Integrating Structured and Unstructured EHR Data for Predicting Mortality by Machine Learning and Latent Dirichlet Allocation Method
Abstract
1. Introduction
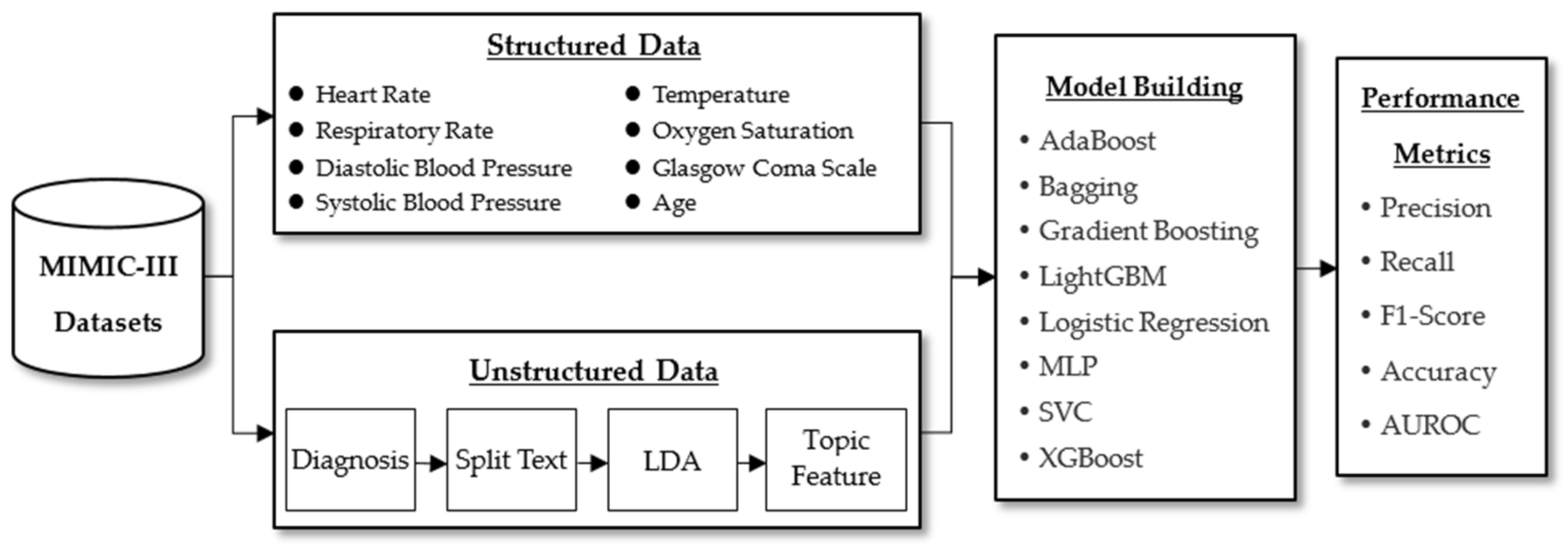
2. Materials and Methods
2.1. Proposed Framework
2.2. Data Collection and Preprocessing
2.3. Baseline Characteristics
2.4. Latent Dirichlet Allocation
2.5. Machine Learning
- AdaBoost is an adaptive method in the sense that incorrect samples from the previous classifier are used to train the next classifier. The AdaBoost method is sensitive to noise and abnormal data. It trains a basic classifier and gives misclassified samples more weight. It is then applied to the next process. This iterative process is repeated until a stopping condition is reached or the error rate is low enough [46,47]. The Python sklearn library was used to implement AdaBoost. Our hyperparameters specified a maximum number of iterations of 50, while others trained the model using the sklearn preset values.
- The Bootstrap Aggregating algorithm, also known as the Bagging algorithm, is an ensemble learning algorithm in the field of machine learning, which was first proposed by Leo Breiman in 1994. The Bagging algorithm can be combined with other classification and regression algorithms to improve accuracy and stability while reducing result variance to avoid overfitting. Bagging is an ensemble method that combines multiple predictors. It helps to prevent model overfitting to data and reduces variance. It has been used in many microarray studies [48,49]. The Python sklearn library was also used to implement bagging. A combined classifier made up of 500 DecisionTreeClassifiers was used. Each classifier has a maximum sampling subset of 100; the self-service sampling method was used for each sampling. Other hyperparameters used sklearn preset values to carry out training.
- Gradient Boosting is an ensemble learning algorithm that can be used to improve the accuracy of various classification prediction models. It trains a model with poor prediction accuracy using the negative gradient information of the model loss function and then combines the trained results with the existing model in a cumulative form [50]. The scikit-learn library was also used in this study to achieve gradient boosting; the maximum number of iterations was set to 100; and other hyperparameters were trained using preset scikit-learn library values.
- The Light Gradient Boosting Machine (LightGBM) is an ensemble method that combines the predictions of multiple decision trees to produce a well-generalized final prediction. LightGBM divides continuous eigenvalues into K intervals and chooses dividing points from those intervals. This method significantly accelerates prediction and reduces memory occupancy without sacrificing prediction accuracy [51]. LightGBM is a decision tree learning algorithm with gradient boosting that has been widely used for feature selection, classification, and regression [52].
- Logistic Regression is a logit model capable of testing statistical interactions and controlling multivariate confidence. It is most commonly used to investigate the risk relationship between disease and exposure [53,54]. In this study, the Python scikit-learn library was used to implement logistic regression, and the hyperparameter optimization method was the SAG linear convergence algorithm. It is a gradient descent method used specifically for large sample data.
- Multilayer Perceptron (MLP) is a feed-forward artificial neural network with a fixed number of computational units or neurons that are fully connected to the next layer [55]. A multilayer perceptron learns and predicts data using the principles of the human nervous system. MLPs are suitable for classifying and predicting tasks with different feature set implementations [56]. The neural network used in this study had five layers: an input layer, three hidden layers, and an output layer. Each hidden layer has 13 neuron nodes, the normalization parameter was set to 1e-5, relu was used as the activation function, and adam was used for training and weight optimization.
- The Support Vector Classifier (SVC) analyzes linear and nonlinear data for classification and regression. SVC aims to recognize categories by the creation of non-linear decision hyperplanes in a higher feature space [57]. SVC is resistant to data bias and variance and produces accurate predictions for binary or multiclass classifications. Additionally, SVC is robust, resists overfitting, and has exceptional generalization capabilities [58].
- eXtreme Gradient Boosting (XGBoost) is a scalable end-to-end tree boosting system that is an optimized implementation of the gradient boosting framework. It is remarkable in that it can handle missing data efficiently, is very flexible, and can build an assembly of weak prediction models into an accurate one [59]. It generates a series of decision trees during training, each building on the previous one to reduce the loss function gradient. Furthermore, a predictive model made up of multiple decision trees can be obtained. The XGBoost algorithm can deal with missing values by including a default orientation for missing values in each tree node and learning the best orientation from the data [60].
2.6. The Synthetic Minority Oversampling Technique (SMOTE)
2.7. Performance Evaluation
3. Results
3.1. Prediction of Mortality in ICU
3.2. Feature Importance
4. Discussion
4.1. Principal Findings
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Prediction | |||
|---|---|---|---|
| Positive | Negative | ||
| Actual | Positive | True Positive, TP | False Negative, FN |
| Negative | False Positive, FP | True Negative, TN | |
- AUROC: The area under the ROC curve (Receiver Operating Characteristic Curve) was used to measure performance of the classifier across all classification thresholds. The AUC measuring standard assigns the same weight to each instance, regardless of the nature of the positive label. The ROC curve is obtained through the FPR value on the abscissa and the TPR value on the ordinate.
- Precision: Also called positive predictive value (PPV). This is the proportion of correct predictions in positive samples; in other words, the proportion of positive samples among all positive samples classified.
- Recall: Also called true-positive rate (TPR). This is the proportion of samples that are predicted to be correct among factual samples, or the proportion of positive samples predicted among all positive samples.
- F1-score: The harmonic mean between precision and recall. Precision and recall are often discussed in identification- and prediction-related algorithms, whereas the F1-score considers both precision and recall and is a comprehensive measure of model performance.
- Accuracy: This is the ratio of the number of samples correctly classified (by the classifier) to the total number of samples for a given test dataset. In other words, it is the overall ratio of the model that predicts the correct quantity.
Appendix B
| Dataset | Method | 3 Days | |||
|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Accuracy | ||
| Structured Data | AdaBoost | 0.1692 ± 0.0116 | 0.8662 ± 0.0149 | 0.2828 ± 0.0156 | 0.8537 ± 0.0061 |
| Bagging | 0.1370 ± 0.0062 | 0.9194 ± 0.0179 | 0.2383 ± 0.0091 | 0.8047 ± 0.0031 | |
| Gradient Boosting | 0.1756 ± 0.0130 | 0.8577 ± 0.0213 | 0.2911 ± 0.0175 | 0.8609 ± 0.0061 | |
| LightGBM | 0.2616 ± 0.0182 | 0.6644 ± 0.0326 | 0.3747 ± 0.0178 | 0.9263 ± 0.0029 | |
| Logistic Regression | 0.1242 ± 0.0082 | 0.8153 ± 0.0457 | 0.2156 ± 0.0140 | 0.8084 ± 0.0012 | |
| MLP | 0.1535 ± 0.0117 | 0.8525 ± 0.0281 | 0.2601 ± 0.0216 | 0.8504 ± 0.0163 | |
| SVC | 0.1244 ± 0.0074 | 0.8176 ± 0.0125 | 0.2158 ± 0.0109 | 0.8023 ± 0.0043 | |
| XGBoost | 0.3284 ± 0.0243 | 0.4240 ± 0.0256 | 0.3688 ± 0.0131 | 0.9518 ± 0.0012 | |
| Structured Data + Unstructured Data | AdaBoost | 0.1752 ± 0.0015 | 0.8754 ± 0.0188 | 0.2920 ± 0.0030 | 0.8624 ± 0.0037 |
| Bagging | 0.1314 ± 0.0013 | 0.9513 ± 0.0155 | 0.2309 ± 0.0023 | 0.8085 ± 0.0036 | |
| Gradient Boosting | 0.1863 ± 0.0060 | 0.8594 ± 0.0220 | 0.3063 ± 0.0093 | 0.8737 ± 0.0046 | |
| LightGBM | 0.3988 ± 0.0091 | 0.6975 ± 0.0299 | 0.5073 ± 0.0130 | 0.9561 ± 0.0018 | |
| Logistic Regression | 0.1264 ± 0.0013 | 0.8324 ± 0.0213 | 0.2194 ± 0.0027 | 0.8108 ± 0.0014 | |
| MLP | 0.1568 ± 0.0080 | 0.8600 ± 0.0540 | 0.2648 ± 0.0088 | 0.8543 ± 0.0181 | |
| SVC | 0.1213 ± 0.0018 | 0.8297 ± 0.0139 | 0.2117 ± 0.0032 | 0.8036 ± 0.0048 | |
| XGBoost | 0.5786 ± 0.0121 | 0.5443 ± 0.0364 | 0.5600 ± 0.0157 | 0.9723 ± 0.0005 | |
| Dataset | Method | 30 Days | |||
|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Accuracy | ||
| Structured Data | AdaBoost | 0.2872 ± 0.0116 | 0.7252 ± 0.0181 | 0.4114 ± 0.0148 | 0.7719 ± 0.0029 |
| Bagging | 0.2880 ± 0.0097 | 0.7709 ± 0.0092 | 0.4193 ± 0.0116 | 0.7653 ± 0.0034 | |
| Gradient Boosting | 0.2904 ± 0.0152 | 0.7262 ± 0.0213 | 0.4149 ± 0.0190 | 0.7748 ± 0.0059 | |
| LightGBM | 0.3349 ± 0.0081 | 0.6875 ± 0.0148 | 0.4504 ± 0.0104 | 0.8156 ± 0.0014 | |
| Logistic Regression | 0.2717 ± 0.0081 | 0.7462 ± 0.0165 | 0.3983 ± 0.0109 | 0.7522 ± 0.0006 | |
| MLP | 0.2846 ± 0.0329 | 0.7520 ± 0.0265 | 0.4113 ± 0.0310 | 0.7610 ± 0.0318 | |
| SVC | 0.2854 ± 0.0107 | 0.7347 ± 0.0205 | 0.4111 ± 0.0143 | 0.7686 ± 0.0027 | |
| XGBoost | 0.3753 ± 0.0129 | 0.5577 ± 0.0157 | 0.4486 ± 0.0128 | 0.8493 ± 0.0024 | |
| Structured Data + Unstructured Data | AdaBoost | 0.2865 ± 0.0026 | 0.7275 ± 0.0145 | 0.4111 ± 0.0050 | 0.7740 ± 0.0012 |
| Bagging | 0.2873 ± 0.0013 | 0.7531 ± 0.0264 | 0.4158 ± 0.0049 | 0.7696 ± 0.0032 | |
| Gradient Boosting | 0.2928 ± 0.0007 | 0.7335 ± 0.0190 | 0.4185 ± 0.0033 | 0.7780 ± 0.0021 | |
| LightGBM | 0.3411 ± 0.0014 | 0.6754 ± 0.0289 | 0.4532 ± 0.0078 | 0.8226 ± 0.0021 | |
| Logistic Regression | 0.2664 ± 0.0046 | 0.7291 ± 0.0205 | 0.3902 ± 0.0077 | 0.7568 ± 0.0039 | |
| MLP | 0.2689 ± 0.0097 | 0.7549 ± 0.0492 | 0.3958 ± 0.0059 | 0.7686 ± 0.0210 | |
| SVC | 0.2685 ± 0.0046 | 0.7250 ± 0.0236 | 0.3918 ± 0.0083 | 0.7649 ± 0.0011 | |
| XGBoost | 0.3816 ± 0.0061 | 0.5435 ± 0.0137 | 0.4483 ± 0.0082 | 0.8543 ± 0.0015 | |
| Dataset | Method | 365 Days | |||
|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Accuracy | ||
| Structured Data | AdaBoost | 0.3005 ± 0.0030 | 0.7125 ± 0.0030 | 0.4227 ± 0.0028 | 0.7750 ± 0.0050 |
| Bagging | 0.2919 ± 0.0022 | 0.7397 ± 0.0117 | 0.4186 ± 0.0041 | 0.7625 ± 0.0029 | |
| Gradient Boosting | 0.3043 ± 0.0021 | 0.7097 ± 0.0014 | 0.4259 ± 0.0023 | 0.7789 ± 0.0013 | |
| LightGBM | 0.3438 ± 0.0057 | 0.6723 ± 0.0073 | 0.4549 ± 0.0063 | 0.8138 ± 0.0028 | |
| Logistic Regression | 0.2763 ± 0.0010 | 0.7156 ± 0.0096 | 0.3986 ± 0.0017 | 0.7504 ± 0.0038 | |
| MLP | 0.2918 ± 0.0141 | 0.7325 ± 0.0389 | 0.4166 ± 0.0100 | 0.7622 ± 0.0213 | |
| SVC | 0.2917 ± 0.0013 | 0.7159 ± 0.0089 | 0.4145 ± 0.0023 | 0.7662 ± 0.0044 | |
| XGBoost | 0.3875 ± 0.0035 | 0.5571 ± 0.0108 | 0.4570 ± 0.0055 | 0.8470 ± 0.0017 | |
| Structured Data + Unstructured Data | AdaBoost | 0.3018 ± 0.0098 | 0.7462 ± 0.0197 | 0.4298 ± 0.0132 | 0.7758 ± 0.0053 |
| Bagging | 0.3054 ± 0.0057 | 0.7681 ± 0.0118 | 0.4370 ± 0.0066 | 0.7760 ± 0.0024 | |
| Gradient Boosting | 0.3065 ± 0.0083 | 0.7463 ± 0.0141 | 0.4345 ± 0.0107 | 0.7801 ± 0.0055 | |
| LightGBM | 0.3602 ± 0.0056 | 0.6997 ± 0.0068 | 0.4755 ± 0.0061 | 0.8253 ± 0.0012 | |
| Logistic Regression | 0.2822 ± 0.0055 | 0.7381 ± 0.0094 | 0.4083 ± 0.0071 | 0.7578 ± 0.0020 | |
| MLP | 0.2826 ± 0.0039 | 0.7518 ± 0.0165 | 0.4107 ± 0.0046 | 0.7657 ± 0.0079 | |
| SVC | 0.2866 ± 0.0058 | 0.7306 ± 0.0055 | 0.4117 ± 0.0068 | 0.7696 ± 0.0042 | |
| XGBoost | 0.3945 ± 0.0033 | 0.5659 ± 0.0112 | 0.4649 ± 0.0060 | 0.8526 ± 0.0017 | |
References
- Marshall, J.C.; Bosco, L.; Adhikari, N.K.; Connolly, B.; Diaz, J.V.; Dorman, T.; Fowler, R.A.; Meyfroidt, G.; Nakagawa, S.; Pelosi, P.; et al. What is an intensive care unit? A report of the task force of the World Federation of Societies of Intensive and Critical Care Medicine. J. Crit. Care 2017, 37, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Mahbub, M.; Srinivasan, S.; Danciu, I.; Peluso, A.; Begoli, E.; Tamang, S.; Peterson, G.D. Unstructured clinical notes within the 24 hours since admission predict short, mid & long-term mortality in adult ICU patients. PLoS ONE 2022, 17, e0262182. [Google Scholar]
- Chen, W.; Long, G.; Yao, L.; Sheng, Q.Z. AMRNN: Attended multi-task recurrent neural networks for dynamic illness severity prediction. World Wide Web 2019, 23, 2753–2770. [Google Scholar] [CrossRef]
- Romana, S.; Bernhard, F. Iatrogenic events contributing to paediatric intensive care unit admission. Swiss Med. Wkly. 2021, 151, 7. [Google Scholar]
- Caicedo-Torres, W.; Gutierrez, K. ISeeU2: Visually interpretable mortality prediction inside the ICU using deep learning and free-text medical notes. Expert Syst. Appl. 2022, 202, 117190. [Google Scholar] [CrossRef]
- Romano, M. The Role of Palliative Care in the Cardiac Intensive Care Unit. Healthcare 2019, 7, 30. [Google Scholar] [CrossRef]
- El-Rashidy, N.; El-Sappagh, S.; Abuhmed, T.; Abdelrazek, S.; El-Bakry, H.M. Intensive Care Unit Mortality Prediction: An Improved Patient-Specific Stacking Ensemble Model. IEEE Access 2020, 8, 133541–133564. [Google Scholar] [CrossRef]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonça, A.; Bruining, H.; Reinhart, C.K.; Suter, P.; Thijs, L.G. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Legall, J.R.; Lemeshow, S.; Saulnier, F. A new simplified acute physiology score (SAPS-II) based on a European North-American multicenter study. Jama J. Am. Med. Assoc. 1993, 270, 2957–2963. [Google Scholar] [CrossRef]
- Baue, A.E.; Durham, R.; Faist, E. Systemic inflammatory response syndrome (SIRS), multiple organ dysfunction syndrome (MODS), multiple organ failure (MOF): Are we winning the battle? Shock 1998, 10, 79–89. [Google Scholar] [CrossRef]
- Ibrahim, Z.M.; Wu, H.H.; Hamoud, A.; Stappen, L.; Dobson, R.J.B.; Agarossi, A. On classifying sepsis heterogeneity in the ICU: Insight using machine learning. J. Am. Med. Inform. Assoc. 2020, 27, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Darabi, S.; Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. TAPER: Time-Aware Patient EHR Representation. IEEE J. Biomed. Health Inform. 2020, 24, 3268–3275. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.G.; Pan, K.; Xie, Y.; Qin, A.K.; Tang, Z.D. Preserving differential privacy in deep neural networks with relevance-based adaptive noise imposition. Neural Netw. 2020, 125, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Sheikhalishahi, S.; Balaraman, V.; Osmani, V. Benchmarking machine learning models on multi-centre eICU critical care dataset. PLoS ONE 2020, 15, e0235424. [Google Scholar] [CrossRef]
- Loreto, M.; Lisboa, T.; Moreira, V.P. Early prediction of ICU readmissions using classification algorithms. Comput. Biol. Med. 2020, 118, 8. [Google Scholar] [CrossRef]
- Baker, S.; Xiang, W.; Atkinson, I. Continuous and automatic mortality risk prediction using vital signs in the intensive care unit: A hybrid neural network approach. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Davidson, S.; Villarroel, M.; Harford, M.; Finnegan, E.; Jorge, J.; Young, D.; Watkinson, P.; Tarassenko, L. Day-to-day progression of vital-sign circadian rhythms in the intensive care unit. Crit. Care 2021, 25, 13. [Google Scholar] [CrossRef]
- Alghatani, K.; Ammar, N.; Rezgui, A.; Shaban-Nejad, A. Predicting Intensive Care Unit Length of Stay and Mortality Using Patient Vital Signs: Machine Learning Model Development and Validation. JMIR Med. Inform. 2021, 9, e21347. [Google Scholar] [CrossRef]
- Sarang, B.; Bhandarkar, P.; Raykar, N.; O’Reilly, G.M.; Soni, K.D.; Wärnberg, M.G.; Khajanchi, M.; Dharap, S.; Cameron, P.; Howard, T.; et al. Associations of On-arrival Vital Signs with 24-hour In-hospital Mortality in Adult Trauma Patients Admitted to Four Public University Hospitals in Urban India: A Prospective Multi-Centre Cohort Study. Inj. Int. J. Care Inj. 2021, 52, 1158–1163. [Google Scholar] [CrossRef]
- Hashir, M.; Sawhney, R. Towards unstructured mortality prediction with free-text clinical notes. J. Biomed. Inform. 2020, 108, 103489. [Google Scholar] [CrossRef]
- Tootooni, M.S.; Pasupathy, K.S.; Heaton, H.A.; Clements, C.M.; Sir, M.Y. CCMapper: An adaptive NLP-based free-text chief complaint mapping algorithm. Comput. Biol. Med. 2019, 113, 13. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.C.; Yao, L.; Shen, J.H.; Janarthanam, R.; Luo, Y. Predicting mortality in critically ill patients with diabetes using machine learning and clinical notes. BMC Med. Inform. Decis. Mak. 2020, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Yin, C.C.; Zeng, J.C.; Yuan, X.H.; Zhang, P. Combining structured and unstructured data for predictive models: A deep learning approach. BMC Med. Inform. Decis. Mak. 2020, 20, 280. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T. Machine Learning; McGraw-Hill: New York, NY, USA, 1997; Volume 1. [Google Scholar]
- Adlung, L.; Cohen, Y.; Mor, U.; Elinav, E. Machine learning in clinical decision making. Med 2021, 2, 642–665. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Purushotham, S.; Meng, C.Z.; Che, Z.P.; Liu, Y. Benchmarking deep learning models on large healthcare datasets. J. Biomed. Inform. 2018, 83, 112–134. [Google Scholar] [CrossRef]
- Cheng, X.; Cao, Q.; Liao, S.S. An overview of literature on COVID-19, MERS and SARS: Using text mining and latent Dirichlet allocation. J. Inf. Sci. 2022, 48, 304–320. [Google Scholar] [CrossRef]
- Xue, J.; Chen, J.X.; Chen, C.; Zheng, C.D.; Li, S.J.; Zhu, T.S. Public discourse and sentiment during the COVID 19 pandemic: Using Latent Dirichlet Allocation for topic modeling on Twitter. PLoS ONE 2020, 15, e0239441. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Breuninger, T.A.; Wawro, N.; Breuninger, J.; Reitmeier, S.; Clavel, T.; Six-Merker, J.; Pestoni, G.; Rohrmann, S.; Rathmann, W.; Peters, A.; et al. Associations between habitual diet, metabolic disease, and the gut microbiota using latent Dirichlet allocation. Microbiome 2021, 9, 61. [Google Scholar] [CrossRef]
- Gangavarapu, T.; Jayasimha, A.; Krishnan, G.S.; Kamath, S.S. Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes. Knowl. Based Syst. 2020, 190, 105321. [Google Scholar] [CrossRef]
- Chiu, C.C.; Wu, C.M.; Chien, T.N.; Kao, L.J.; Qiu, J.T. Predicting the Mortality of ICU Patients by Topic Model with Machine-Learning Techniques. Healthcare 2022, 10, 1087. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Zheng, Y.; Zhang, R.; Jiang, Y.; Poon, C.C.Y. Using a Multi-Task Recurrent Neural Network With Attention Mechanisms to Predict Hospital Mortality of Patients. IEEE J. Biomed. Health Inf. 2020, 24, 486–492. [Google Scholar] [CrossRef]
- Guo, C.H.; Lu, M.L.; Chen, J.F. An evaluation of time series summary statistics as features for clinical prediction tasks. BMC Med. Inform. Decis. Mak. 2020, 20, 48. [Google Scholar] [CrossRef]
- Sayed, M.; Riano, D.; Villar, J. Predicting Duration of Mechanical Ventilation in Acute Respiratory Distress Syndrome Using Supervised Machine Learning. J. Clin. Med. 2021, 10, 3824. [Google Scholar] [CrossRef]
- Kozlowski, D.; Semeshenko, V.; Molinari, A. Latent Dirichlet allocation model for world trade analysis. PLoS ONE 2021, 16, e0245393. [Google Scholar] [CrossRef]
- Li, Y.; Rapkin, B.; Atkinson, T.M.; Schofield, E.; Bochner, B.H. Leveraging Latent Dirichlet Allocation in processing free-text personal goals among patients undergoing bladder cancer surgery. Qual. Life Res. 2019, 28, 1441–1455. [Google Scholar] [CrossRef]
- Celard, P.; Vieira, A.S.; Iglesias, E.L.; Borrajo, L. LDA filter: A Latent Dirichlet Allocation preprocess method for Weka. PLoS ONE 2020, 15, e0241701. [Google Scholar] [CrossRef]
- Chen, J.H.; Goldstein, M.K.; Asch, S.M.; Mackey, L.; Altman, R.B. Predicting inpatient clinical order patterns with probabilistic topic models vs conventional order sets. J. Am. Med. Inform. Assoc. 2017, 24, 472–480. [Google Scholar] [CrossRef]
- Pivovarov, R.; Perotte, A.J.; Grave, E.; Angiolillo, J.; Wiggins, C.H.; Elhadad, N. Learning probabilistic phenotypes from heterogeneous EHR data. J. Biomed. Inform. 2015, 58, 156–165. [Google Scholar] [CrossRef]
- Choi, Y.; Chiu, C.Y.-I.; Sontag, D. Learning low-dimensional representations of medical concepts. AMIA Summits Transl. Sci. Proc. 2016, 2016, 41–50. [Google Scholar] [PubMed]
- Gabriel, R.A.; Kuo, T.-T.; McAuley, J.; Hsu, C.-N. Identifying and characterizing highly similar notes in big clinical note datasets. J. Biomed. Inform. 2018, 82, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Ma, Z.; Chen, J.; Xiao, M.; Huang, L.F. Automatic Medical Code Assignment via Deep Learning Approach for Intelligent Healthcare. IEEE J. Biomed. Health Inform. 2020, 24, 2506–2515. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Choi, J.Y.; Ro, Y.M. Region based stellate features combined with variable selection using AdaBoost learning in mammographic computer-aided detection. Comput. Biol. Med. 2015, 63, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Choi, J.W.; Shin, E.H. Machine learning model for predicting malaria using clinical information. Comput. Biol. Med. 2021, 129, 104151. [Google Scholar] [CrossRef]
- Ali, S.; Majid, A.; Javed, S.G.; Sattar, M. Can-CSC-GBE: Developing Cost-sensitive Classifier with Gentleboost Ensemble for breast cancer classification using protein amino acids and imbalanced data. Comput. Biol. Med. 2016, 73, 38–46. [Google Scholar] [CrossRef]
- Sarmah, C.K.; Samarasinghe, S. Microarray gene expression: A study of between-platform association of Affymetrix and cDNA arrays. Comput. Biol. Med. 2011, 41, 980–986. [Google Scholar] [CrossRef]
- Ramos-Gonzalez, J.; Lopez-Sanchez, D.; Castellanos-Garzon, J.A.; de Paz, J.F.; Corchado, J.M. A CBR framework with gradient boosting based feature selection for lung cancer subtype classification. Comput. Biol. Med. 2017, 86, 98–106. [Google Scholar] [CrossRef]
- Song, J.Z.; Liu, G.X.; Jiang, J.Q.; Zhang, P.; Liang, Y.C. Prediction of Protein-ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. Int. J. Mol. Sci. 2021, 22, 939. [Google Scholar] [CrossRef]
- Li, L.J.; Lin, Y.K.; Yu, D.X.; Liu, Z.Y.; Gao, Y.J.; Qiao, J.P. A Multi-Organ Fusion and LightGBM Based Radiomics Algorithm for High-Risk Esophageal Varices Prediction in Cirrhotic Patients. IEEE Access 2021, 9, 15041–15052. [Google Scholar] [CrossRef]
- Cuadrado-Godia, E.; Jamthikar, A.D.; Gupta, D.; Khanna, N.N.; Araki, T.; Maniruzzaman, M.; Saba, L.; Nicolaides, A.; Sharma, A.; Omerzu, T.; et al. Ranking of stroke and cardiovascular risk factors for an optimal risk calculator design: Logistic regression approach. Comput. Biol. Med. 2019, 108, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Ergun, U.; Serhatioglu, S.; Hardalac, F.; Guler, I. Classification of carotid artery stenosis of patients with diabetes by neural network and logistic regression. Comput. Biol. Med. 2004, 34, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.S.; Kurita, T.; Ahn, B.C. Critical texture pattern feature assessment for characterizing colonies of induced pluripotent stem cells through machine learning techniques. Comput. Biol. Med. 2018, 94, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Guler, E.C.; Sankur, B.; Kahya, Y.P.; Raudys, S. Visual classification of medical data using MLP mapping. Comput. Biol. Med. 1998, 28, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, S.; Fogarty, S.; Tremeer, M.; Ross, K.; Richards, B.; Bergmeir, C.; Xu, S.; Stub, D.; Smith, K.; Tacey, M.; et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med. 2018, 15, e1002709. [Google Scholar] [CrossRef]
- Akbari, G.; Nikkhoo, M.; Wang, L.; Chen, C.P.; Han, D.S.; Lin, Y.H.; Chen, H.B.; Cheng, C.H. Frailty Level Classification of the Community Elderly Using Microsoft Kinect-Based Skeleton Pose: A Machine Learning Approach. Sensors 2021, 21, 4017. [Google Scholar] [CrossRef]
- Hou, N.; Li, M.; He, L.; Xie, B.; Wang, L.; Zhang, R.; Yu, Y.; Sun, X.; Pan, Z.; Wang, K. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: A machine learning approach using XGboost. J. Transl. Med. 2020, 18, 462. [Google Scholar] [CrossRef]
- Luo, X.Q.; Yan, P.; Duan, S.B.; Kang, Y.X.; Deng, Y.H.; Liu, Q.; Wu, T.; Wu, X. Development and Validation of Machine Learning Models for Real-Time Mortality Prediction in Critically Ill Patients With Sepsis-Associated Acute Kidney Injury. Front. Med. 2022, 9, 853102. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Classifying imbalanced data using SMOTE based class-specific kernelized ELM. Int. J. Mach. Learn. Cybern. 2021, 12, 1255–1280. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, Z.W.; Chen, C.; Wei, Q.Q.; Gu, H.M.; Yu, B. DeepStack-DTIs: Predicting Drug-Target Interactions Using LightGBM Feature Selection and Deep-Stacked Ensemble Classifier. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 311–330. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Mpanya, D.; Celik, T.; Klug, E.; Ntsinjana, H. Machine learning and statistical methods for predicting mortality in heart failure. Heart Fail. Rev. 2021, 26, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Javan, S.L.; Sepehri, M.M.; Javan, M.L.; Khatibi, T. An intelligent warning model for early prediction of cardiac arrest in sepsis patients. Comput. Methods Programs Biomed. 2019, 178, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Blagus, R.; Lusa, L. Joint use of over-and under-sampling techniques and cross-validation for the development and assessment of prediction models. BMC Bioinform. 2015, 16, 363. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, L.; Liu, F.; Wang, X.; Chen, J.; Chou, K.-C. Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS ONE 2015, 10, e0121501. [Google Scholar] [CrossRef]
- Liu, B.; Fang, L.; Liu, F.; Wang, X.; Chou, K.-C. iMiRNA-PseDPC: MicroRNA precursor identification with a pseudo distance-pair composition approach. J. Biomol. Struct. Dyn. 2016, 34, 223–235. [Google Scholar] [CrossRef]
- Upadhyay, D.; Manero, J.; Zaman, M.; Sampalli, S. Gradient Boosting Feature Selection With Machine Learning Classifiers for Intrusion Detection on Power Grids. IEEE Trans. Netw. Serv. Manag. 2021, 18, 1104–1116. [Google Scholar] [CrossRef]
- Adler, A.I.; Painsky, A. Feature Importance in Gradient Boosting Trees with Cross-Validation Feature Selection. Entropy 2022, 24, 687. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2001. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Liu, D.; Wu, Y.L.; Li, X.; Qi, L. Medi-Care AI: Predicting medications from billing codes via robust recurrent neural networks. Neural Netw. 2020, 124, 109–116. [Google Scholar] [CrossRef]
- Savkov, A.; Carroll, J.; Koeling, R.; Cassell, J. Annotating patient clinical records with syntactic chunks and named entities: The Harvey Corpus. Lang. Resour. Eval. 2016, 50, 523–548. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.F.; Kumar, S.; Sen, A.; Sinha, A. Impact of the Hospital Readmission Reduction Program on hospital readmission and mortality: An economic analysis. Prod. Oper. Manag. 2022, 31, 2341–2360. [Google Scholar] [CrossRef]
- Senot, C. Continuity of care and risk of readmission: An investigation into the healthcare journey of heart failure patients. Prod. Oper. Manag. 2019, 28, 2008–2030. [Google Scholar] [CrossRef]
- Lin, Y.W.; Zhou, Y.Q.; Faghri, F.; Shawl, M.J.; Campbell, R.H. Analysis and prediction of unplanned intensive care unit readmission using recurrent neural networks with long shortterm memory. PLoS ONE 2019, 14, e0218942. [Google Scholar]



| Distinct Patients | 46,520 |
|---|---|
| Age, years, median [Q1–Q3] | 65.8 [52.8–77.8] |
| Gender, male, percent of unit stays | 26,121 (56.15%) |
| Distinct hospital admissions | 58,976 |
| Elective | 7706 (13.07%) |
| Emergency | 42,071 (71.34%) |
| Newborn | 7863 (13.33%) |
| Urgent | 1336 (2.27%) |
| Hospital mortality, percent of unit stays | 5854 (9.93%) |
| Hospital length of stay, median days [Q1–Q3] | 10.13 [3.74–11.80] |
| Distinct ICU stays | 61,532 |
| Coronary Care Unit | 7726 (12.56%) |
| Cardiac Surgery Recovery Unit | 9312 (15.13%) |
| Medical Intensive Care Unit | 21,088 (34.27%) |
| Neonatal Intensive Care Unit | 8100 (13.16%) |
| Surgical Intensive Care Unit | 8891 (14.45%) |
| Trauma Surgical Intensive Care Unit | 6415 (10.43%) |
| ICU length of stay, median days [Q1–Q3] | 4.92 [1.11–4.48] |
| Total | Survivors | Non-Survivors | |
|---|---|---|---|
| General | |||
| Number | 27,550 (100%) | 24,364 (88.44%) | 3186 (11.56%) |
| Gender (male) | 15,441 (56.05%) | 13,764 (89.14%) | 1677 (10.86%) |
| Length of stay | |||
| Hospital (days) [Q1–Q3] | 10.39 [4.36–12.42] | 10.29 [4.44–12.35] | 11.20 [3.88–12.54] |
| ICU (days) [Q1–Q3] | 4.48 [1.55–4.61] | 4.18 [1.53–4.33] | 6.77 [1.70–6.13] |
| Admission Type | |||
| Elective | 3537 (12.84%) | 3434 (14.09%) | 103 (3.23%) |
| Emergency | 23,283 (84.51%) | 20,293 (83.29%) | 2990 (93.85%) |
| Urgent | 730 (2.65%) | 637 (2.61%) | 93 (2.92%) |
| Insurance | |||
| Government | 844 (3.06%) | 799 (3.28%) | 45 (1.41%) |
| Medicaid | 2301 (8.35%) | 2078 (8.52%) | 223 (7.00%) |
| Medicare | 14,750 (53.54%) | 12,618 (51.79%) | 2132 (66.92%) |
| Private | 9303 (33.77%) | 8570 (35.17%) | 733 (23.01%) |
| Self-Pay | 352 (1.28%) | 300 (1.23%) | 52 (1.63%) |
| Variable value (First 24 h) | |||
| Heart Rate | 85.72 ± 15.91 | 85.11 ± 15.52 | 90.38 ± 17.93 |
| Respiratory Rate | 18.94 ± 4.01 | 18.70 ± 3.84 | 20.75 ± 4.76 |
| Diastolic Blood Pressure | 60.33 ± 12.21 | 60.66 ± 12.09 | 57.79 ± 12.79 |
| Systolic Blood Pressure | 118.01 ± 18.60 | 118.45 ± 18.30 | 114.63 ± 20.45 |
| Temperature | 98.23 ± 2.03 | 98.28 ± 1.73 | 97.90 ± 3.54 |
| Oxygen Saturation | 97.21 ± 2.11 | 97.28 ± 1.90 | 96.68 ± 3.28 |
| Glasgow Coma Scale | 12.31 ± 3.21 | 12.66 ± 2.91 | 9.64 ± 4.00 |
| Age | 64.00 ± 17.71 | 63.13 ± 17.75 | 70.66 ± 15.81 |
| SUBJECT_ID | HADM_ID | Diagnosis |
|---|---|---|
| 00412 | 109897 | AORTIC STENOSIS; MITRAL REGURGITATION; CAD\AORTIC VALVE REPLACEMENT; MITRAL VALVE REPLACEMENT; CORONARY ARTERY BYPASS GRAFT; TRICUSPID VALVE REPLACEMENT/SDA |
| 00969 | 137250 | BATTERY DEPLETION; HEART FAILURE\IMPLANTABLE CARDIOVERTER DEFIBRILLATOR EXPLANT; PACEMAKER IMPLANT; DIURISIS POST PROCEDURE/SDA |
| 14229 | 145873 | MARFAN’S SYNDROME\BENTALL PROCEDURE; TOTAL VALVE SPARING ROOT REPLACEMENT VS; HOMOGRAFT ROOT REPLACEMENT; REPLACEMENT OF ARCH, PROXIMAL ROOT/SDA |
| 22416 | 130625 | DESCENDING AORTIC ANEURYSM; COARCTATION OF DESCENDING AORTA\ DISTAL ARCH REPLACEMENT; DESCENDING THORACIC AORTIC REPLACEMENT; AORTA TO SUBCLAVIAN BYPASS/SDA |
| 23360 | 104836 | POLYCHONDRITIS WITH AIRWAY MANISFESTATION\ STERNATOMY CARDIOPULMONARY; BYPASS; ANTERIOR TRACHEAL SPLITTING; TY STENT PLACEMENT; LAPAROTOMY/SDA |
| 28352 | 154475 | PULMONARY VEIN INJURY\THORACOSCOPIC MAZE PROCEDURE LEFT; MINI MAZE; BILATERAL MINI THORACOTOMY; PULMONARY VEIN ISOLATION; RESECTION OF LEFT ATRIAL APPENDAGE/SDA |
| 45688 | 144761 | RIGHT VENTRICULAR LEAD MALFUNCTION; INAPPROPRIATE IMPLANTABLE CARDIOVERTER-DEFIBRILLATOR FRING\RIGHT VENTRICULAR IMPLANTABLE CARDIOVERTER-DEFIBRILLATOR LEAD EXTRACTION/SDA |
| 51821 | 182983 | MEDIASTINAL ADENOPATHY\FLEXIBLE BRONCHOSCOPY; LINEAR ENDOBRONCHIAL ULTRASOUND (EBUS); FLUOROSCOPY; TRANSBRONCHIAL BIOPSY; TRANSBRONCHIAL NEEDLE ASPIRATION; BRONCHIAL ALVEOLAR LAVARGE |
| 92284 | 193856 | AIRWAY OBSTRUCTION\FLEXIBLE BRONCHOSCOPY; RADIAL ENDOBRONCHIAL ULTRASOUND (EBUS); BRONCHIAL AVEOLAR LAVAGE/ BRUSH; POSSIBLE TRANSBRONCHIAL BIOPSY (LEFT UPPER LOBE); FLUOROSCOPY |
| Variable | Topic | Keywords |
|---|---|---|
| TOPIC1 | Coronary Artery Disease | coronari, arteri, diseas, graft, bypass, sda, syndrom, effus, cath, avr, acut, etoh, pericardi, cerebr, pleural, cholang, mvr, leav, vascular, angioplasty. |
| TOPIC2 | Aortic Valve Replacement | aortic, sda, valv, replac, stenosi, mitral, cancer, subarachnoid, hemorrhag, esophag, procedur, ascend, regurgit, aorta, maze, airway, redo, bental, repair, invas, obstruct |
| TOPIC3 | Heart Failure | failur, heart, congest, acut, infarct, myocardi, renal, cath, liver, pancreat, elev, cardiac, dehydr, rule, cholecyst, hyperkalemia, leukemia, lacer, hyperglycemia, block, chronic, implant |
| TOPIC4 | Pneumonia | pneumonia, telemetri, fractur, stroke, ischem, atrial, attack, transient, angina, dyspnea, fibril, hip, unstabl, cath, chronic, segment, diseas, pulm, obst, ablat, cardiomyopathi, pelvic, septal |
| TOPIC5 | SDA | sda, right, aneurysm, leav, accid, motor, vehicl, short, breath, tachycardia, cellul, ventricular, abdomin, lung, injuri, hepat, bilater, metastat, perfor, colon, spinal |
| TOPIC6 | Chest Pain | pain, chest, hemorrhag, intracrani, fever, abdomin, hypotens, fall, cath, telemetri, dissect, insuffici, stroke, cardiac, femur, strike, epidur, neck, pedestrian, skull, cervic |
| TOPIC7 | Bleed Mass | bleed, upper, lower, mass, pulmonari, obstruct, bowel, head, brain, weak, emboli, bradycardia, hypertens, stemi, small, edema, hemoptysi, cirrhosi, vomit |
| TOPIC8 | Sepsis | sepsi, infect, asthma, exacerb, copd, sda, urinari, tract, tumor, brain, catheter, overdos, leav, anemia, pyelonephr, syncop, bscess, foot, ulcer, disord |
| TOPIC9 | Subdural Hematoma | hematoma, subdur, respiratori, diabet, seizur, ketoacidosi, trauma, failur, blunt, sda, withdraw, distress, hyponatremia, wind, hernia, remot |
| TOPIC10 | Altered Mental Status | status, mental, alter, arrest, cardiac, carotid, hypoxia, chang, leg, angiogram, stenosi, transplant, kidney, chf, accid, extrem, cerebrovascular, fib, stent, thrombosi, ischemia |
| Model | Parameters |
|---|---|
| AdaBoost | base_estimator = DecistionTreeClassifer, random_state = 1, n_estimators=50, learning_rate = 1.0, algorithm = ‘SAMME.R’ |
| Bagging | base_estimator = None, n_estimators = 500, max_samples = 100, bootstrap = True, bootstrap_features = False, oob_score = False, warm_start = False, n_jobs = 1, random_state = None, verbose = 0 |
| Gradient Boosting | n_estimators = 100, learning_rate = 1.0, max_depth = 1, random_state = 0 |
| LightGBM | boosting_type = ‘gbdt’, num_leaves = 31, max_depth = −1, learning_rate = 0.1, n_estimators = 100, subsample_for_bin = 200,000, min_child_samples = 20, subsample = 1.0, subsample_freq = 0, colsample_bytree = 1.0, reg_alpha = 0.0, reg_lambda = 0.0 n_jobs = −1, importance_type = ‘split’, |
| Logistic Regression | solver = ‘sag’, penalty = ’l2′, max_iter = ’max_iter’ |
| MLP | solver = ‘adam’, alpha = 1e-5, hidden_layer_sizes = (13,13,13), max_iter = 1000 |
| SVC | C = 1.0, kernel = ‘rbf’, degree = 3, gamma = ‘auto’, coef0 = 0.0, shrinking = True, probability = False, tol = 0.001, cache_size = 200, class_weight = None, verbose = False, max_iter = −1 |
| XGBoost | n_estimators = 100, booster = ’gbtree’, eta = 0.3, min_child_weight = 1, max_depth = 3, gamma = 0, max_delta_step = 0, subsample = 1, colsample_bytree = 1, colsample_byleve = 1, lambda = 1, learning_rate = 0.1, n_jobs = 1, base_score = 0.5, max_delta_step = 0, min_child_weight = 1 |
| 3 Days | 30 Days | 365 Days | |
|---|---|---|---|
| Number of patients | 27,550 | 27,550 | 27,550 |
| Number of survivors | 26,640 | 24,522 | 24,364 |
| Number of non-survivors | 910 | 3028 | 3186 |
| Mortality ratio | 3.30% | 10.99% | 11.56% |
| SMOTE increase | 2900% | 900% | 900% |
| Number of survivors | 26,640 | 24,522 | 24,364 |
| Number of non-survivors | 26,390 | 24,224 | 22,302 |
| Mortality ratio | 49.76% | 49.69% | 47.79% |
| 3 Days | 30 Days | 365 Days | ||
|---|---|---|---|---|
| Structured Data | AdaBoost | 0.8530 ± 0.0041 | 0.7514 ± 0.0095 | 0.7478 ± 0.0058 |
| Bagging | 0.8568 ± 0.0073 | 0.7627 ± 0.0054 | 0.7526 ± 0.0049 | |
| Gradient Boosting | 0.8598 ± 0.0082 | 0.7634 ± 0.0126 | 0.7588 ± 0.0053 | |
| LightGBM | 0.8159 ± 0.0149 | 0.7594 ± 0.0062 | 0.7523 ± 0.0035 | |
| Logistic Regression | 0.8110 ± 0.0221 | 0.7396 ± 0.0076 | 0.7353 ± 0.0063 | |
| MLP | 0.8494 ± 0.0163 | 0.7571 ± 0.0097 | 0.7493 ± 0.0060 | |
| SVC | 0.8097 ± 0.0040 | 0.7487 ± 0.0103 | 0.7443 ± 0.0056 | |
| XGBoost | 0.7070 ± 0.0115 | 0.7215 ± 0.0070 | 0.7201 ± 0.0041 | |
| Structured Data + Unstructured Data | AdaBoost | 0.8686 ± 0.0076 | 0.7531 ± 0.0066 | 0.7629 ± 0.0114 |
| Bagging | 0.8713 ± 0.0059 | 0.7644 ± 0.0098 | 0.7725 ± 0.0048 | |
| Gradient Boosting | 0.8820 ± 0.0119 | 0.7815 ± 0.0073 | 0.7754 ± 0.0091 | |
| LightGBM | 0.8361 ± 0.0143 | 0.7780 ± 0.0118 | 0.7705 ± 0.0036 | |
| Logistic Regression | 0.8298 ± 0.0109 | 0.7618 ± 0.0102 | 0.7502 ± 0.0051 | |
| MLP | 0.8679 ± 0.0168 | 0.7693 ± 0.0112 | 0.7540 ± 0.0042 | |
| SVC | 0.8142 ± 0.0082 | 0.7518 ± 0.0110 | 0.7512 ± 0.0040 | |
| XGBoost | 0.7655 ± 0.0174 | 0.7379 ± 0.0067 | 0.7345 ± 0.0044 |
| Dataset | Variable Importance | 3 Days | 30 Days | 365 Days |
|---|---|---|---|---|
| Structured Data | 1 | X7 | X7 | X7 |
| 2 | X8 | X8 | X8 | |
| 3 | X1 | X1 | X1 | |
| 4 | X4 | X4 | X4 | |
| 5 | X3 | X5 | X5 | |
| Structured Data + Unstructured Data | 1 | TOPIC6 | X7 | X7 |
| 2 | X7 | TOPIC1 | TOPIC1 | |
| 3 | TOPIC1 | TOPIC6 | TOPIC6 | |
| 4 | TOPIC7 | X8 | X8 | |
| 5 | TOPIC8 | TOPIC10 | TOPIC8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, C.-C.; Wu, C.-M.; Chien, T.-N.; Kao, L.-J.; Li, C.; Chu, C.-M. Integrating Structured and Unstructured EHR Data for Predicting Mortality by Machine Learning and Latent Dirichlet Allocation Method. Int. J. Environ. Res. Public Health 2023, 20, 4340. https://doi.org/10.3390/ijerph20054340
Chiu C-C, Wu C-M, Chien T-N, Kao L-J, Li C, Chu C-M. Integrating Structured and Unstructured EHR Data for Predicting Mortality by Machine Learning and Latent Dirichlet Allocation Method. International Journal of Environmental Research and Public Health. 2023; 20(5):4340. https://doi.org/10.3390/ijerph20054340
Chicago/Turabian StyleChiu, Chih-Chou, Chung-Min Wu, Te-Nien Chien, Ling-Jing Kao, Chengcheng Li, and Chuan-Mei Chu. 2023. "Integrating Structured and Unstructured EHR Data for Predicting Mortality by Machine Learning and Latent Dirichlet Allocation Method" International Journal of Environmental Research and Public Health 20, no. 5: 4340. https://doi.org/10.3390/ijerph20054340
APA StyleChiu, C.-C., Wu, C.-M., Chien, T.-N., Kao, L.-J., Li, C., & Chu, C.-M. (2023). Integrating Structured and Unstructured EHR Data for Predicting Mortality by Machine Learning and Latent Dirichlet Allocation Method. International Journal of Environmental Research and Public Health, 20(5), 4340. https://doi.org/10.3390/ijerph20054340








