg02048416 | DOK1 | 2 | 2.89 × 10−5 | −0.330 |
| cg01901332 | ARRB1 | 11 | 1.71 × 10−8 | −0.366 | cg10352046 | 12 | 2.99 × 10−5 | −0.330 | |
| cg24090911 | AHRR | 5 | 2.02 × 10−8 | −0.364 | cg16487464 | 8 | 3.10 × 10−5 | 0.329 | |
| cg16841366 | 2 | 2.60 × 10−8 | −0.361 | cg27072224 | DNAJC15 | 13 | 3.12 × 10−5 | 0.329 | |
| cg19554457 | NUDT4 | 12 | 3.29 × 10−8 | −0.359 | cg21038620 | LAT | 16 | 3.31 × 10−5 | −0.328 |
| cg14372879 | 1 | 5.72 × 10−8 | −0.353 | cg02882774 | NDUFA6 | 22 | 3.57 × 10−5 | −0.327 | |
| cg25305703 | 8 | 6.03 × 10−8 | −0.352 | cg09061824 | FLOT1 | 6 | 3.60 × 10−5 | −0.326 | |
| cg13681954 | 2 | 6.48 × 10−8 | −0.352 | cg08445320 | 14 | 3.86 × 10−5 | 0.325 | ||
| cg17738628 | 15 | 7.22 × 10−8 | −0.350 | cg27228559 | CASC2 | 10 | 4.13 × 10−5 | −0.324 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Carrión, R.; Sorlí, J.V.; Asensio, E.M.; Pascual, E.C.; Portolés, O.; Alvarez-Sala, A.; Francès, F.; Ramírez-Sabio, J.B.; Pérez-Fidalgo, A.; Villamil, L.V.; et al. DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet. Int. J. Environ. Res. Public Health 2023, 20, 3635. https://doi.org/10.3390/ijerph20043635
Fernández-Carrión R, Sorlí JV, Asensio EM, Pascual EC, Portolés O, Alvarez-Sala A, Francès F, Ramírez-Sabio JB, Pérez-Fidalgo A, Villamil LV, et al. DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet. International Journal of Environmental Research and Public Health. 2023; 20(4):3635. https://doi.org/10.3390/ijerph20043635
Chicago/Turabian StyleFernández-Carrión, Rebeca, José V. Sorlí, Eva M. Asensio, Eva C. Pascual, Olga Portolés, Andrea Alvarez-Sala, Francesc Francès, Judith B. Ramírez-Sabio, Alejandro Pérez-Fidalgo, Laura V. Villamil, and et al. 2023. "DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet" International Journal of Environmental Research and Public Health 20, no. 4: 3635. https://doi.org/10.3390/ijerph20043635
APA StyleFernández-Carrión, R., Sorlí, J. V., Asensio, E. M., Pascual, E. C., Portolés, O., Alvarez-Sala, A., Francès, F., Ramírez-Sabio, J. B., Pérez-Fidalgo, A., Villamil, L. V., Tinahones, F. J., Estruch, R., Ordovas, J. M., Coltell, O., & Corella, D. (2023). DNA-Methylation Signatures of Tobacco Smoking in a High Cardiovascular Risk Population: Modulation by the Mediterranean Diet. International Journal of Environmental Research and Public Health, 20(4), 3635. https://doi.org/10.3390/ijerph20043635











