Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future
Abstract
:1. Introduction
2. Why Do We Need Radiomics in the Breast Cancer Care?
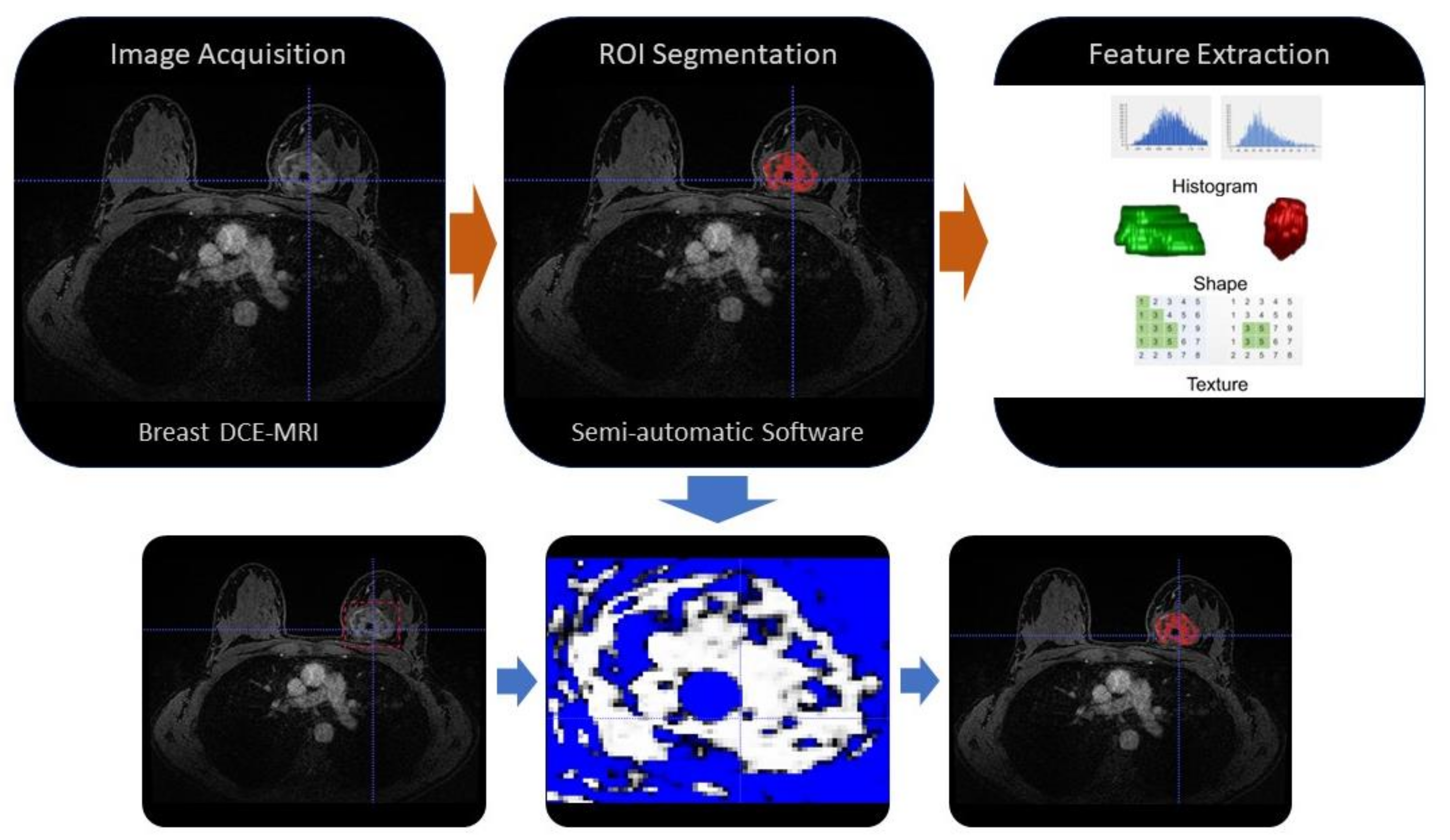
3. The Workflow of a Radiomic Study
4. Radiomics Application in Breast Cancer
5. Discrimination between Benign and Malignant Breast Lesions
6. Prediction of Breast Cancer’s Molecular Subtypes
7. Prediction of Response to Neoadjuvant Chemotherapy
8. Prediction of Lymph Node Metastases
9. What Next?
10. Role of Artificial Intelligence and Big Data in Radiomics
11. Standardization and Curation of Radiomics Data
12. Radiomics Data Sharing
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pesapane, F.; Codari, M.; Sardanelli, F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur. Radiol. Exp. 2018, 2, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, A.S.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Suter, M.B.; Rotili, A.; Penco, S.; Nigro, O.; Cremonesi, M.; Bellomi, M.; Jereczek-Fossa, B.A.; Pinotti, G.; Cassano, E. Will traditional biopsy be substituted by radiomics and liquid biopsy for breast cancer diagnosis and characterisation? Med. Oncol. 2020, 37, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chen, C.; Liu, Z.; Zhang, L.; Zhou, X.; Cheng, M.; Ji, F.; Zhu, T.; Lei, C.; Zhang, J.; et al. Multiparametric MRI-based radiomics analysis for the prediction of breast tumor regression patterns after neoadjuvant chemotherapy. Transl. Oncol. 2020, 13, 100831. [Google Scholar] [CrossRef]
- Choudhery, S.; Gomez-Cardona, D.; Favazza, C.P.; Hoskin, T.L.; Haddad, T.C.; Goetz, M.P.; Boughey, J.C. MRI Radiomics for Assessment of Molecular Subtype, Pathological Complete Response, and Residual Cancer Burden in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Acad. Radiol. 2020, 20, 30607-3. [Google Scholar] [CrossRef]
- Tan, H.; Gan, F.; Wu, Y.; Zhou, J.; Tian, J.; Lin, Y.; Wang, M. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Carcinoma Using Radiomics Features Based on the Fat-Suppressed T2 Sequence. Acad. Radiol. 2020, 27, 1217–1225. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2019, 49, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Pinker, K.; Chin, J.; Melsaether, A.N.; Morris, E.A.; Moy, L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology 2018, 287, 732–747. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef]
- Tan, W.; Yang, M.; Yang, H.; Zhou, F.; Shen, W. Predicting the response to neoadjuvant therapy for early-stage breast cancer: Tumor-, blood-, and imaging-related biomarkers. Cancer Manag. Res. 2018, 10, 4333–4347. [Google Scholar] [CrossRef] [Green Version]
- Tirada, N.; Aujero, M.; Khorjekar, G.; Richards, S.; Chopra, J.; Dromi, S.; Ioffe, O. Breast Cancer Tissue Markers, Genomic Profiling, and Other Prognostic Factors: A Primer for Radiologists. Radiographics 2018, 38, 1902–1920. [Google Scholar] [CrossRef] [Green Version]
- Gradishar, W.J.; Anderson, B.O.; Balassanian, R.; Blair, S.L.; Burstein, H.J.; Cyr, A.; Elias, A.D.; Farrar, W.B.; Forero, A.; Giordano, S.H.; et al. Breast Cancer, Version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2018, 16, 310–320. [Google Scholar] [CrossRef]
- Prud’homme, C.; Deschamps, F.; Allorant, A.; Massard, C.; Hollebecque, A.; Yevich, S.; Ngo-Camus, M.; Gravel, G.; Nicotra, C.; Michiels, S.; et al. Image-guided tumour biopsies in a prospective molecular triage study (MOSCATO-01): What are the real risks? Eur. J. Cancer 2018, 103, 108–119. [Google Scholar] [CrossRef]
- Dercle, L.; Ammari, S.; Bateson, M.; Durand, P.B.; Haspinger, E.; Massard, C.; Jaudet, C.; Varga, A.; Deutsch, E.; Soria, J.C.; et al. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci. Rep. 2017, 7, 7952. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Tselikas, L.; Sun, R.; Ammari, S.; Dercle, L.; Yevich, S.; Hollebecque, A.; Ngo-Camus, M.; Nicotra, C.; Deutsch, E.; Deschamps, F.; et al. Role of image-guided biopsy and radiomics in the age of precision medicine. Chin. Clin. Oncol. 2019, 8, 57. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallieres, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef]
- Bundred, N.J. Prognostic and predictive factors in breast cancer. Cancer Treat. Rev. 2001, 27, 137–142. [Google Scholar] [CrossRef]
- Curigliano, G.; Burstein, H.J.; Winer, E.P.; Gnant, M.; Dubsky, P.; Loibl, S.; Colleoni, M.; Regan, M.M.; Piccart-Gebhart, M.; Senn, H.J.; et al. De-escalating and escalating treatments for early-stage breast cancer: The St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann. Oncol. 2019, 30, 1181. [Google Scholar] [CrossRef]
- Viale, G.; Hanlon Newell, A.E.; Walker, E.; Harlow, G.; Bai, I.; Russo, L.; Dell’Orto, P.; Maisonneuve, P. Ki-67 (30-9) scoring and differentiation of Luminal A- and Luminal B-like breast cancer subtypes. Breast Cancer Res. Treat. 2019, 178, 451–458. [Google Scholar] [CrossRef] [Green Version]
- Cain, E.H.; Saha, A.; Harowicz, M.R.; Marks, J.R.; Marcom, P.K.; Mazurowski, M.A. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res. Treat. 2018, 173, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Rotili, A.; Trimboli, R.M.; Penco, S.; Pesapane, F.; Tantrige, P.; Cassano, E.; Sardanelli, F. Double reading of diffusion-weighted magnetic resonance imaging for breast cancer detection. Breast Cancer Res. Treat. 2020, 180, 111–120. [Google Scholar] [CrossRef]
- Davnall, F.; Yip, C.S.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yip, S.S.F.; Parmar, C.; Kim, J.; Huynh, E.; Mak, R.H.; Aerts, H. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur. J. Radiol. 2017, 97, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Rahmim, A.; Salimpour, Y.; Jain, S.; Blinder, S.A.; Klyuzhin, I.S.; Smith, G.S.; Mari, Z.; Sossi, V. Application of texture analysis to DAT SPECT imaging: Relationship to clinical assessments. Neuroimage Clin. 2016, 12, e1–e9. [Google Scholar] [CrossRef] [Green Version]
- Pesapane, F.; Patella, F.; Fumarola, E.M.; Panella, S.; Ierardi, A.M.; Pompili, G.G.; Franceschelli, G.; Angileri, S.A.; Magenta Biasina, A.; Carrafiello, G. Intravoxel Incoherent Motion (IVIM) Diffusion Weighted Imaging (DWI) in the Periferic Prostate Cancer Detection and Stratification. Med. Oncol. 2017, 34, 35. [Google Scholar] [CrossRef]
- Patella, F.; Franceschelli, G.; Petrillo, M.; Sansone, M.; Fusco, R.; Pesapane, F.; Pompili, G.; Ierardi, A.M.; Saibene, A.M.; Moneghini, L.; et al. A multiparametric analysis combining DCE-MRI- and IVIM-derived parameters to improve differentiation of parotid tumors: A pilot study. Future Oncol. 2018, 14, 2893–2903. [Google Scholar] [CrossRef]
- King, A.D.; Chow, K.K.; Yu, K.H.; Mo, F.K.; Yeung, D.K.; Yuan, J.; Bhatia, K.S.; Vlantis, A.C.; Ahuja, A.T. Head and neck squamous cell carcinoma: Diagnostic performance of diffusion-weighted MR imaging for the prediction of treatment response. Radiology 2013, 266, 531–538. [Google Scholar] [CrossRef]
- Peng, S.L.; Chen, C.F.; Liu, H.L.; Lui, C.C.; Huang, Y.J.; Lee, T.H.; Chang, C.C.; Wang, F.N. Analysis of parametric histogram from dynamic contrast-enhanced MRI: Application in evaluating brain tumor response to radiotherapy. NMR Biomed. 2013, 26, 443–450. [Google Scholar] [CrossRef]
- Aerts, H.J. The Potential of Radiomic-Based Phenotyping in Precision Medicine: A Review. JAMA Oncol. 2016, 2, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, H.; Ko, E.S. Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review. Korean J. Radiol. 2020, 21, 779–792. [Google Scholar] [CrossRef]
- Bianchini, L.; Santinha, J.; Loucao, N.; Figueiredo, M.; Botta, F.; Origgi, D.; Cremonesi, M.; Cassano, E.; Papanikolaou, N.; Lascialfari, A. A multicenter study on radiomic features from T2-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics. Magn. Reson. Med. 2020, 85, 1713–1726. [Google Scholar] [CrossRef]
- Mao, N.; Yin, P.; Li, Q.; Wang, Q.; Liu, M.; Ma, H.; Dong, J.; Che, K.; Wang, Z.; Duan, S.; et al. Radiomics nomogram of contrast-enhanced spectral mammography for prediction of axillary lymph node metastasis in breast cancer: A multicenter study. Eur. Radiol. 2020, 30, 6732–6739. [Google Scholar] [CrossRef]
- Hao, W.; Gong, J.; Wang, S.; Zhu, H.; Zhao, B.; Peng, W. Application of MRI Radiomics-Based Machine Learning Model to Improve Contralateral BI-RADS 4 Lesion Assessment. Front. Oncol. 2020, 10, 531476. [Google Scholar] [CrossRef]
- Lo Gullo, R.; Daimiel, I.; Rossi Saccarelli, C.; Bitencourt, A.; Gibbs, P.; Fox, M.J.; Thakur, S.B.; Martinez, D.F.; Jochelson, M.S.; Morris, E.A.; et al. Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers. Eur. Radiol. 2020, 30, 6721–6731. [Google Scholar] [CrossRef]
- Demircioglu, A.; Grueneisen, J.; Ingenwerth, M.; Hoffmann, O.; Pinker-Domenig, K.; Morris, E.; Haubold, J.; Forsting, M.; Nensa, F.; Umutlu, L. A rapid volume of interest-based approach of radiomics analysis of breast MRI for tumor decoding and phenotyping of breast cancer. PLoS ONE 2020, 15, e0234871. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, Y.; Liu, W.; Bai, J.; Zheng, J.; Yang, X.; Zhou, L. Radiomics Based on Multimodal MRI for the Differential Diagnosis of Benign and Malignant Breast Lesions. J. Magn. Reson. Imaging 2020, 52, 596–607. [Google Scholar] [CrossRef]
- Liu, Z.; Feng, B.; Li, C.; Chen, Y.; Chen, Q.; Li, X.; Guan, J.; Chen, X.; Cui, E.; Li, R.; et al. Preoperative prediction of lymphovascular invasion in invasive breast cancer with dynamic contrast-enhanced-MRI-based radiomics. J. Magn. Reson. Imaging 2019, 50, 847–857. [Google Scholar] [CrossRef]
- Xie, T.; Wang, Z.; Zhao, Q.; Bai, Q.; Zhou, X.; Gu, Y.; Peng, W.; Wang, H. Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Front. Oncol. 2019, 9, 505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Cheng, Z.; Huang, Y.; He, L.; Chen, X.; Ma, Z.; Huang, X.; Liang, C.; Liu, Z. An MRI-based Radiomics Classifier for Preoperative Prediction of Ki-67 Status in Breast Cancer. Acad. Radiol. 2018, 25, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Wu, Y.; Bao, F.; Zhou, J.; Wan, J.; Tian, J.; Lin, Y.; Wang, M. Mammography-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in breast cancer. Br. J. Radiol. 2020, 93, 20191019. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, P.D.; Steding, O.; Raudner, M.W.; Euller, G.; Clauser, P.; Baltzer, P.A.T. Combined texture analysis and machine learning in suspicious calcifications detected by mammography: Potential to avoid unnecessary stereotactical biopsies. Eur. J. Radiol. 2020, 132, 109309. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wang, Z.; Zhang, K.; Yang, P.; Ma, H.; Shi, Y.; Liu, M.; Wang, Q.; Cui, J.; Mao, N.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for Identifying Benign and Malignant Breast Lesions of Sub-1 cm. Front. Oncol. 2020, 10, 573630. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, H.; Bai, Y.; Li, J.; Lu, Q.; Chen, R.; Zhang, M.; Feng, Q.; Wang, M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019, 121, 108718. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Wei, W.; Liu, Z.; Xiong, Q.; Yang, C.; Yang, M.; Zhang, L.; Zhu, T.; Zhuang, X.; Liu, C.; et al. Mammography-based radiomic analysis for predicting benign BI-RADS category 4 calcifications. Eur. J. Radiol. 2019, 121, 108711. [Google Scholar] [CrossRef]
- Ou, X.; Zhang, J.; Wang, J.; Pang, F.; Wang, Y.; Wei, X.; Ma, X. Radiomics based on (18) F-FDG PET/CT could differentiate breast carcinoma from breast lymphoma using machine-learning approach: A preliminary study. Cancer Med. 2019. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, Y.; Chang, K.T.; Lee, K.E.; Wang, O.; Li, J.; Lin, Y.; Pan, Z.; Chang, P.; Chow, D.; et al. Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J. Magn. Reson. Imaging 2019, 51, 798–809. [Google Scholar] [CrossRef]
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine learning in breast MRI. J. Magn. Reson. Imaging 2020, 52, 998–1018. [Google Scholar] [CrossRef]
- Granzier, R.W.Y.; van Nijnatten, T.J.A.; Woodruff, H.C.; Smidt, M.L.; Lobbes, M.B.I. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: A systematic review. Eur. J. Radiol. 2019, 121, 108736. [Google Scholar] [CrossRef] [Green Version]
- American Cancer Society. Breast Cancer Survival Rates by Stage. Available online: http://www.cancer.org/cancer/breastcancer/detailedguide/breast-cancer-survival-by-stage (accessed on 15 March 2021).
- NCCN.org. Breast Cancer Screening and Diagnosis Version 3. In NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines); National Comprehensive Cancer Network: Plymouth Meeting, PA, USA, 2018. [Google Scholar]
- Mann, R.M.; Balleyguier, C.; Baltzer, P.A.; Bick, U.; Colin, C.; Cornford, E.; Evans, A.; Fallenberg, E.; Forrai, G.; Fuchsjager, M.H.; et al. Breast MRI: EUSOBI recommendations for women’s information. Eur. Radiol. 2015, 25, 3669–3678. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Xie, F.; Liu, L.; Peng, Y.; Cai, H.; Li, L. Discrimination of malignant and benign breast masses using automatic segmentation and features extracted from dynamic contrast-enhanced and diffusion-weighted MRI. Oncol. Lett. 2018, 16, 1521–1528. [Google Scholar] [CrossRef] [Green Version]
- Bickelhaupt, S.; Jaeger, P.F.; Laun, F.B.; Lederer, W.; Daniel, H.; Kuder, T.A.; Wuesthof, L.; Paech, D.; Bonekamp, D.; Radbruch, A.; et al. Radiomics Based on Adapted Diffusion Kurtosis Imaging Helps to Clarify Most Mammographic Findings Suspicious for Cancer. Radiology 2018, 287, 761–770. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Mendel, K.R.; Lan, L.; Sheth, D.; Giger, M.L. Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma. Radiology 2019, 291, 15–20. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Valdora, F.; Mariscotti, G.; Durando, M.; Nori, J.; La Forgia, D.; Rosenberg, I.; Caumo, F.; Gandolfo, N.; Houssami, N.; et al. An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts. Breast 2018, 40, 92–96. [Google Scholar] [CrossRef]
- Massafra, R.; Bove, S.; Lorusso, V.; Biafora, A.; Comes, M.C.; Didonna, V.; Diotaiuti, S.; Fanizzi, A.; Nardone, A.; Nolasco, A.; et al. Radiomic Feature Reduction Approach to Predict Breast Cancer by Contrast-Enhanced Spectral Mammography Images. Diagnostics 2021, 11, 684. [Google Scholar] [CrossRef]
- Luo, W.Q.; Huang, Q.X.; Huang, X.W.; Hu, H.T.; Zeng, F.Q.; Wang, W. Predicting Breast Cancer in Breast Imaging Reporting and Data System (BI-RADS) Ultrasound Category 4 or 5 Lesions: A Nomogram Combining Radiomics and BI-RADS. Sci. Rep. 2019, 9, 11921. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Li, H.; Wang, S.; Zheng, B.; Zhang, J.; Li, L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS ONE 2017, 12, e0171683. [Google Scholar] [CrossRef]
- Xie, T.; Zhao, Q.; Fu, C.; Bai, Q.; Zhou, X.; Li, L.; Grimm, R.; Liu, L.; Gu, Y.; Peng, W. Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging. Eur. Radiol. 2019, 29, 2535–2544. [Google Scholar] [CrossRef]
- La Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic Analysis in Contrast-Enhanced Spectral Mammography for Predicting Breast Cancer Histological Outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018, 19, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, N.K.; Dhesy-Thind, S. Clinical practice guidelines in breast cancer. Curr. Oncol. 2018, 25, S151–S160. [Google Scholar] [CrossRef] [PubMed]
- Krop, I.; Ismaila, N.; Andre, F.; Bast, R.C.; Barlow, W.; Collyar, D.E.; Hammond, M.E.; Kuderer, N.M.; Liu, M.C.; Mennel, R.G.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2017, 35, 2838–2847. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Q.; Zhou, X.; Liu, Z.; Lei, C.; Yang, C.; Yang, M.; Zhang, L.; Zhu, T.; Zhuang, X.; Liang, C.; et al. Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin. Transl. Oncol. 2019, 22, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Drukker, K.; Li, H.; Antropova, N.; Edwards, A.; Papaioannou, J.; Giger, M.L. Most-enhancing tumor volume by MRI radiomics predicts recurrence-free survival "early on" in neoadjuvant treatment of breast cancer. Cancer Imaging 2018, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Teruel, J.R.; Heldahl, M.G.; Goa, P.E.; Pickles, M.; Lundgren, S.; Bathen, T.F.; Gibbs, P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014, 27, 887–896. [Google Scholar] [CrossRef]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef]
- Fan, M.; Wu, G.; Cheng, H.; Zhang, J.; Shao, G.; Li, L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Radiol. 2017, 94, 140–147. [Google Scholar] [CrossRef]
- Ahmed, A.; Gibbs, P.; Pickles, M.; Turnbull, L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J. Magn. Reson. Imaging 2013, 38, 89–101. [Google Scholar] [CrossRef]
- Parikh, J.; Selmi, M.; Charles-Edwards, G.; Glendenning, J.; Ganeshan, B.; Verma, H.; Mansi, J.; Harries, M.; Tutt, A.; Goh, V. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology 2014, 272, 100–112. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, F.; Ma, H.; Shi, Y.; Dong, J.; Yang, P.; Zhang, K.; Guo, N.; Zhang, R.; Cui, J.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for the Prediction of Neoadjuvant Chemotherapy-Insensitive Breast Cancers. Front. Oncol. 2021, 11, 605230. [Google Scholar] [CrossRef]
- Galimberti, V.; Cole, B.F.; Viale, G.; Veronesi, P.; Vicini, E.; Intra, M.; Mazzarol, G.; Massarut, S.; Zgajnar, J.; Taffurelli, M.; et al. Axillary dissection versus no axillary dissection in patients with breast cancer and sentinel-node micrometastases (IBCSG 23-01): 10-year follow-up of a randomised, controlled phase 3 trial. Lancet Oncol. 2018, 19, 1385–1393. [Google Scholar] [CrossRef]
- Galimberti, V.; Cole, B.F.; Zurrida, S.; Viale, G.; Luini, A.; Veronesi, P.; Baratella, P.; Chifu, C.; Sargenti, M.; Intra, M.; et al. Axillary dissection versus no axillary dissection in patients with sentinel-node micrometastases (IBCSG 23-01): A phase 3 randomised controlled trial. Lancet Oncol. 2013, 14, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, U.; Paganelli, G.; Viale, G.; Luini, A.; Zurrida, S.; Galimberti, V.; Intra, M.; Veronesi, P.; Maisonneuve, P.; Gatti, G.; et al. Sentinel-lymph-node biopsy as a staging procedure in breast cancer: Update of a randomised controlled study. Lancet Oncol. 2006, 7, 983–990. [Google Scholar] [CrossRef]
- Charalampoudis, P.; Markopoulos, C.; Kovacs, T. Controversies and recommendations regarding sentinel lymph node biopsy in primary breast cancer: A comprehensive review of current data. Eur. J. Surg. Oncol. 2018, 44, 5–14. [Google Scholar] [CrossRef]
- Dihge, L.; Bendahl, P.O.; Ryden, L. Nomograms for preoperative prediction of axillary nodal status in breast cancer. Br. J. Surg. 2017, 104, 1494–1505. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.J.; Youk, J.H.; Choi, H.; Song, J.S. Dynamic contrast-enhanced and diffusion-weighted MRI of invasive breast cancer for the prediction of sentinel lymph node status. J. Magn. Reson. Imaging 2020, 51, 615–626. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef]
- Han, L.; Zhu, Y.; Liu, Z.; Yu, T.; He, C.; Jiang, W.; Kan, Y.; Dong, D.; Tian, J.; Luo, Y. Radiomic nomogram for prediction of axillary lymph node metastasis in breast cancer. Eur. Radiol. 2019, 29, 3820–3829. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, N.; Zhao, Y.; Chen, S.; Li, S.; Xu, M.; Chai, R. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Cancer using Radiomics Features of DCE-MRI. Sci. Rep. 2019, 9, 2240. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, J.; Spuhler, K.; Gao, Y.; Serrano Sosa, M.; Moriarty, M.; Hussain, S.; He, X.; Liang, C.; Huang, C. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J. Magn. Reson. Imaging 2019, 49, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wang, T.; Yang, L.; Wang, Y.; Li, H.; Zhou, X.; Zhao, W.; Ren, J.; Li, X.; Tian, J.; et al. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Cancer Using Mammography-Based Radiomics Method. Sci. Rep. 2019, 9, 4429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.H.; Wang, J.X.; Ye, X.H.; Deng, J.; Hang, J.; Yang, B. Ultrasound-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in early-stage invasive breast cancer. Eur. J. Radiol. 2019, 119, 108658. [Google Scholar] [CrossRef]
- Yang, X.; Wu, L.; Ye, W.; Zhao, K.; Wang, Y.; Liu, W.; Li, J.; Li, H.; Liu, Z.; Liang, C. Deep Learning Signature Based on Staging CT for Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer. Acad. Radiol. 2020, 27, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.M.; Wang, H.T.; Yu, T. The Application of Radiomics in Breast MRI: A Review. Technol. Cancer Res. Treat. 2020, 19, 1533033820916191. [Google Scholar] [CrossRef] [PubMed]
- Da-Ano, R.; Masson, I.; Lucia, F.; Dore, M.; Robin, P.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Reinhold, C.; Castelli, J.; et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci. Rep. 2020, 10, 10248. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [Green Version]
- Abajian, A.; Murali, N.; Savic, L.J.; Laage-Gaupp, F.M.; Nezami, N.; Duncan, J.S.; Schlachter, T.; Lin, M.; Geschwind, J.F.; Chapiro, J. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. J. Vasc. Interv. Radiol. 2018, 29, 850–857.e1. [Google Scholar] [CrossRef]
- El-Sayed, M.E.; Rakha, E.A.; Reed, J.; Lee, A.H.; Evans, A.J.; Ellis, I.O. Predictive value of needle core biopsy diagnoses of lesions of uncertain malignant potential (B3) in abnormalities detected by mammographic screening. Histopathology 2008, 53, 650–657. [Google Scholar] [CrossRef]
- Miller, D.D.; Brown, E.W. Artificial Intelligence in Medical Practice: The Question to the Answer? Am. J. Med. 2018, 131, 129–133. [Google Scholar] [CrossRef]
- Samuel, A.L. Some studies in machine learning using the game of checkers. IBM J. Res. Dev. 1959, 3, 210–229. [Google Scholar] [CrossRef]
- Lee, J.G.; Jun, S.; Cho, Y.W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J. Am. Coll. Radiol. 2018, 15, 504–508. [Google Scholar] [CrossRef]
- Pesapane, F.; Volonte, C.; Codari, M.; Sardanelli, F. Artificial intelligence as a medical device in radiology: Ethical and regulatory issues in Europe and the United States. Insights Imaging 2018, 9, 745–753. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Wang, L.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical Image Synthesis with Deep Convolutional Adversarial Networks. IEEE Trans. Biomed. Eng. 2018, 65, 2720–2730. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Mayer-Schonberger, V.; Ingelsson, E. Big Data and medicine: A big deal? J. Intern. Med. 2018, 283, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Yi, P.H.; Hui, F.K.; Ting, D.S.W. Artificial Intelligence and Radiology: Collaboration Is Key. J. Am. Coll. Radiol. 2018, 15, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Swensen, S.J.; Johnson, C.D. Radiologic quality and safety: Mapping value into radiology. J. Am. Coll. Radiol. 2005, 2, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F. How scientific mobility can help current and future radiology research: A radiology trainee’s perspective. Insights Imaging 2019, 10, 85. [Google Scholar] [CrossRef] [Green Version]
- Kruse, C.S.; Goswamy, R.; Raval, Y.; Marawi, S. Challenges and Opportunities of Big Data in Health Care: A Systematic Review. JMIR Med. Inform. 2016, 4, e38. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.B.; Towbin, A.J.; Pryor, R.M.; Donnelly, L.F. Improving consistency in radiology reporting through the use of department-wide standardized structured reporting. Radiology 2013, 267, 240–250. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, L.H.; Panicek, D.M.; Berk, A.R.; Li, Y.; Hricak, H. Improving communication of diagnostic radiology findings through structured reporting. Radiology 2011, 260, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Moeskops, P.; Viergever, M.A.; Mendrik, A.M.; de Vries, L.S.; Benders, M.J.; Isgum, I. Automatic Segmentation of MR Brain Images With a Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1252–1261. [Google Scholar] [CrossRef] [Green Version]
- Kruskal, J.B.; Berkowitz, S.; Geis, J.R.; Kim, W.; Nagy, P.; Dreyer, K. Big Data and Machine Learning-Strategies for Driving This Bus: A Summary of the 2016 Intersociety Summer Conference. J. Am. Coll. Radiol. 2017, 14, 811–817. [Google Scholar] [CrossRef]
- Kansagra, A.P.; Yu, J.P.; Chatterjee, A.R.; Lenchik, L.; Chow, D.S.; Prater, A.B.; Yeh, J.; Doshi, A.M.; Hawkins, C.M.; Heilbrun, M.E.; et al. Big Data and the Future of Radiology Informatics. Acad. Radiol. 2016, 23, 30–42. [Google Scholar] [CrossRef]
- Ranschaert, E.R.; Sergey, M.; Algra, P.R. Artificial Intelligence in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Krittanawong, C. The rise of artificial intelligence and the uncertain future for physicians. Eur. J. Intern. Med. 2018, 48, e13–e14. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.M. Reporting of artificial intelligence prediction models. Lancet 2019, 393, 1577–1579. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Woodruff, H.C.; de Jong, E.E.C.; van Timmeren, J.E.; Jochems, A.; Dubois, L.; Lambin, P. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother. Oncol. 2018, 127, 349–360. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Kemp, J.L.; Mahoney, M.C.; Mathews, V.P.; Wintermark, M.; Yee, J.; Brown, S.D. Patient-centered Radiology: Where Are We, Where Do We Want to Be, and How Do We Get There? Radiology 2017, 285, 601–608. [Google Scholar] [CrossRef]
- Castelvecchi, D. Can we open the black box of AI? Nature 2016, 538, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Chevrier, R.; Foufi, V.; Gaudet-Blavignac, C.; Robert, A.; Lovis, C. Use and Understanding of Anonymization and De-Identification in the Biomedical Literature: Scoping Review. J. Med. Internet Res. 2019, 21, e13484. [Google Scholar] [CrossRef] [Green Version]
- The European Parliament and the Council of The European Union. Directive (EU) 2016/1148 of the European Parliament and of the Council Concerning Measures for a High Common Level of Security of Network and Information Systems across the Union. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?toc=OJ:L:2016:194:TOC&uri=uriserv:OJ.L_.2016.194.01.0001.01.ENG (accessed on 15 March 2021).
- Tsang, L.; Kracov, D.A.; Mulryne, J.; Strom, L.; Perkins, N.; Dickinson, R.; Wallace, V.M.; Jones, B. The Impact of Artificial Intelligence on Medical Innovation in the European Union and United States. Available online: https://www.arnoldporter.com/~/media/files/perspectives/publications/2017/08/the-impact-of-artificial-inteelligence-on-medical-innovation.pdf (accessed on 15 March 2021).
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- The Cancer Imaging Archive (TCIA). Available online: http://www.cancerimagingarchive.net (accessed on 15 March 2021).
- Fonseca, C.G.; Backhaus, M.; Bluemke, D.A.; Britten, R.D.; Chung, J.D.; Cowan, B.R.; Dinov, I.D.; Finn, J.P.; Hunter, P.J.; Kadish, A.H.; et al. The Cardiac Atlas Project—An imaging database for computational modeling and statistical atlases of the heart. Bioinformatics 2011, 27, 2288–2295. [Google Scholar] [CrossRef]
- Jimenez-Del-Toro, O.; Muller, H.; Krenn, M.; Gruenberg, K.; Taha, A.A.; Winterstein, M.; Eggel, I.; Foncubierta-Rodriguez, A.; Goksel, O.; Jakab, A.; et al. Cloud-Based Evaluation of Anatomical Structure Segmentation and Landmark Detection Algorithms: VISCERAL Anatomy Benchmarks. IEEE Trans. Med. Imaging 2016, 35, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- UK. Available online: http://www.ukbiobank.ac.uk/ (accessed on 15 March 2021).
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, D.A.; Rosman, G.; Rus, D.; Meireles, O.R. Artificial Intelligence in Surgery: Promises and Perils. Ann. Surg. 2018, 268, 70–76. [Google Scholar] [CrossRef] [PubMed]
| Modality/Technique | Author | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion |
|---|---|---|---|---|---|---|
| CESM | Lin et al., 2020 [35] | Identification of benign and malignant BC lesions <1 cm | Radiomics features extracted from low-energy and recombined images on CC position | 139 patients | The radiomics nomogram combined with Radiomic-score, BI-RADS category and age showed AUC of 0.940. | The radiomics nomogram incorporated with CESM-based radiomics features, BI-RADS category and age could identify benign and malignant BC <1 cm |
| CESM | Mao et al. 2020 [36] | Pre-operative prediction of ALN metastasis | LASSO logistic regression was established for feature selection and utilized to construct radiomics signature | 394 patients | ROC curves of 0.774, 0.767 and 0.79 in the training, internal validation and external validation sets, respectively. | Authors identified the cutoff score in the radiomics nomogram as −1.49, which corresponded to a total point of 49 that could diagnose ALN metastasis with a sensitivity of >95%. |
| MRI | Tan et al., 2020 [6] | Value of radiomics feature extracted on the fat-suppressed T2WI for preoperative predicting ALN metastasis in BC | 17 texture features, 5 first-order statistical features, patient age, tumor size, HER2 status and thrombus | 329 BCs | Sensitivity, specificity, accuracy and are under the curve value of radiomics signature 65.22%, 81.08%, 75.00% and 0.819. | The MRI-based radiomics signature and nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. |
| Choudhery et al., 2020 [5] | Assessment of BC molecular subtype, pCR and Residual Cancer Burden in BC Patients Treated with NACT | Morphological and three-dimensional MRI textural features were computed, including unfiltered and filtered image data, with different spatial scaling factors | 259 BCs | Differences in minimum signal intensity and entropy among the tumor subtypes were significant. Sphericity in HER2+ tumors and entropy in luminal tumors were significantly associated with pCR. Multiple features demonstrated significant association with pathological complete response and residual cancer burden in TNBC with SD of intensity achieving the highest AUC for pCR in TNBC. | MRI radiomics features are associated with different molecular subtypes of breast cancer, pathological complete response and residual cancer burden. | |
| Hao et al., 2020 [37] | Contralateral BI-RADS 4 lesion assessment | 1046 radiomic features | 178 BCs | DCE-T1WI and T2WI imaging features signatures yielded an AUC of 0.77, which was better than the AUC of each signature alone. | The MRI radiomics-based ML model based on T2WI and DCE-T1WI features provided complementary information in discriminating benign and malignant contralateral BI-RADS 4 lesions. | |
| Lo Gullo et al., 2020 [38] | Assessment of sub-centimetric breast masses in BRCA patients | Radiomics features calculated using open-source CERR software | 96 BRCA carrier | The ML model combining 5 parameters including clinical factors, GLCM-based correlation from the pre-DCE phases and first-order coefficient of variation from the 1st post-DCE phase, achieved a diagnostic accuracy of 81.5%. | Radiomics analysis improved diagnostic accuracy compared with qualitative morphological assessment alone. | |
| Demircioglu et al., 2020 [39] | Molecular subtype, hormonal receptor status, Ki67- and HER2-expression, metastasis of lymph nodes and lymph vessel involvement as well as grading | 13.118 radiomic features extracted with a VOI-based approach | 98 BCs | PR and ER status predictions yielded AUCs of 0.67–0.69, Ki67 0.81 and HER2 Expressions 0.62. Involvement of the ALN could be predicted with an AUC of 0.80, while lymph node metastasis yielded an AUC of 0.71. | A rapid approach to VOI-based tumor-annotations for radiomics provides consisternt results to other studies in the same field. | |
| Zhang et al., 2020 [40] | Differentiation between benign and malignant lesions | Radiomics features extracted from T2WI, T1WI, DKI, ADC maps and DCE pharmacokinetic parameter maps | 207 BCs | The AUC of the optimal radiomics model, including T2 WI, DKI and quantitative DCE-MRI parameter maps was 0.921, with an accuracy of 0.833. | The model based on radiomics features from T2WI, DKI and quantitative DCE parameter maps has a high discriminatory ability for benign and malignant BC lesions. | |
| Zhou et al., 2020 [41] | Differentiation between benign and malignant BC lesions | 99 texture and histogram parameters | 133 patients | The highest accuracy of 91% was achieved when using the smallest bounding box of peritumoral tissues in segmentation. | Using the smallest bounding box containing proximal peritumor tissue as input had higher accuracy compared to using tumor alone or larger boxes. | |
| Liu et al., 2019 [42] | Assess lymphovascular invasion status | Radiomic signature composed of two features | 149 BCs | The value of AUC for a model combining both radiomic signature and ALN status (0.763) was higher than that for MRI ALN status alone and similar to that for the radiomics signature. | The DCE-MRI-based radiomics signature in combination with ALN status was effective in predicting the lymph and vascular invasion status of patients with BC before surgery. | |
| Xie et al., 2019 [43] | Subtype classification of breast cancer | 2498 features extracted from the DCE and DWI, together with DCE images, changing over 6 time points and DWI images changing over 3 b-values | 134 invasive ductal carcinoma | Highest accuracy of 91% for comparing triple negative to non-triple negative cancers. | Whole-tumor radiomics on MRI provides a non-invasive approach for BC subtype classification. | |
| Liang et al., 2018 [44] | Preoperative Ki-67 status | Radiomic features based on T2W and DCE-T1WI | 318 BC | The T2W image-based radiomics classifier showed significant discrimination for Ki-67 status, with AUC of 0.74 in the validation dataset. | The T2WI-based radiomics classifier was a significant predictor of Ki-67 status in patients with breast cancer while DCE-T1WI radiomic features were not able to discriminate Ki-67 status in the validation dataset. | |
| Digital mammography | Tan et al., 2020 [45] | Pre-operative prediction of ALN metastasis | Radiomic signature nomogram combined with receptor status and molecular subtype | 216 BCs | The radiomics nomogram, comprising PR status, molecular subtype and radiomics signature, showed excellent calibration and better performance for the metastatic ALN detection (AUC 0.883 and 0.863 in the primary and validation cohorts), better than each independent clinical feature and radiomics signature. | The mammography-based radiomics nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. |
| Digital Mammography | Stelzer et al., 2020 [46] | Distinguish malignant from benign classification | 249 image features from gray-value histogram, co-occurrence and run-length matrices | 226 patients | A high sensitivity threshold criterion was identified in the training dataset and successfully applied to the testing dataset, demonstrating the potential to avoid 37.1-45.7 % of unnecessary biopsies at the cost of one false-negative. | Combined texture analysis and ML could be used for risk stratification in suspicious mammographic calcifications. |
| Zhou et al., 2019 [47] | HER-2 status | 186 radiomic features | 306 l BCs | In the testing set the AUC of the radiomic model in assessing HER-2 status was 0.787. | Radiomics features could help in the preoperative evaluation of HER-2 status in patients with BC. | |
| Lei et al., 2019 [48] | Prediction of benign BI-RADS 4 calcifications | 8286 radiomic features extracted from the craniocaudal and mediolateral oblique scans | 212 calcifications | Six radiomic features and the menopausal state included in a radiomic nomogram could discriminate benign from malignant calcifications with an AUC of 0.80 in the validation cohort. | The mammography-based radiomic nomogram is a potential tool to distinguish benign calcifications from malignant calcifications. | |
| PET/CT | Ou et al., 2020 [49] | Differentiating breast carcinoma from breast lymphoma | Radiomic features extracted with a local software | 44 BCs | AUCs of 0.867 and 0.806 for PET radiomic and clinical model, AUCs of 0.891 and 0.759 for CT based radiomic model on training and validation data. | Models based on clinical, and radiomic features of 18 F-FDG PET/CT images could accurately discriminate BC from breast lymphoma. |
| Reference | Modality/Techique | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion |
|---|---|---|---|---|---|---|
| Reig et al., 2020 [50] | MRI | Review focused on machine learning techniques in breast MRI | Pre-processing, neural networks, deep learning, machine learning, segmentation, texture analysis | Breast malignant and benign pathology. | The Author discuss the possible future directions of machine learning in the current workflow of breast lesions assessed with MRI. | |
| Granzier et al., 2019 [51] | MRI | Systematic review, response prediction of neoadjuvant therapy | Various radiomic feature models, evaluated with the Radiomics Quality Score (RQS) | Studies ranging between 35-414 BC | AUC values ranged from 0.83 to 0.85. The best performing multivariate prediction model, based on logistic regression analysis, showed AUC of 0.94. | The systematic review revealed large heterogeneity for each step of the MRI-based radiomics workflow. Consequently, the results are difficult to compare. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesapane, F.; Rotili, A.; Agazzi, G.M.; Botta, F.; Raimondi, S.; Penco, S.; Dominelli, V.; Cremonesi, M.; Jereczek-Fossa, B.A.; Carrafiello, G.; et al. Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future. Curr. Oncol. 2021, 28, 2351-2372. https://doi.org/10.3390/curroncol28040217
Pesapane F, Rotili A, Agazzi GM, Botta F, Raimondi S, Penco S, Dominelli V, Cremonesi M, Jereczek-Fossa BA, Carrafiello G, et al. Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future. Current Oncology. 2021; 28(4):2351-2372. https://doi.org/10.3390/curroncol28040217
Chicago/Turabian StylePesapane, Filippo, Anna Rotili, Giorgio Maria Agazzi, Francesca Botta, Sara Raimondi, Silvia Penco, Valeria Dominelli, Marta Cremonesi, Barbara Alicja Jereczek-Fossa, Gianpaolo Carrafiello, and et al. 2021. "Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future" Current Oncology 28, no. 4: 2351-2372. https://doi.org/10.3390/curroncol28040217
APA StylePesapane, F., Rotili, A., Agazzi, G. M., Botta, F., Raimondi, S., Penco, S., Dominelli, V., Cremonesi, M., Jereczek-Fossa, B. A., Carrafiello, G., & Cassano, E. (2021). Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future. Current Oncology, 28(4), 2351-2372. https://doi.org/10.3390/curroncol28040217







