Metabolome and Transcriptome Profiling Reveal That Four Terpenoid Hormones Dominate the Growth and Development of Sanghuangporus baumii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Materials and Collection
2.2. Widely Targeted Metabolome Analysis
2.3. Transcriptome Sequencing and Data Analysis
2.4. Validation of Gene Expression from Transcriptome Data
2.5. Adding or Spraying Exogenous Terpenoid Hormones in the Growth and Development Stage of S. baumii
2.6. Determination of the Transcript Level of Gene and Hormone Content
3. Results
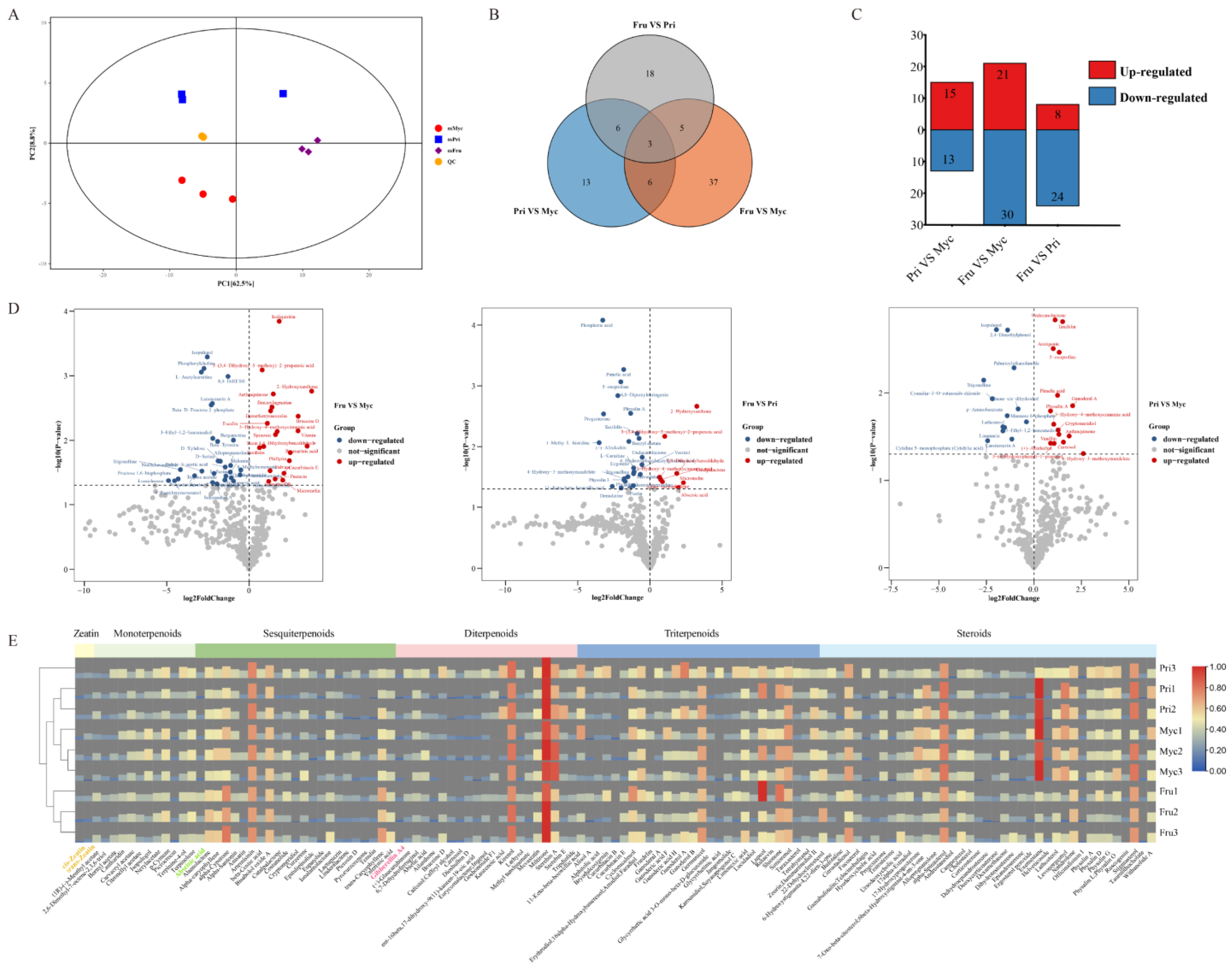
3.1. Metabolome Analysis of Different Developmental Stages of S. baumii
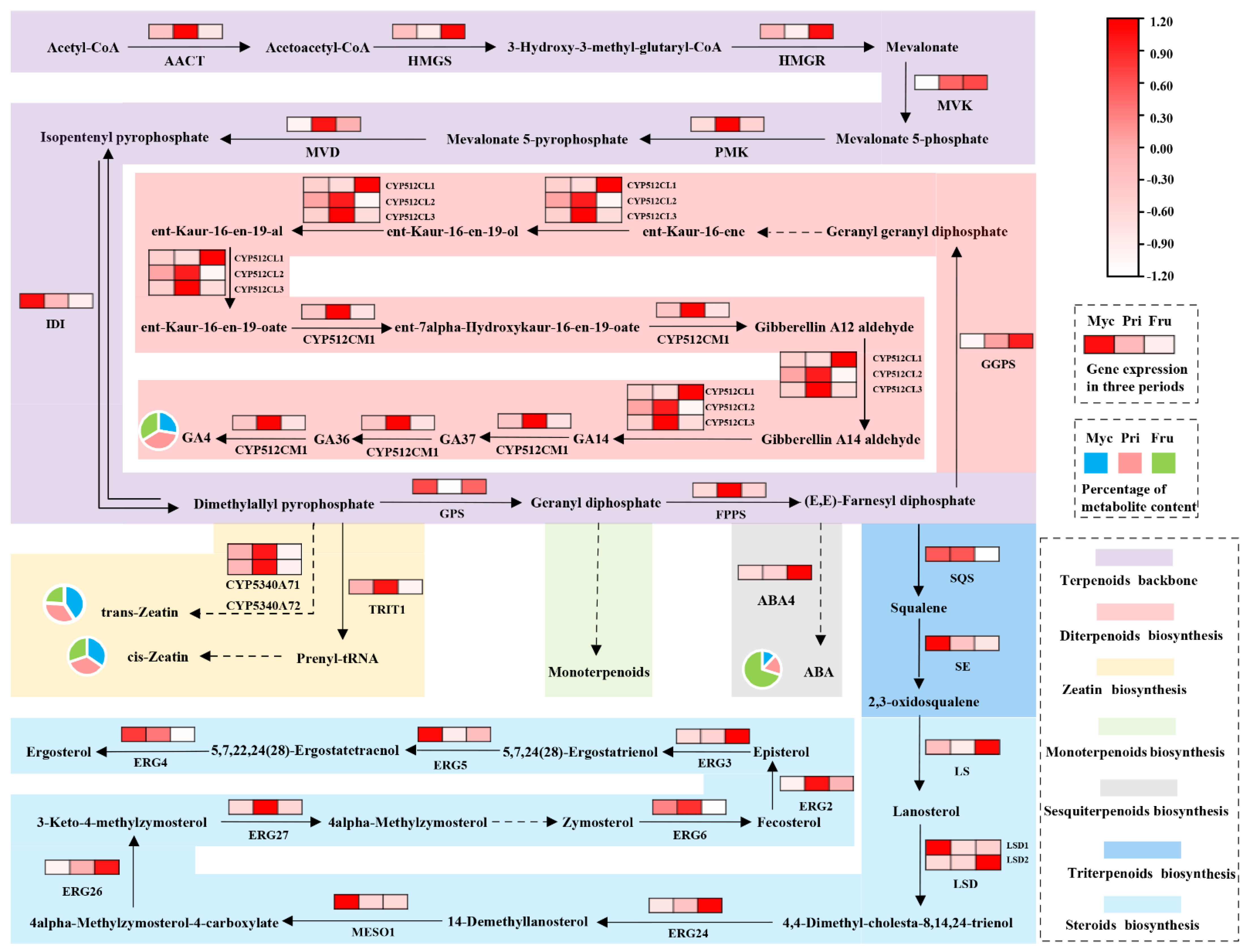
3.2. Terpenoid Analysis Related to Growth and Development of S. baumii
3.3. Differentially Expressed Genes between the Three Different Developmental Stages of S. baumii
3.4. Genes Related to Terpenoid Biosynthesis
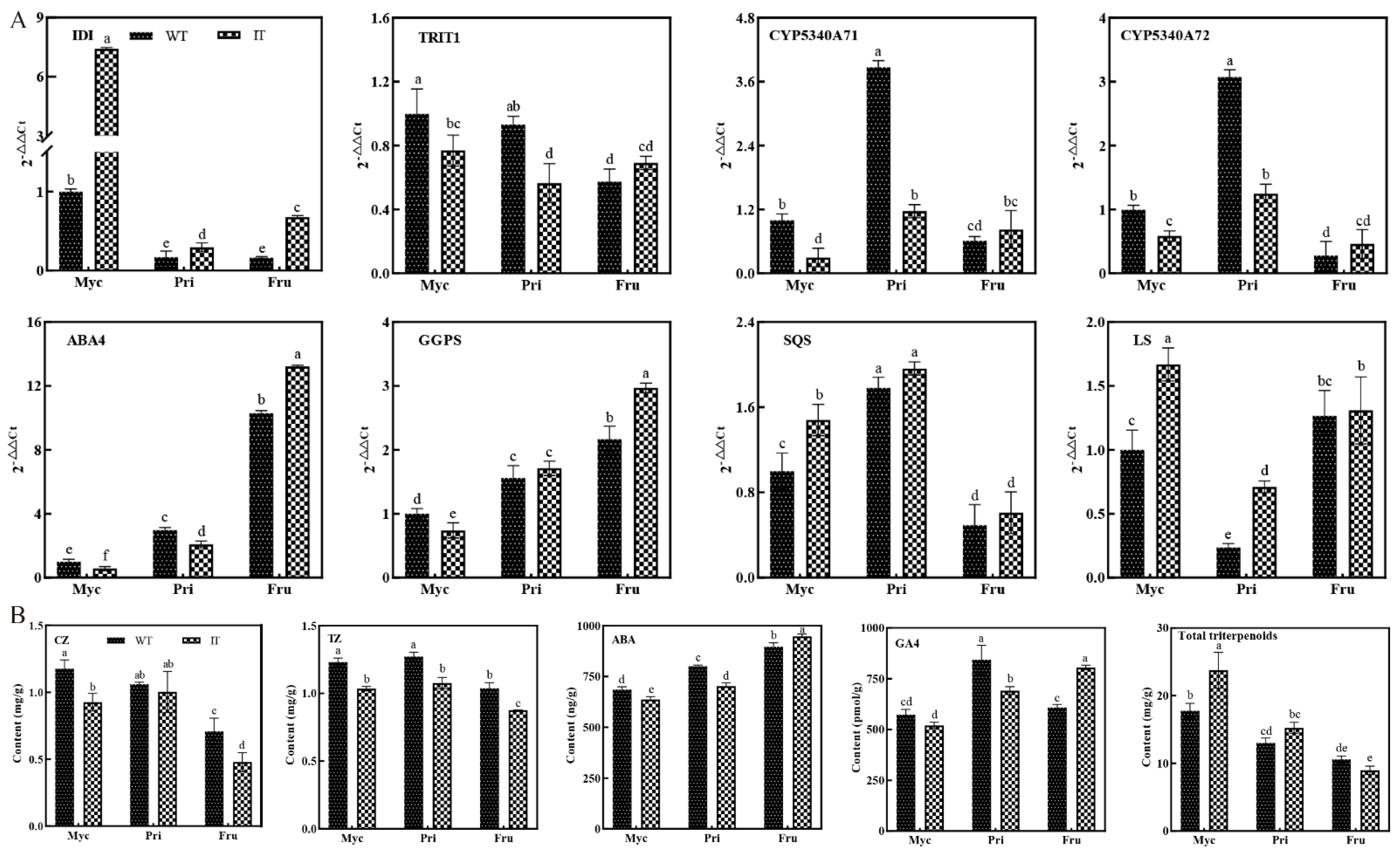
3.5. Validation of Gene Expression Patterns by qRT−PCR
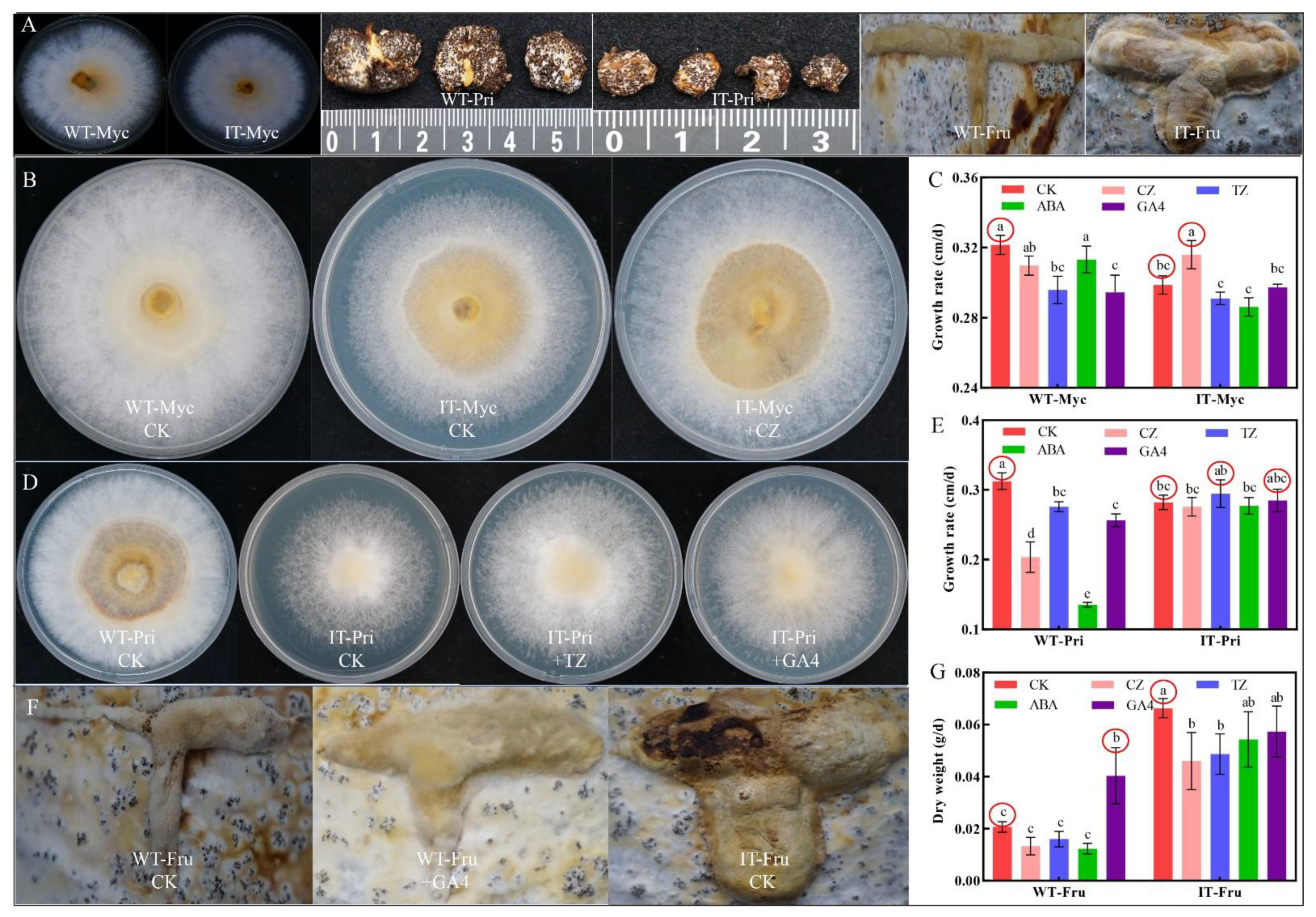
3.6. Effects of Four Exogenous Terpenoid Hormones on the Growth and Development of WT and IT S. baumii
3.7. Differences in Gene Expression and Terpenoid Content between WT and IT S. baumii
4. Discussion
4.1. Application of Metabolomesin S. baumii
4.2. Application of Transcriptomesin S. baumii
4.3. Correlation of Terpenoid Metabolites and Relative Genes of S. baumii
4.4. Exploration of Growth and Development Mechanisms of S. baumii
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, J.; Wang, K.; Zuo, S.; Chen, L.; Ding, Z.Y.; El-Shazly, M.; Zhang, B.B. Unsaturated fatty acid promotes the production of triterpenoids in submerged fermentation of Sanghuangporus baumii. Food Biosci. 2020, 37, 100712. [Google Scholar] [CrossRef]
- Zheng, N.; Ming, Y.F.; Chu, J.Z.; Yang, S.D.; Wu, G.C.; Li, W.H.; Zhang, R.; Cheng, X.H. Optimization of extraction process and the antioxidant activity of phenolics from Sanghuangporus baumii. Molecules 2021, 26, 3850. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.C.; Sun, T.T.; Wang, S.X.; Zou, L. Cloning, molecular properties and differential expression analysis of the isopentenyl diphosphate isomerase gene in Sanghuangporus baumii. Biotechnol. Biotechnol. Equip. 2020, 34, 623–630. [Google Scholar] [CrossRef]
- Qian, Q.; Yang, Y.H.; Zhang, W.B.; Hu, Y.L.; Li, Y.G.; Yu, H.; Hou, X.L. A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 2021, 231, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Wen, X.F.; Zhang, Y.Y.; Zou, P.P.; Cheng, L.; Gan, R.Y.; Li, X.; Liu, D.Y.; Geng, F. Quantitative proteomic and metabolomic analysis of Dictyophora indusiata fruiting bodies during post-harvest morphological development. Food Chem. 2020, 339, 127884. [Google Scholar] [CrossRef]
- Gong, C.S.; Zhu, H.J.; Lu, X.Q.; Yang, D.D.; Zhao, S.J.; Umer, M.J.; He, N.; Yuan, P.L.; Anees, M.; Diao, W.N.; et al. An integrated transcriptome and metabolome approach reveals the accumulation of taste-related metabolites and gene regulatory networks during watermelon fruit development. Planta 2021, 254, 35. [Google Scholar] [CrossRef]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.H.; Jiang, L.X.; Li, N.; Yu, X.; Zhao, P.; Li, T.; Xu, J.W. Overexpression of the squalene epoxidase gene alone and in combination with the 3-hydroxy-3-methylglutaryl coenzyme a gene increases ganoderic acid production in Ganoderma lingzhi. J. Agric. Food Chem. 2017, 65, 4683–4690. [Google Scholar] [CrossRef]
- Kaori, M.; Petr, T.; Miho, M.K.; Tomohiko, K.; Shusei, S.; Danuse, T.; Satoshi, T.; Göran, S.; Tatsuo, K. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [Green Version]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.D.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.; Hussain, R.M.; Rehman, N.U.; She, G.B.; Li, P.H.; Wan, X.C.; Guo, L.; Zhao, J. De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in Blue Anise Sage (Salvia guaranitica L.). DNA Res. 2018, 25, 597–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Zhou, R.; Lin, J.; Wang, Q.; Wang, P.; Wang, W.; Wang, L.; Li, Z. Integrated transcriptomics and nontargeted metabolomics analysis reveal key metabolic pathways in Ganoderma lucidum in response to ethylene. J. Fungi 2022, 8, 456. [Google Scholar] [CrossRef] [PubMed]
- Zha, H.H.; Cai, Y.P.; Yin, Y.D.; Wang, Z.Z.; Li, K.; Zhu, Z.J. SWATHtoMRM: Development of high-coverage targeted metabolomics method using SWATH technology for biomarker discovery. Anal. Chem. 2018, 90, 4062–4070. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Peter, O.; Hiroyuki, T.; Takanori, S.; Katsuya, O.; Hitoshi, O.; Satoru, K. Paclobutrazol elevates auxin and abscisic acid, reduces gibberellins and zeatin and modulates their transporter genes in Marubakaido apple (Malus prunifolia Borkh. var. ringo Asami) rootstocks. Plant Physiol. Biochem. 2020, 155, 502–511. [Google Scholar] [CrossRef]
- Liu, W.; Song, Q.; Cao, Y.; Xie, N.; Li, Z.; Jiang, Y.; Zheng, J.; Tu, P.; Song, Y.; Li, J. From 1H NMR-based non-targeted to LC–MS-based targeted metabolomics strategy for in-depth chemome comparisons among four Cistanche species. J. Pharm. Biomed. Anal. 2019, 162, 16–27. [Google Scholar] [CrossRef]
- Justin, T.; Fischedick, S.R.; Johnson, R.E.B.; Ketchum, R.B.; Croteau, B.; Markus, L. NMR spectroscopic search module for Spektraris, an online resource for plant natural product identification–Taxane diterpenoids from Taxus × media cell suspension cultures as a case study. Phytochemistry 2015, 113, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Fernández, M.A.; Tornos, M.P.; García, M.D.; Heras, B.; Villar, A.M.; Sáenz, M.T. Anti-inflammatory activity of abietic acid, a diterpene isolated from Pimenta racemosa var. grissea. J. Pharm. Pharmacol. 2001, 53, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Kara, M.; Öztas, E.; Boran, T.; Karaman, E.F.; Veskoukis, A.S.; Tsatsakis, A.M. Ameliorative effects of the sesquiterpenoid valerenic acid on oxidative stress induced in HepG2 cells after exposure to the fungicide benomyl. Antioxidants 2021, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Grieb, B.; Thyagarajan, A.; Sliva, D. Ganoderic acids suppress growth and invasive behavior of breast cancer cells by modulating AP-1 and NF-kappaB signaling. Int. J. Mol. Med. 2008, 21, 577–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaprakasha, G.K.; Jadegoud, Y.; Nagana, G.G.A.; Patil, B.S. Bioactive compounds from sour orange inhibit colon cancer cell proliferation and induce cell cycle arrest. J. Agric. Food Chem. 2010, 58, 180–186. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Huang, T.Y.; Chen, M.J. Therapeutic ROS targeting of GADD45γ in the induction of G2/M arrest in primary human colorectal cancer cell lines by cucurbitacin E. Cell Death Dis. 2014, 5, e1198. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Jung, S.H.; Lee, Y.J.; Han, J.Y.; Choi, Y.E.; Hong, H.D.; Jeon, H.Y.; Hwang, J.Y.; Na, S.H.; Kim, Y.M.; et al. Dammarenediol-II prevents VEGF-mediated microvascular permeability in diabetic mice. Phytother. Res. 2015, 29, 1910–1916. [Google Scholar] [CrossRef]
- Hajjaj, H.; Mace, C.; Roberts, M.; Niederberger, P.; Fay, L.B. Effect of 26-oxygenosterolsfrom Ganoderma lucidum and their activity as cholesterol synthesis inhibitors. Appl. Environ. Microbiol. 2005, 71, 3653–3658. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Bu, H.; Zhu, J.; Wang, Y.; Oliver, K.M.; Hu, F.; Huang, B.; Li, Z.; Peng, F. Integration of untargeted metabolomics with transcriptomics provides insights into beauvericin biosynthesis in Cordyceps chanhua under H2O2-induced oxidative stress. J. Fungi 2022, 8, 484. [Google Scholar] [CrossRef]
- Ali, M.; Li, P.H.; She, G.B.; Chen, D.F.; Wan, X.C.; Zhao, J. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in gardensage (Salvia officinalis). Sci. Rep. 2017, 7, 16074. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.M.Y.; Fan, W.L.; Wang, W.F.; Chen, T.C.; Tang, Y.C.; Chu, F.H.; Chang, T.T.; Wang, S.Y.; Li, M.Y.; Chen, Y.H.; et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc. Natl. Acad. Sci. USA 2014, 111, E4743–E4752. [Google Scholar] [CrossRef] [Green Version]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, T.; Shu, D.; Yang, J.; Ding, Z.T.; Tan, H. Sequencing and transcriptional analysis of the biosynthesis gene cluster of abscisic acid-producing Botrytis cinerea. Int. J. Mol. Sci. 2014, 15, 17396–17410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zheng, P.C.; Liu, P.P.; Song, X.W.; Guo, F.; Li, Y.Y.; Ni, D.J.; Jiang, C.J. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019, 272, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Barreto, L.C.; Herken, D.M.D.; Silva, B.M.R.; MunnéBosch, S.; Garcia, Q.S. ABA and GA4 dynamic modulates secondary dormancy and germination in Syngonanthus verticillatus seeds. Planta 2020, 251, 86. [Google Scholar] [CrossRef]
- Zhang, D.; Ren, L.; Yue, J.H.; Wang, L.; Zhuo, L.H.; Shen, X.H. GA4 and IAA were involved in the morphogenesis and development of flowers in Agapanthus praecox ssp. orientalis. J. Plant Physiol. 2014, 171, 966–976. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.G.; Hong, Y.; Lee, E.J. Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J. Plant Physiol. 2019, 239, 52–60. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Tong, X.; Liu, R.; Zou, L. Metabolome and Transcriptome Profiling Reveal That Four Terpenoid Hormones Dominate the Growth and Development of Sanghuangporus baumii. J. Fungi 2022, 8, 648. https://doi.org/10.3390/jof8070648
Liu Z, Tong X, Liu R, Zou L. Metabolome and Transcriptome Profiling Reveal That Four Terpenoid Hormones Dominate the Growth and Development of Sanghuangporus baumii. Journal of Fungi. 2022; 8(7):648. https://doi.org/10.3390/jof8070648
Chicago/Turabian StyleLiu, Zengcai, Xinyu Tong, Ruipeng Liu, and Li Zou. 2022. "Metabolome and Transcriptome Profiling Reveal That Four Terpenoid Hormones Dominate the Growth and Development of Sanghuangporus baumii" Journal of Fungi 8, no. 7: 648. https://doi.org/10.3390/jof8070648
APA StyleLiu, Z., Tong, X., Liu, R., & Zou, L. (2022). Metabolome and Transcriptome Profiling Reveal That Four Terpenoid Hormones Dominate the Growth and Development of Sanghuangporus baumii. Journal of Fungi, 8(7), 648. https://doi.org/10.3390/jof8070648




