Exploring the Relationship between Melioidosis Morbidity Rate and Local Environmental Indicators Using Remotely Sensed Data
Abstract
1. Introduction
2. Data and Methods
2.1. Study Area
2.2. Conceptual Framework
2.3. Melioidosis Data
2.4. Spatial Data
2.4.1. LST
2.4.2. Vegetation
2.4.3. Soil Moisture
2.4.4. Rainfall
2.5. Data Preparation and Pre-Processing
2.6. Spatial Statistics
2.6.1. Spatial Autocorrelation
2.6.2. Global Poisson Regression (GPR)
2.6.3. Local Poisson Regression
3. Results
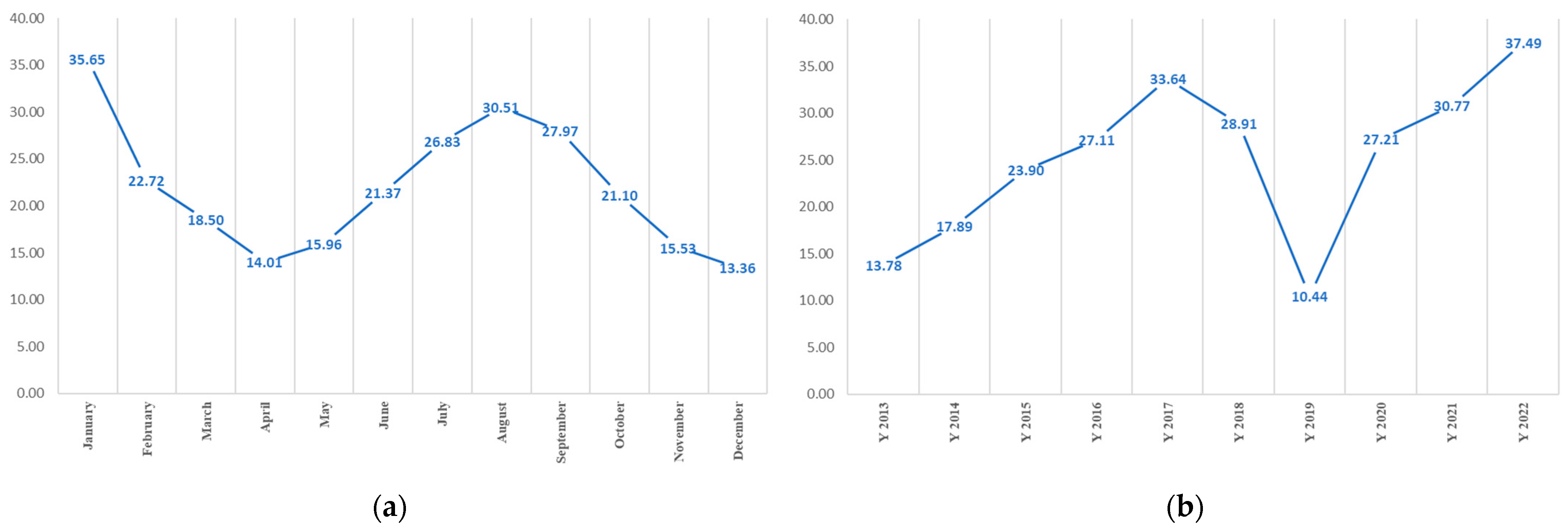
3.1. Melioidosis Morbidity Rate
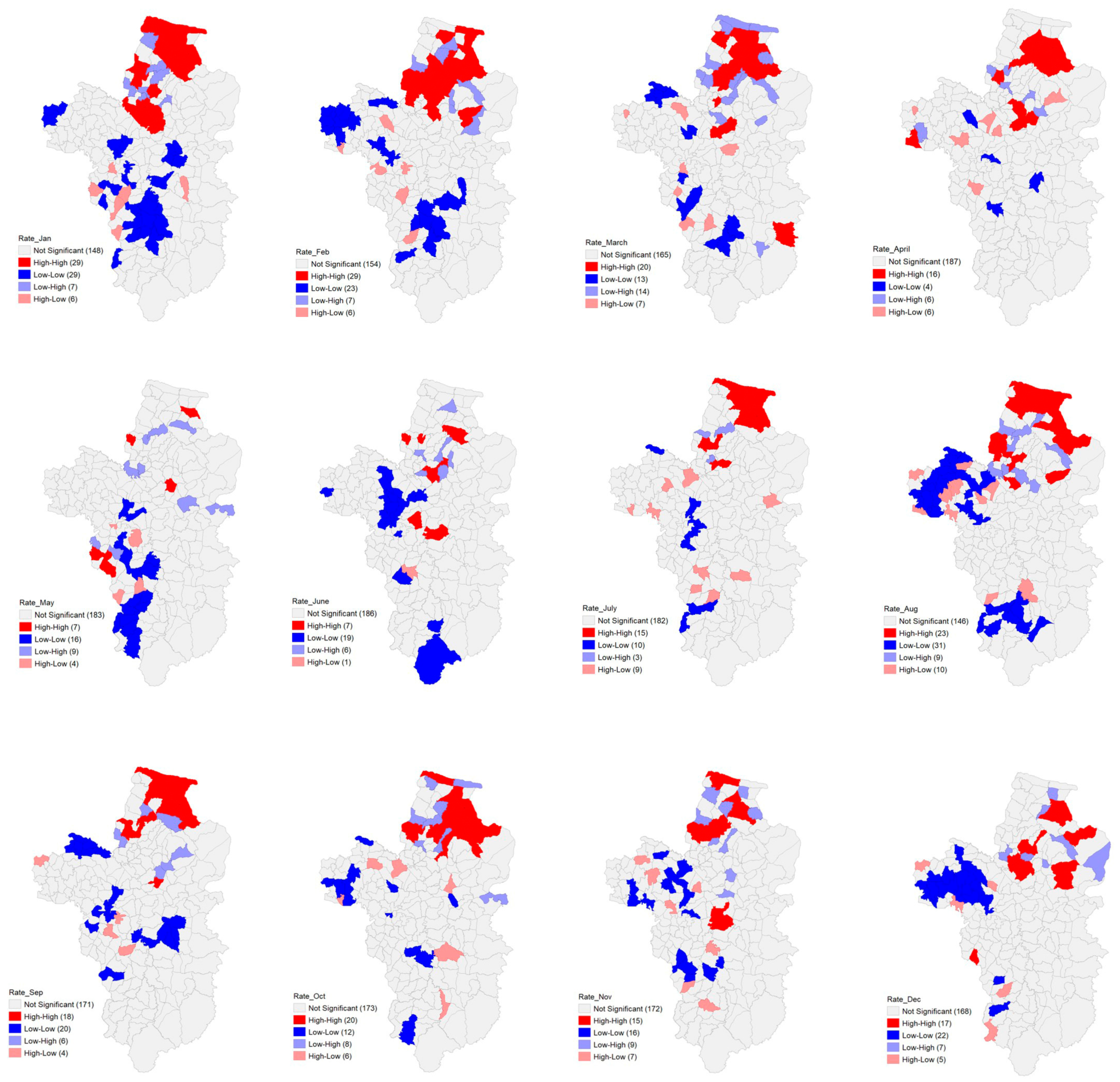
3.2. Spatial Autocorrelation
3.3. GPR Model
3.4. Local Poisson Regression
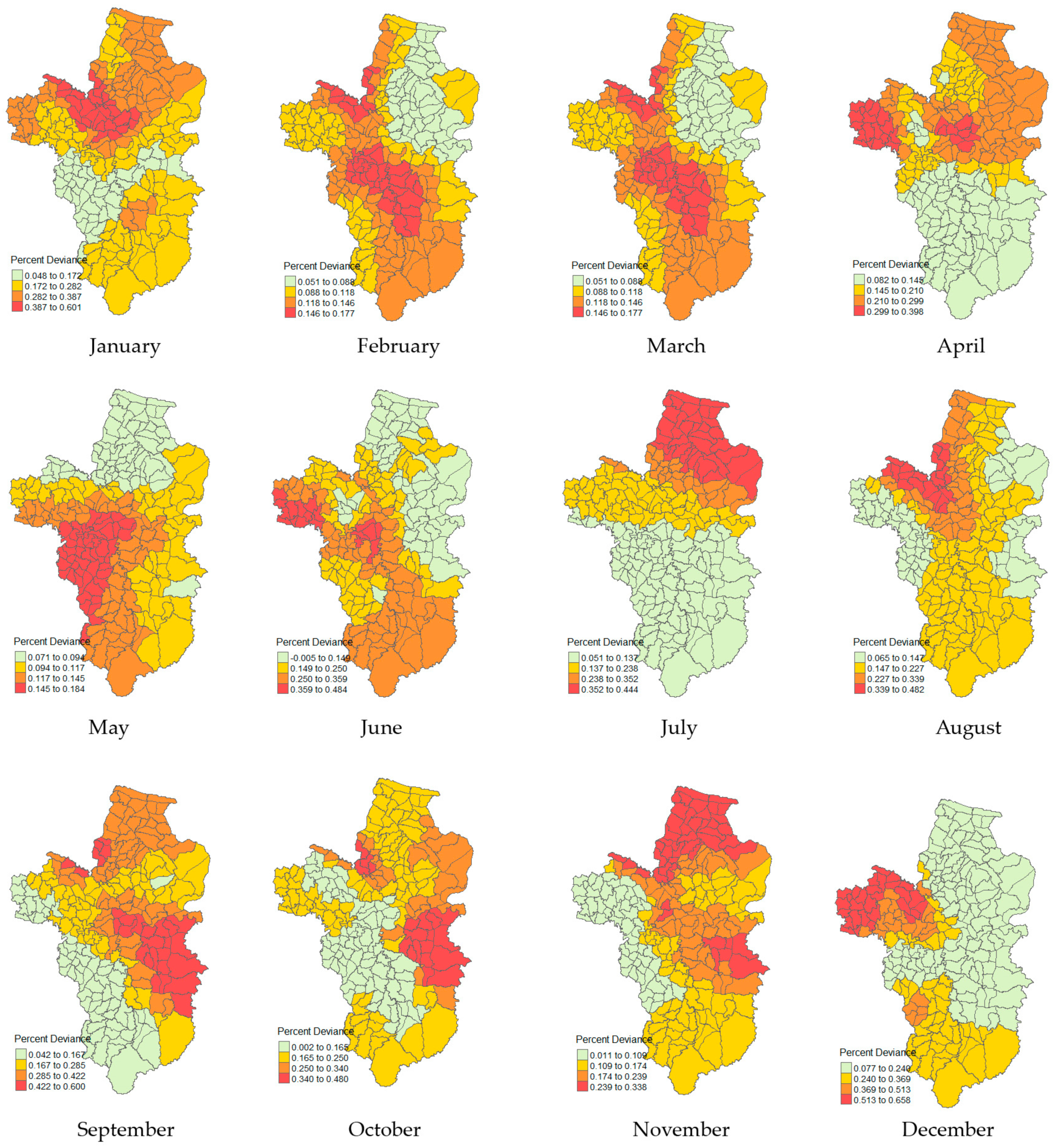
3.5. Local Percent Deviance
3.6. Comparison between GPR and GWPR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Currie, B.J.; Dance, D.A.; Cheng, A.C. The global distribution of Burkholderia pseudomallei and melioidosis: An update. Trans. R. Soc. Trop. Med. Hyg. 2008, 102 (Suppl. S1), S1–S4. [Google Scholar] [CrossRef]
- Corkeron, M.L.; Norton, R.; Nelson, P.N. Spatial analysis of melioidosis distribution in a suburban area. Epidemiol. Infect. 2010, 138, 1346–1352. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Golding, N.; Dance, D.A.; Messina, J.P.; Pigott, D.M.; Moyes, C.L.; Rolim, D.B.; Bertherat, E.; Day, N.P.; Peacock, S.J.; et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 2016, 1, 15008. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; Virk, H.S.; Torres, A.G.; Currie, B.J.; Peacock, S.J.; Dance, D.A.B.; Limmathurotsakul, D. Melioidosis. Nat. Rev. Dis. Primers 2018, 4, 17107. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pang, L.; Sim, S.H.; Goh, K.T.; Ravikumar, S.; Win, M.S.; Tan, G.; Cook, A.R.; Fisher, D.; Chai, L.Y. Association of melioidosis incidence with rainfall and humidity, Singapore, 2003–2012. Emerg. Infect. Dis. 2015, 21, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Bulterys, P.L.; Bulterys, M.A.; Phommasone, K.; Luangraj, M.; Mayxay, M.; Kloprogge, S.; Miliya, T.; Vongsouvath, M.; Newton, P.N.; Phetsouvanh, R.; et al. Climatic drivers of melioidosis in Laos and Cambodia: A 16-year case series analysis. Lancet Planet Health 2018, 2, e334–e343. [Google Scholar] [CrossRef] [PubMed]
- Kaewpan, A.; Duangurai, T.; Rungruengkitkun, A.; Muangkaew, W.; Kanjanapruthipong, T.; Jitprasutwit, N.; Ampawong, S.; Sukphopetch, P.; Chantratita, N.; Pumirat, P. Burkholderia pseudomallei pathogenesis in human skin fibroblasts: A Bsa type III secretion system is involved in the invasion, multinucleated giant cell formation, and cellular damage. PLoS ONE 2022, 17, e0261961. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Dance, D.A.; Wuthiekanun, V.; Kaestli, M.; Mayo, M.; Warner, J.; Wagner, D.M.; Tuanyok, A.; Wertheim, H.; Yoke Cheng, T.; et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl. Trop. Dis. 2013, 7, e2105. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Chen, S.C.; Kao, C.M.; Chen, Y.L. Effects of soil pH, temperature and water content on the growth of Burkholderia pseudomallei. Folia Microbiol. 2003, 48, 253–256. [Google Scholar] [CrossRef]
- Palasatien, S.; Lertsirivorakul, R.; Royros, P.; Wongratanacheewin, S.; Sermswan, R.W. Soil physicochemical properties related to the presence of Burkholderia pseudomallei. Trans. R. Soc. Trop. Med. Hyg. 2008, 102 (Suppl. S1), S5–S9. [Google Scholar] [CrossRef]
- Tong, S.; Yang, S.; Lu, Z.; He, W. Laboratory investigation of ecological factors influencing the environmental presence of Burkholderia pseudomallei. Microbiol. Immunol. 1996, 40, 451–453. [Google Scholar] [CrossRef] [PubMed]
- McMichael, A.J.; Lindgren, E. Climate change: Present and future risks to health, and necessary responses. J. Intern. Med. 2011, 270, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gasparrini, A.; Armstrong, B.G.; Tawatsupa, B.; Tobias, A.; Lavigne, E.; Coelho, M.; Pan, X.; Kim, H.; Hashizume, M.; et al. Heat Wave and Mortality: A Multicountry, Multicommunity Study. Environ. Health Perspect. 2017, 125, 087006. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.Y.A.; Fisher, D. Earth, wind, rain, and melioidosis. Lancet Planet. Health 2018, 2, e329–e330. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Al-Montakim, M.N.; Hasan, M.A.; Mitu, M.M.P.; Gazi, M.Y.; Uddin, M.M.; Mia, M.B. Relationship between Urban Environmental Components and Dengue Prevalence in Dhaka City-An Approach of Spatial Analysis of Satellite Remote Sensing, Hydro-Climatic, and Census Dengue Data. Int. J. Environ. Res. Public Health 2023, 20, 3858. [Google Scholar] [CrossRef] [PubMed]
- Sewe, M.O.; Ahlm, C.; Rocklöv, J. Remotely Sensed Environmental Conditions and Malaria Mortality in Three Malaria Endemic Regions in Western Kenya. PLoS ONE 2016, 11, e0154204. [Google Scholar] [CrossRef] [PubMed]
- Dhewantara, P.W.; Hu, W.; Zhang, W.; Yin, W.W.; Ding, F.; Mamun, A.A.; Soares Magalhães, R.J. Climate variability, satellite-derived physical environmental data and human leptospirosis: A retrospective ecological study in China. Environ. Res. 2019, 176, 108523. [Google Scholar] [CrossRef] [PubMed]
- Goodrick, I.; Stewart, J.; Todd, G. Soil characteristics influencing the spatial distribution of melioidosis in Far North Queensland, Australia. Epidemiol. Infect. 2018, 146, 1602–1607. [Google Scholar] [CrossRef]
- Kanjaras, P.; Bumrerraj, S.; Seng, R.; Noradee, S.; Nithikathkul, C. Geospatial Analysis and Modeling of Melioidosis Prevention and Control in Si Sa Ket Province, Thailand. J. Geoinform. 2023, 19, 57–65. [Google Scholar]
- Wongbutdee, J.; Saengnill, W.; Jittimanee, J.; Panomket, P.; Saenwang, P. The Association between the Mapping Distribution of Melioidosis Incidences and Meteorological Factors in an Endemic Area: Ubon Ratchathani, Thailand (2009–2018). Chiang Mai Univ. (CMU) J. Nat. Sci. 2021, 20, 1–12. [Google Scholar] [CrossRef]
- Wongbutdee, J.; Jittimanee, J.; Saengnill, W. Spatiotemporal distribution and geostatistically interpolated mapping of the melioidosis risk in an endemic zone in Thailand. Geospat. Health 2023, 18, 1189. [Google Scholar] [CrossRef] [PubMed]
- Wuthiekanun, V.; Mayxay, M.; Chierakul, W.; Phetsouvanh, R.; Cheng, A.C.; White, N.J.; Day, N.P.; Peacock, S.J. Detection of Burkholderia pseudomallei in soil within the Lao People’s Democratic Republic. J. Clin. Microbiol. 2005, 43, 923–924. [Google Scholar] [CrossRef] [PubMed]
- Limmathurotsakul, D.; Wuthiekanun, V.; Chantratita, N.; Wongsuvan, G.; Amornchai, P.; Day, N.P.J.; Peacock, S.J. Burkholderia pseudomallei Is Spatially Distributed in Soil in Northeast Thailand. PLoS Neglected Trop. Dis. 2010, 4, e694. [Google Scholar] [CrossRef] [PubMed]
- Saengnill, W.; Charoenjit, K.; Hrimpeng, K.; Jittimanee, J. Mapping the probability of detecting Burkholderia pseudomallei in rural rice paddy soil based on indicator kriging and spatial soil factor analysis. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Wimberly, M.C.; Nekorchuk, D.M.; Kankanala, R.R. Cloud-based applications for accessing satellite Earth observations to support malaria early warning. Sci. Data 2022, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gurgel, H.; Xu, L.; Yang, L.; Dong, J. Improving Dengue Forecasts by Using Geospatial Big Data Analysis in Google Earth Engine and the Historical Dengue Information-Aided Long Short Term Memory Modeling. Biology 2022, 11, 169. [Google Scholar] [CrossRef]
- Ghosh, S.; Kumar, D.; Kumari, R. Google earth engine based computational system for the earth and environment monitoring applications during the COVID-19 pandemic using thresholding technique on SAR datasets. Phys. Chem. Earth 2022, 127, 103163. [Google Scholar] [CrossRef] [PubMed]
- Nafiz Rahaman, S.; Shehzad, T.; Sultana, M. Effect of Seasonal Land Surface Temperature Variation on COVID-19 Infection Rate: A Google Earth Engine-Based Remote Sensing Approach. Environ. Health Insights 2022, 16, 11786302221131467. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Epidemiology, Thailand. National Disease Surveillance (Report 506): Melioidosis. Available online: http://doe.moph.go.th/surdata/y62/rate_Melioidosis_62.rtf (accessed on 11 January 2024).
- Bureau of Epidemiology, Thailand. National Disease Surveillance (Report 506): Melioidosis. Available online: http://doe.moph.go.th/surdata/y63/mcd_Melioidosis_63.rtf (accessed on 11 January 2024).
- Tennekes, M. tmap: Thematic Maps in R. J. Stat. Softw. 2018, 84, 1–39. [Google Scholar] [CrossRef]
- Didan, K.; Barreto-Muñoz, A. MODIS Collection 6.1 (C61) VegetationIndex Product UserGuide. Available online: https://lpdaac.usgs.gov/documents/621/MOD13_User_Guide_V61.pdf (accessed on 11 March 2024).
- Gao, B.-c. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Ceccato, P.; Flasse, S.; Tarantola, S.; Jacquemoud, S.; Grégoire, J.-M. Detecting vegetation leaf water content using reflectance in the optical domain. Remote Sens. Environ. 2001, 77, 22–33. [Google Scholar] [CrossRef]
- Funk, C.; Peterson, P.; Landsfeld, M.; Pedreros, D.; Verdin, J.; Shukla, S.; Husak, G.; Rowland, J.; Harrison, L.; Hoell, A.; et al. The climate hazards infrared precipitation with stations--a new environmental record for monitoring extremes. Sci. Data 2015, 2, 150066. [Google Scholar] [CrossRef] [PubMed]
- Chaithong, T. Flash Flood Susceptibility Assessment Based on Morphometric Aspects and Hydrological Approaches in the Pai River Basin, Mae Hong Son, Thailand. Water 2022, 14, 3174. [Google Scholar] [CrossRef]
- Puttanapong, N.; Martinez, A.; Bulan, J.A.; Addawe, M.; Durante, R.L.; Martillan, M. Predicting Poverty Using Geospatial Data in Thailand. ISPRS Int. J. Geo-Inf. 2022, 11, 293. [Google Scholar] [CrossRef]
- Rojpratak, S.; Supharatid, S. Regional-scale flood impacts on a small mountainous catchment in Thailand under a changing climate. J. Water Clim. Chang. 2023, 14, 4782–4801. [Google Scholar] [CrossRef]
- Nakaya, T.; Fotheringham, A.S.; Brunsdon, C.; Charlton, M. Geographically weighted Poisson regression for disease association mapping. Stat. Med. 2005, 24, 2695–2717. [Google Scholar] [CrossRef] [PubMed]
- Fotheringham, A.S.; Brunsdon, C.; Charlton, M. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships. John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Hadayeghi, A.; Shalaby, A.; Persaud, B. Development of Planning-Level Transportation Safety Models using Full Bayesian Semiparametric Additive Techniques. J. Transp. Saf. Secur. 2010, 2, 45–68. [Google Scholar] [CrossRef]
- Hantrakun, V.; Kongyu, S.; Klaytong, P.; Rongsumlee, S.; Day, N.P.J.; Peacock, S.J.; Hinjoy, S.; Limmathurotsakul, D. Clinical Epidemiology of 7126 Melioidosis Patients in Thailand and the Implications for a National Notifiable Diseases Surveillance System. Open Forum Infect. Dis. 2019, 6, ofz498. [Google Scholar] [CrossRef]
- Dai, D.; Chen, Y.S.; Chen, P.S.; Chen, Y.L. Case cluster shifting and contaminant source as determinants of melioidosis in Taiwan. Trop. Med. Int. Health 2012, 17, 1005–1013. [Google Scholar] [CrossRef]
- Seng, R.; Saiprom, N.; Phunpang, R.; Baltazar, C.J.; Boontawee, S.; Thodthasri, T.; Silakun, W.; Chantratita, N. Prevalence and genetic diversity of Burkholderia pseudomallei isolates in the environment near a patient’s residence in Northeast Thailand. PLoS Neglected Trop. Dis. 2019, 13, e0007348. [Google Scholar] [CrossRef]
- Currie, B.J.; Jacups, S.P. Intensity of rainfall and severity of melioidosis, Australia. Emerg. Infect. Dis. 2003, 9, 1538–1542. [Google Scholar] [CrossRef] [PubMed]
- Kaestli, M.; Grist, E.P.M.; Ward, L.; Hill, A.; Mayo, M.; Currie, B.J. The association of melioidosis with climatic factors in Darwin, Australia: A 23-year time-series analysis. J. Infect. 2016, 72, 687–697. [Google Scholar] [CrossRef]
- Mu, J.J.; Cheng, P.Y.; Chen, Y.S.; Chen, P.S.; Chen, Y.L. The occurrence of melioidosis is related to different climatic conditions in distinct topographical areas of Taiwan. Epidemiol. Infect. 2014, 142, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Jiee, S.; Lim, K.; Choon Vui, D.; Marius, D.; Illyana, N.; Jantim, A. Extreme Weather and Melioidosis: An endemic tropical disease in Penampang district of Sabah, Malaysia. J. Health Res. 2023, 37, 297–305. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Wongratanacheewin, S.; Teerawattanasook, N.; Wongsuvan, G.; Chaisuksant, S.; Chetchotisakd, P.; Chaowagul, W.; Day, N.P.; Peacock, S.J. Increasing incidence of human melioidosis in Northeast Thailand. Am. J. Trop. Med. Hyg. 2010, 82, 1113–1117. [Google Scholar] [CrossRef]
- John Braun, W.; Rousson, V. An autocorrelation criterion for bandwidth selection in nonparametric regression. J. Stat. Comput. Simul. 2000, 68, 89–101. [Google Scholar] [CrossRef]
- Tavares, J.P.; Costa, A.C. Spatial Modeling and Analysis of the Determinants of Property Crime in Portugal. ISPRS Int. J. Geo-Inf. 2021, 10, 731. [Google Scholar] [CrossRef]
- Zeleke, A.J.; Miglio, R.; Palumbo, P.; Tubertini, P.; Chiari, L. Spatiotemporal heterogeneity of SARS-CoV-2 diffusion at the city level using geographically weighted Poisson regression model: The case of Bologna, Italy. Geospat. Health 2022, 17, 1145. [Google Scholar] [CrossRef] [PubMed]
- Paksanont, S.; Sintiprungrat, K.; Yimthin, T.; Pumirat, P.; Peacock, S.J.; Chantratita, N. Effect of temperature on Burkholderia pseudomallei growth, proteomic changes, motility and resistance to stress environments. Sci. Rep. 2018, 8, 9167. [Google Scholar] [CrossRef]
- Birnie, E.; Biemond, J.J.; Wiersinga, W.J. Drivers of melioidosis endemicity: Epidemiological transition, zoonosis, and climate change. Curr. Opin. Infect. Dis. 2022, 35, 196–204. [Google Scholar] [CrossRef]
- Kaestli, M.; Mayo, M.; Harrington, G.; Ward, L.; Watt, F.; Hill, J.V.; Cheng, A.C.; Currie, B.J. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Neglected Trop. Dis. 2009, 3, e364. [Google Scholar] [CrossRef] [PubMed]
- Ribolzi, O.; Rochelle-Newall, E.; Dittrich, S.; Auda, Y.; Newton, P.N.; Rattanavong, S.; Knappik, M.; Soulileuth, B.; Sengtaheuanghoung, O.; Dance, D.A.; et al. Land use and soil type determine the presence of the pathogen Burkholderia pseudomallei in tropical rivers. Environ. Sci. Pollut. Res. Int. 2016, 23, 7828–7839. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.; Assig, K.; Tellapragada, C.; Wagner, G.E.; Choudhary, M.; Göhler, A.; Eshwara, V.K.; Steinmetz, I.; Mukhopadhyay, C. Environmental Factors Associated with Soil Prevalence of the Melioidosis Pathogen Burkholderia pseudomallei: A Longitudinal Seasonal Study from South West India. Front. Microbiol. 2022, 13, 902996. [Google Scholar] [CrossRef] [PubMed]
- Kaestli, M.; Mayo, M.; Harrington, G.; Watt, F.; Hill, J.; Gal, D.; Currie, B.J. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl. Environ. Microbiol. 2007, 73, 6891–6897. [Google Scholar] [CrossRef] [PubMed]
- Chuah, C.J.; Tan, E.K.H.; Sermswan, R.W.; Ziegler, A.D. Hydrological connectivity and Burkholderia pseudomallei prevalence in wetland environments: Investigating rice-farming community’s risk of exposure to melioidosis in North-East Thailand. Environ. Monit. Assess. 2017, 189, 287. [Google Scholar] [CrossRef]
- Shaharudin, R.; Ahmad, N.; Kamaluddin, M.A.; Veloo, Y. Detection of burkholderia pseudomallei from post-flood soil samples in kelantan, malaysia. Southeast Asian J. Trop. Med. Public Health 2016, 47, 951–956. [Google Scholar]

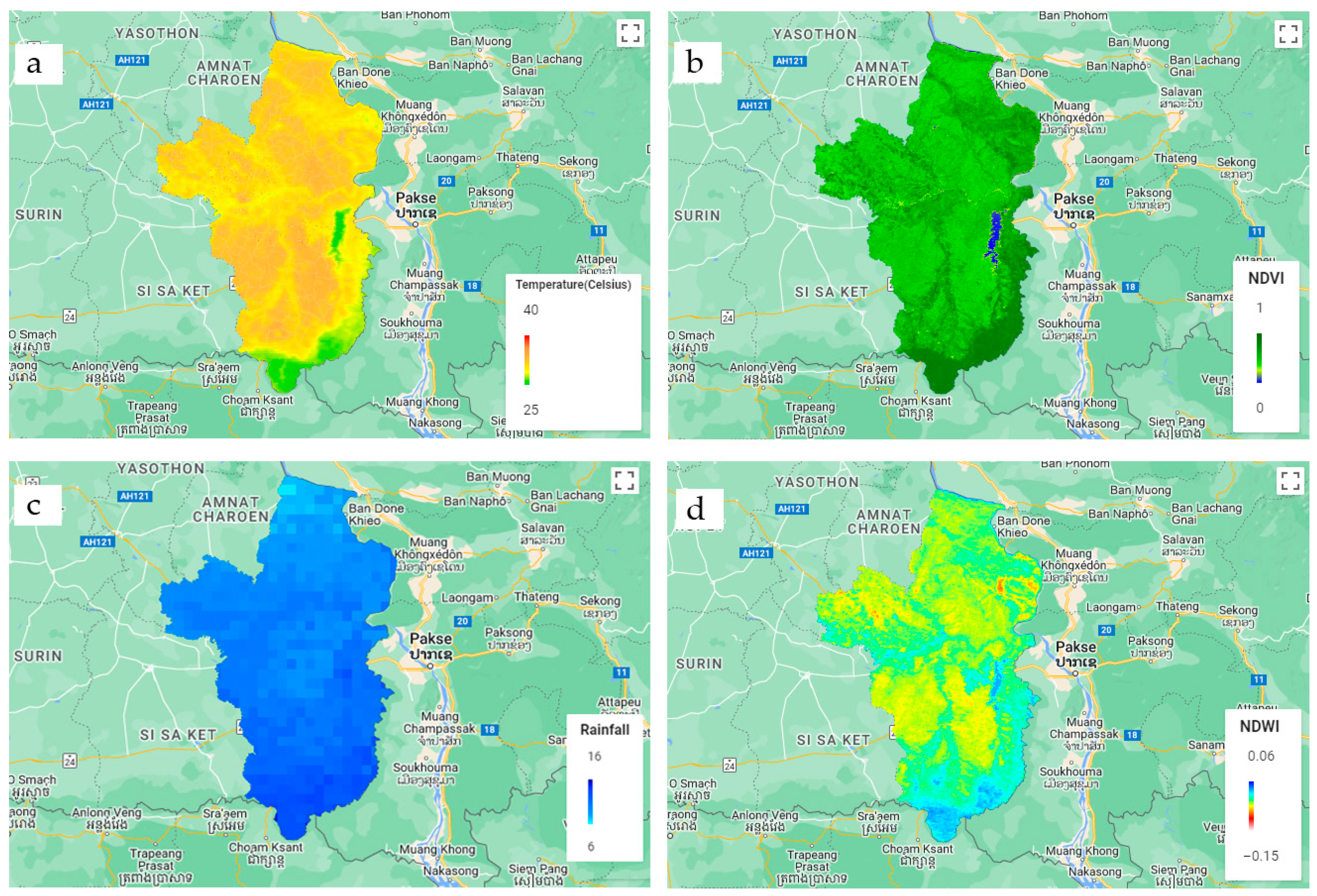

| ID | Data | Satellite | Data Full Name | Resolution |
|---|---|---|---|---|
| 1 | LST | MODIS | MOD11A1.006 Terra Land Surface Temperature and Emissivity Daily Global 1 km | 1 km |
| 2 | NDVI | MODIS | MOD13Q1.006 Terra Vegetation Indices 16-Day | 250 m |
| 3 | NDWI | MODIS | MODIS Terra Daily NDWI | 463.313 m |
| 4 | Rainfall | CHIRPS PENTAD | Climate Hazards Group InfraRed Precipitation with Station Data | 5566 m |
| Monthly | Moran’s I | Mean | S.D. | z-Value | Pseudo p-Value |
|---|---|---|---|---|---|
| January | 0.462 | –0.004 | 0.039 | 11.673 | 0.002 |
| February | 0.317 | –0.007 | 0.040 | 8.021 | 0.002 |
| March | 0.162 | –0.004 | 0.039 | 4.178 | 0.004 |
| April | 0.187 | –0.007 | 0.042 | 4.639 | 0.002 |
| May | 0.068 | –0.008 | 0.038 | 1.990 | 0.042 |
| June | 0.076 | –0.005 | 0.042 | 1.913 | 0.034 |
| July | 0.246 | –0.006 | 0.038 | 6.570 | 0.002 |
| August | 0.351 | –0.0003 | 0.040 | 8.780 | 0.002 |
| September | 0.253 | –0.004 | 0.040 | 6.390 | 0.002 |
| October | 0.244 | –0.003 | 0.040 | 6.083 | 0.002 |
| November | 0.187 | –0.006 | 0.038 | 4.994 | 0.002 |
| December | 0.257 | –0.001 | 0.039 | 6.610 | 0.002 |
| Monthly | Intercept | LST | NDVI | NDWI | Rainfall |
|---|---|---|---|---|---|
| January | –4.546 | 0.135 | 1.82 | –2.329 | 0.077 |
| February | –2.289 | 0.046 | –0.055 | 1.34 | 0.16 |
| March | 2.804 | –0.082 | –0.193 | –6.077 | –0.025 |
| April | 1.762 | –0.054 | –3.555 | –6.519 | 0.005 |
| May | –0.546 | 0.004 | –1.581 | –10.219 | –0.003 |
| June | –2.625 | 0.073 | 3.209 | 4.1483 | –0.001 |
| July | 5.14 | –0.11 | –3.195 | 31.425 | 0.0008 |
| August | –2.808 | 0.045 | 0.72 | 34.014 | 0.003 |
| September | 2.068 | 0.033 | 0.705 | –8.659 | –0.003 |
| October | –2.948 | 0.056 | 6.686 | 1.9126 | –0.006 |
| November | 1.991 | –0.018 | –0.823 | 4.952 | –0.023 |
| December | 6.755 | –0.208 | 0.088 | –10.567 | 0.130 |
| Monthly | GPR | GWPR | Moran’s I | z-Score | p-Value | ||
|---|---|---|---|---|---|---|---|
| AICc | Deviance | AICc | Deviance | ||||
| January | 1197.00 | 0.144 | 432.808 | 0.526 | 0.008 | 0.318 | 0.374 |
| February | 924.00 | 0.157 | 403.175 | 0.395 | –0.056 | –1.275 | 0.898 |
| March | 859.00 | 0.053 | 430.776 | 0.231 | –0.019 | –0.373 | 0.645 |
| April | 828.00 | 0.025 | 431.394 | 0.280 | –0.035 | –0.753 | 0.774 |
| May | 850.00 | 0.037 | 436.957 | 0.145 | –0.025 | –0.520 | 0.698 |
| June | 952.00 | 0.018 | 441.450 | 0.299 | –0.104 | –2.435 | 0.992 |
| July | 974.00 | 0.204 | 406.892 | 0.325 | –0.072 | –1.656 | 0.951 |
| August | 1087.00 | 0.121 | 421.122 | 0.416 | –0.048 | –1.071 | 0.858 |
| September | 1075.00 | 0.087 | 439.833 | 0.442 | –0.092 | –2.154 | 0.984 |
| October | 899.00 | 0.132 | 408.941 | 0.378 | –0.047 | –1.059 | 0.855 |
| November | 837.00 | 0.022 | 418.079 | 0.275 | –0.035 | –0.747 | 0.772 |
| December | 751.00 | 0.156 | 402.712 | 0.422 | 0.014 | 0.475 | 0.317 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wongbutdee, J.; Jittimanee, J.; Daendee, S.; Thongsang, P.; Saengnill, W. Exploring the Relationship between Melioidosis Morbidity Rate and Local Environmental Indicators Using Remotely Sensed Data. Int. J. Environ. Res. Public Health 2024, 21, 614. https://doi.org/10.3390/ijerph21050614
Wongbutdee J, Jittimanee J, Daendee S, Thongsang P, Saengnill W. Exploring the Relationship between Melioidosis Morbidity Rate and Local Environmental Indicators Using Remotely Sensed Data. International Journal of Environmental Research and Public Health. 2024; 21(5):614. https://doi.org/10.3390/ijerph21050614
Chicago/Turabian StyleWongbutdee, Jaruwan, Jutharat Jittimanee, Suwaporn Daendee, Pongthep Thongsang, and Wacharapong Saengnill. 2024. "Exploring the Relationship between Melioidosis Morbidity Rate and Local Environmental Indicators Using Remotely Sensed Data" International Journal of Environmental Research and Public Health 21, no. 5: 614. https://doi.org/10.3390/ijerph21050614
APA StyleWongbutdee, J., Jittimanee, J., Daendee, S., Thongsang, P., & Saengnill, W. (2024). Exploring the Relationship between Melioidosis Morbidity Rate and Local Environmental Indicators Using Remotely Sensed Data. International Journal of Environmental Research and Public Health, 21(5), 614. https://doi.org/10.3390/ijerph21050614





