Tackling Neonatal Sepsis—Can It Be Predicted?
Abstract
1. Introduction
2. Materials and Methods
3. Results
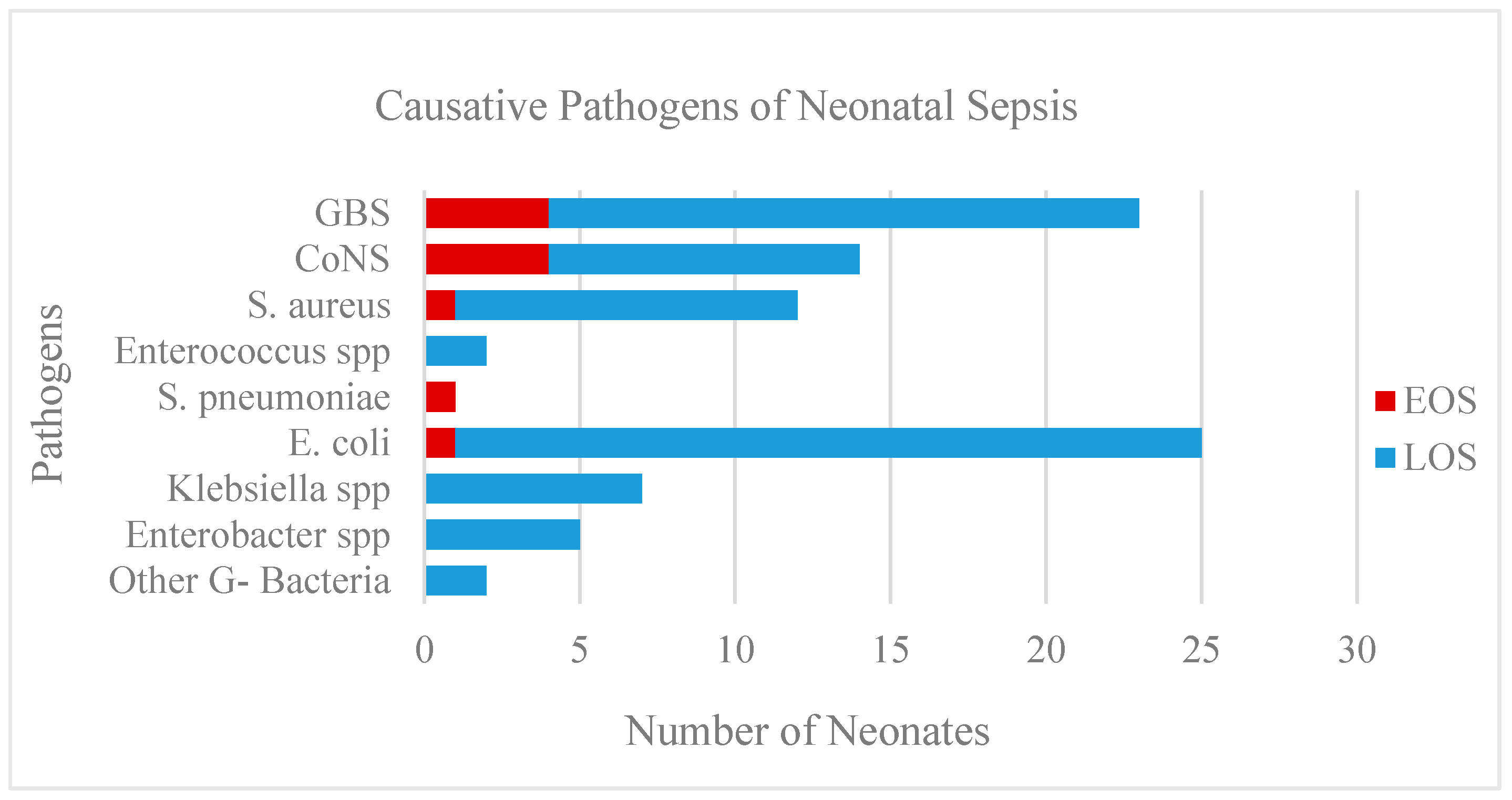
3.1. Pathogens
3.2. Perinatal Factors
3.3. Clinical Presentation of Neonatal Sepsis
3.4. Laboratory Markers of Infection
3.5. The Model for Predicting Neonatal Sepsis
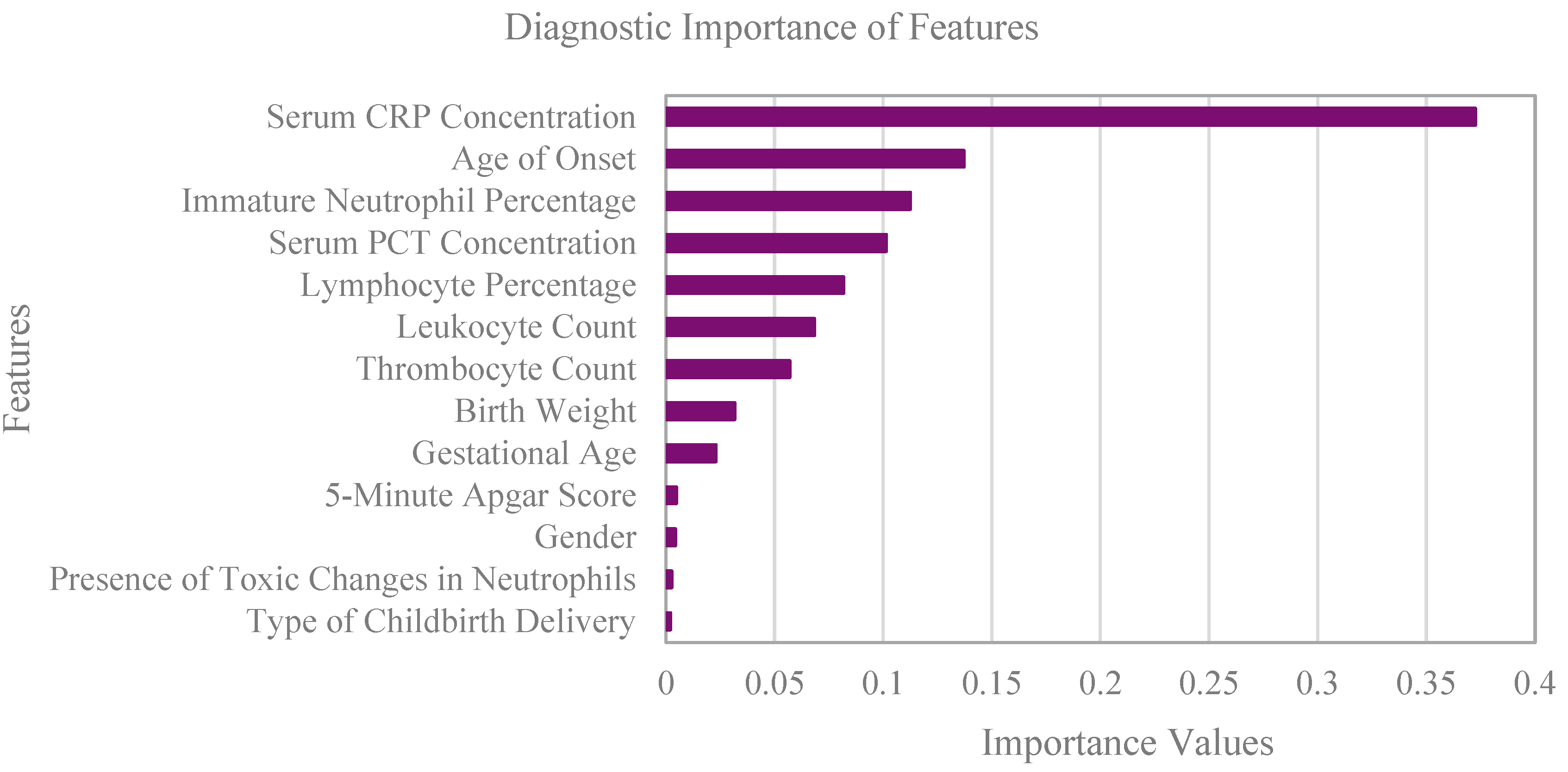
3.6. Feature Selection for Model Training
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shane, A.L.; Sánchez, P.J. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Glaser, M.A.; Hughes, L.M.; Jnah, A.; Cnattingius, S.; Joseph, K.S. Neonatal sepsis: A review of pathophysiology and current management strategies. Adv. Neonatal Care 2020, 21, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Giannoni, E.; Agyeman, P.K.A.; Stocker, M.; Benjamin, D.K.; Becker, K.C. Neonatal sepsis of early onset, and hospital-acquired and community-acquired late onset: A prospective population-based cohort study. J. Pediatr. 2018, 201, 106–114. [Google Scholar] [CrossRef]
- Stoll, B.J.; Hansen, N.I.; Sánchez, P.J.; Cnattingius, S.; Joseph, K.S. Early onset neonatal sepsis: The burden of group B Streptococcal and E. coli disease continues. Pediatrics 2011, 127, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Boghossian, N.S.; Page, G.P.; Bell, E.F.; Stoll, B.J.; Murray, J.C.; Cotten, C.M.; Shankaran, S.; Walsh, M.C.; Laptook, A.R.; Newman, N.S.; et al. Late-onset sepsis in very low birth weight infants from singleton and multiple-gestation births. J. Pediatr. 2013, 162, 1120–1124. [Google Scholar] [CrossRef]
- Shane, A.L.; Sánchez, P.J.; Stoll, B.J. Neonatal sepsis. Lancet 2017, 390, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Chirico, G.; Loda, C. Laboratory aid to the diagnosis and therapy of infection in the neonate. Pediatr. Rep. 2011, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Arcagok, B.C.; Karabulut, B. Platelet to lymphocyte ratio in neonates: A predictor of early onset neonatal sepsis. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019055. [Google Scholar] [CrossRef]
- Sahu, P.; Raj Stanly, E.A.; Simon Lewis, L.E.; Prabhu, K.; Rao, M.; Kunhikatta, V. Prediction modelling in the early detection of neonatal sepsis. World J. Pediatr. 2022, 18, 160–175. [Google Scholar] [CrossRef]
- Neamțu, B.M.; Visa, G.; Maniu, I.; Ognean, M.; Pérez-Elvira, R.; Dragomir, A.; Agudo, M.; Șofariu, C.; Gheonea, M.; Pitic, A.; et al. A decision-tree approach to assist in forecasting the outcomes of the neonatal brain injury. Int. J. Environ. Res. Public Health 2021, 18, 4807. [Google Scholar] [CrossRef]
- Fleischmann, C.; Reichert, F.; Cassini, A.; Horner, R.; Harder, T.; Markwart, R.; Tröndle, M.; Savova, Y.; Kissoon, N.; Schlattmann, P.; et al. Global incidence and mortality of neonatal sepsis: A systematic review and meta-analysis. Arch. Dis. Child. 2021, 106, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Beretta, L.; Santaniello, A. Nearest neighbor imputation algorithms: A critical evaluation. BMC Med. Inform. Decis. Mak. 2016, 16 (Suppl. 3), 74. [Google Scholar] [CrossRef] [PubMed]
- Walker, M. Identifying and Fixing Missing Values. In Data Cleaning and Exploration with Machine Learning, 1st ed.; Sugarman, D., Manikandan, K., Eds.; Pakt Publishing Ltd.: Birmingham, UK, 2022; pp. 82–102. [Google Scholar]
- American College of Obstetricians and Gynecologists. Prevention of group B streptococcal early-onset disease in newborns: ACOG committee opinion, number 797. Obstet. Gynecol. 2020, 135, e51–e72. [Google Scholar] [CrossRef] [PubMed]
- Thavarajah, H.; Flatley, C. The relationship between the five minute Apgar score, mode of birth and neonatal outcomes. J. Matern. Fetal Neonatal Med. 2017, 31, 1335–1341. [Google Scholar] [CrossRef]
- Person, M.K.; Esposito, D.H.; Holman, R.C.; Mehal, J.M.; Stoll, B.J. Risk factors for infectious disease death among infants in the United States. Pediatr. Infect. Dis. J. 2014, 33, e280-5. [Google Scholar] [CrossRef]
- Razaz, N.; Cnattingius, S.; Joseph, K.S.; Mehta, S.; Patole, S. Association between Apgar scores of 7 to 9 and neonatal mortality and morbidity: Population based cohort study of term infants in Sweden. BMJ 2019, 40, 33–34. [Google Scholar] [CrossRef]
- Cohen-Wolkowiez, M.; Moran, C.; Benjamin, D.K.; Cotten, C.M.; Clark, R.; Smith, P.B. Early and late onset sepsis in late preterm infants. Pediatr. Infect. Dis. J. 2009, 28, 1052–1056. [Google Scholar] [CrossRef]
- Young Infants Clinical Signs Study Group. Clinical signs that predict severe illness in children under age 2 months: A multicentre study. Lancet 2008, 371, 135–142. [Google Scholar] [CrossRef]
- Camacho-Gonzalez, A.; Spearman, P.W.; Stoll, B.J. Neonatal infectious diseases: Evaluation of neonatal sepsis. Pediatr. Clin. N. Am. 2013, 60, 367–389. [Google Scholar] [CrossRef]
- Sushanth; Avabratha, K.S.; Tauro, K.J.; Benjamin, D.K.; Becker, K.C. Hyperleukocytosis in a neonate: A diagnostic dilemma. Indian J. Med. Paediatr. Oncol. 2010, 31, 86–88. [Google Scholar]
- Celik, I.H.; Hanna, M.; Canpolat, F.E.; Pammi, M. Diagnosis of neonatal sepsis: The past, present and future. Pediatr. Res. 2022, 91, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Hornik, C.P.; Benjamin, D.K.; Becker, K.C.; Li, J.; Clark, R.; Cohen-Wolkowiez, M.; Smith, P.B. Use of the complete blood cell count in early-onset neonatal sepsis. Pediatr. Infect. Dis. J. 2012, 31, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Hornik, C.P.; Benjamin, D.K.; Becker, K.C.; Li, J.; Clark, R.; Cohen-Wolkowiez, M.; Smith, P.B. Use of the complete blood cell count in late-onset neonatal sepsis. Pediatr. Infect. Dis. J. 2012, 31, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Can, E.; Hamilcikan, Ş.; Can, C. The value of neutrophil to lymphocyte ratio and platelet to lymphocyte ratio for detecting early-onset neonatal sepsis. J. Pediatr. Hematol. Oncol. 2018, 40, e229–e232. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C. Diagnostic markers of infection in neonates. Arch. Dis. Child Fetal Neonatal Ed. 2004, 89, F229–F235. [Google Scholar] [CrossRef]
- Haque, K.N. Definitions of bloodstream infection in the newborn. Pediatr. Crit. Care Med. 2005, 6, S45–S49. [Google Scholar] [CrossRef]
- Makkar, M.; Gupta, C.; Pathak, R.; Garg, S.; Mahajan, N.C. Performance evaluation of hematologic scoring system in early diagnosis of neonatal sepsis. J. Clin. Neonatol. 2013, 2, 25–29. [Google Scholar] [CrossRef]
- Ismail, A.Q.; Gandhi, A. Using CRP in neonatal practice. J. Matern. Fetal Neonatal Med. 2015, 28, 3–6. [Google Scholar] [CrossRef]
- Chiesa, C.; Natale, F.; Pascone, R.; Osborne, J.F.; Pacifico, L. C-reactive protein and procalcitonin: Reference intervals for preterm and term newborns during the early neonatal period. Clin. Chim. Acta 2011, 412, 1053–1059. [Google Scholar] [CrossRef]
- Hofer, N.; Müller, W.; Resch, B. Non-infectious conditions and gestational age influence C-reactive protein values in newborns during the first 3 days of life. Clin. Chem. Lab. Med. 2011, 49, 297–302. [Google Scholar] [CrossRef]
- Davidson, J.; Tong, S.; Hauck, A.; Lawson, D.S.; Jaggers, J.; Kaufman, J.; da Cruz, E. Alkaline phosphatase activity after cardiothoracic surgery in infants and correlation with post-operative support and inflammation: A prospective cohort study. Crit. Care 2012, 16, R160. [Google Scholar] [CrossRef]
- Olegário, J.G.; Silva, M.V.; Machado, J.R.; Rocha, L.P.; Reis, M.A.; Guimarães, C.S.D.O.; Corrêa, R.R.M. Pulmonary innate immune response and melatonin receptors in the perinatal stress. Clin. Dev. Immunol. 2013, 2013, 340959. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, K.A.; Anderson-Berry, A.L.; Delair, S.F.; Davies, H.D. Early-onset neonatal sepsis. Clin. Microbiol. Rev. 2014, 27, 21–47. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, A.; Michelow, I.C.; Chapin, K.C.; Mehta, S.; Patole, S. Viral respiratory tract infections in the neonatal intensive care unit: The VIRIoN-I study. J. Pediatr. 2014, 165, 690–696. [Google Scholar] [CrossRef]
- Bizzarro, M.J.; Jiang, Y.; Hussain, N.; Gruen, J.R.; Bhandari, V.; Zhang, H. The impact of environmental and genetic factors on neonatal late-onset sepsis. J. Pediatr. 2011, 158, 234–238. [Google Scholar] [CrossRef]
- Meem, M.; Modak, J.K.; Mortuza, R.; Shane, A.L.; Sánchez, P.J. Biomarkers for diagnosis of neonatal infections: A systematic analysis of their potential as a point-of-care diagnostics. J. Glob. Health 2011, 1, 201–209. [Google Scholar]
- Kaiser Permanente Research. Neonatal Early-Onset Sepsis Calculator. Available online: https://neonatalsepsiscalculator.kaiserpermanente.org/InfectionProbabilityCalculator.aspx (accessed on 17 December 2022).
- Akangire, G.; Simpson, E.; Weiner, J.; Noel-Macdonnell, J.; Petrikin, J.; Sheehan, M. Implementation of the neonatal sepsis calculator in early-onset sepsis and maternal chorioamnionitis. Adv. Neonatal Care 2020, 20, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, M.; Mehta, S.; Patole, S. Sepsis calculator for neonatal early onset sepsis—A systematic review and meta-analysis. J. Matern. Fetal Neonatal Med. 2021, 34, 1832–1840. [Google Scholar] [CrossRef]
- Li, X.; Li, T.; Wang, J.; Dong, G.; Zhang, M.; Xu, Z.; Hu, Y.; Xie, B.; Yang, J.; Wang, Y. Higher blood urea nitrogen level is independently linked with the presence and severity of neonatal sepsis. Ann. Med. 2021, 53, 2192–2198. [Google Scholar] [CrossRef] [PubMed]
- Coggins, S.A.; Laskin, B.; Harris, M.C.; Grundmeier, R.W.; Passarella, M.; McKenna, K.J.; Srinivasan, L. Acute kidney injury associated with late-onset neonatal sepsis: A matched cohort study. J. Pediatr. 2021, 231, 185–192. [Google Scholar] [CrossRef]

| Proven Sepsis | Probable Sepsis | Suspected Sepsis | |
|---|---|---|---|
| Blood culture results | + | - | - |
| Clinical signs of infection | + | + | uncharacteristic |
| Laboratory markers of infection | + | + | uncharacteristic |
| Perinatal Risk Factors | Proven Sepsis | N’/N (%) | Probable Sepsis | N’/N (%) | Suspected Sepsis | N’/N (%) | Control Group | N’/N (%) |
|---|---|---|---|---|---|---|---|---|
| Gestational Age, Me (IQR) [gestation weeks] | 39 (36–40) | 90/91 (99) | 39 (38–40) | 160/161 (99) | 38 (37–40) | 149/151 (99) | 39 (38–40) | 94/94 (100) |
| Birth Weight, Me (IQR) [g] | 3200 (2460–3645) | 91/91 (100) | 3400 (2910–3780) | 161/161 (100) | 3170 (2743–3525) | 151/151 (100) | 3290 (3010–3628) | 94/94 (100) |
| Head Circumference, Me (IQR) [cm] | 34 (32–36) | 88/91 (97) | 35 (33–36) | 156/161 (97) | 34 (33–35) | 149/151 (99) | 34 (33–35) | 92/94 (98) |
| Male gender, n/N (%) * | 63/91 (69) | 63/91 (69) | 103/161 (64) | 103/161 (64) | 86/151 (57) | 86/151 (57) | 57/94 (61) | 57/94 (61) |
| Vaginal Delivery, n/N’ (%) | 59/89 (66) | 59/91 (65) | 128/158 (81) | 128/161 (80) | 97/145 (67) | 97/151 (64) | 69/94 (73) | 69/94 (73) |
| Apgar 5, Me (range) | 9 (2–10) | 82/91 (90) | 9 (2–10) | 152/161 (94) | 9 (3–10) | 138/151 (91) | 9 (5–10) | 85/94 (90) |
| Meconium-Stained Amniotic Fluid, n/N’ (%) | 10/65 (15) | 65/91 (71) | 23/114 (20) | 114/161 (71) | 24/104 (23) | 104/151 (69) | 2/84 (2) | 10/94 (11) |
| Maternal Risk Factors (ATB/Steroids/PROM), n/N’ (%) | 14/52 (27) | 52/91 (57) | 21/107 (20) | 107/161 (66) | 26/68 (38) | 68/151 (45) | 11/70 (16) | 70/94 (74) |
| Age of Onset, Me (IQR) [days] | 14 (9–21) | 91/91 (100) | 11 (4–19) | 161/161 (100) | 2 (1–13) | 151/151 (100) | 7 (3–16) | 94/94 (100) |
| Clinical Presentation | Proven Sepsis | Probable Sepsis | Suspected Sepsis |
|---|---|---|---|
| Temperature > 38.5 or < 36.0, n/N (%) [°C] | 25/91 (27) | 35/161 (22) | 8/151 (5) |
| Heart Rate > 160, n/N (%) [beats/min] | 43/91 (47) | 52/161 (32) | 27/151 (18) |
| Respiration Rate > 60, n/N (%) [breaths/min] | 33/91 (36) | 66/161 (41) | 39/151 (26) |
| Inappetence, n/N (%) | 40/91 (44) | 75/161 (47) | 35/151 (23) |
| Capillary Refill > 3, n/N (%) [s] | 26/91 (29) | 38/161 (24) | 8/151 (5) |
| Irritability, n/N (%) | 42/91 (46) | 86/161 (53) | 37/151 (25) |
| Lethargy, n/N (%) | 21/91 (23) | 58/161 (36) | 28/151 (19) |
| Jaundice, n/N (%) | 15/91 (16) | 43/161 (27) | 24/151 (16) |
| Laboratory Markers | Proven Sepsis | N’/N (%) | Probable Sepsis | N’/N (%) | Suspected Sepsis | N’/N (%) | Control Group | N’/N (%) |
|---|---|---|---|---|---|---|---|---|
| Leukocyte Count, Me (IQR) [×109/L] | 11 (6.3–16.8) | 91/91 (100) | 13.2 (8.0–18.7) | 150/151 (99) | 12.9 (9.0–18.8) | 160/161 (99) | 10.4 (8.8–11.9) | 94/94 (100) |
| Immature Neutrophils, Me (IQR) [%] | 8 (3–14) | 87/91 (96) | 4 (1–10) | 151/161 (94) | 1 (0–5) | 132/151 (87) | 0 (0–2) | 94/94 (100) |
| Mature Neutrophils, Me (IQR) [%] | 56 (42–68) | 87/91 (96) | 57 (44–68) | 155/161 (96) | 54 (39–64) | 136/151 (90) | 34 (26–46) | 94/94 (100) |
| Lymphocytes, Me (IQR) [%] | 23 (11–31) | 87/91 (96) | 22 (17–33) | 155/161 (96) | 29 (20–42) | 136/151 (90) | 47 (34–56) | 94/94 (100) |
| I:M, Me (IQR) | 0.13 (0.05–0.29) | 87/91 (96) | 0.09 (0.01–0.19) | 151/161 (94) | 0.02 (0.00–0.07) | 132/151 (87) | 0.00 (0.00–0.06) | 94/94 (100) |
| I:T, Me (IQR) | 0.11 (0.05–0.22) | 87/91 (96) | 0.07 (0.00–0.15) | 151/161 (94) | 0.02 (0.00–0.07) | 132/151 (87) | 0.00 (0.00–0.06) | 94/94 (100) |
| Toxic Changes in Neutrophils, n/N’ (%) | 13/90 (14) | 90/91 (99) | 20/154 (13) | 154/161 (96) | 10/133 (8) | 143/151 (95) | 7/90 (8) | 90/94 (96) |
| Thrombocyte Count, Me (IQR) [×103/L] | 272 (191–356) | 90/91 (99) | 296 (210–402) | 158/161 (98) | 276 (215–360) | 149/151 (99) | 322 (259–376) | 94/94 (100) |
| Serum CRP concentration, Me (IQR) [mg/L] | 29 (12–79) | 91/91 (100) | 34 (15–61) | 161/161 (100) | 0 (0–10) | 151/151 (100) | 0 (0–0) | 94/94 (100) |
| Serum PCT concentration, Me (IQR) [μg/L] | 5.6 (1.5–27.9) | 91/91 (100) | 1.9 (0.5–10.0) | 161/161 (100) | 0.4 (0.2–1.0) | 114/151 (75) | 0.1 (0.1–0.2) | 32/94 (34) |
| CA (%) | Se (%) | Sp (%) | PPV (%) | NPV (%) | F1 (%) | AUC (%) | |
|---|---|---|---|---|---|---|---|
| Random Forest | 83 | 81 | 87 | 86 | 79 | 84 | 84 |
| Logistic Regression | 81 | 76 | 86 | 86 | 74 | 80 | 81 |
| Decision Tree Classifier | 82 | 80 | 84 | 84 | 80 | 82 | 82 |
| Support Vector Machine | 79 | 75 | 83 | 83 | 75 | 78 | 79 |
| K-Nearest Neighbours | 62 | 35 | 78 | 78 | 35 | 49 | 63 |
| CA (%) | Se (%) | Sp (%) | PPV (%) | NPV (%) | F1 (%) |
|---|---|---|---|---|---|
| 86 | 82 | 89 | 89 | 82 | 85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
But, Š.; Celar, B.; Fister, P. Tackling Neonatal Sepsis—Can It Be Predicted? Int. J. Environ. Res. Public Health 2023, 20, 3644. https://doi.org/10.3390/ijerph20043644
But Š, Celar B, Fister P. Tackling Neonatal Sepsis—Can It Be Predicted? International Journal of Environmental Research and Public Health. 2023; 20(4):3644. https://doi.org/10.3390/ijerph20043644
Chicago/Turabian StyleBut, Špela, Brigita Celar, and Petja Fister. 2023. "Tackling Neonatal Sepsis—Can It Be Predicted?" International Journal of Environmental Research and Public Health 20, no. 4: 3644. https://doi.org/10.3390/ijerph20043644
APA StyleBut, Š., Celar, B., & Fister, P. (2023). Tackling Neonatal Sepsis—Can It Be Predicted? International Journal of Environmental Research and Public Health, 20(4), 3644. https://doi.org/10.3390/ijerph20043644




