Artificial Intelligence in Clinical Oncology: From Productivity Enhancement to Creative Discovery
Simple Summary
Abstract
1. Introduction
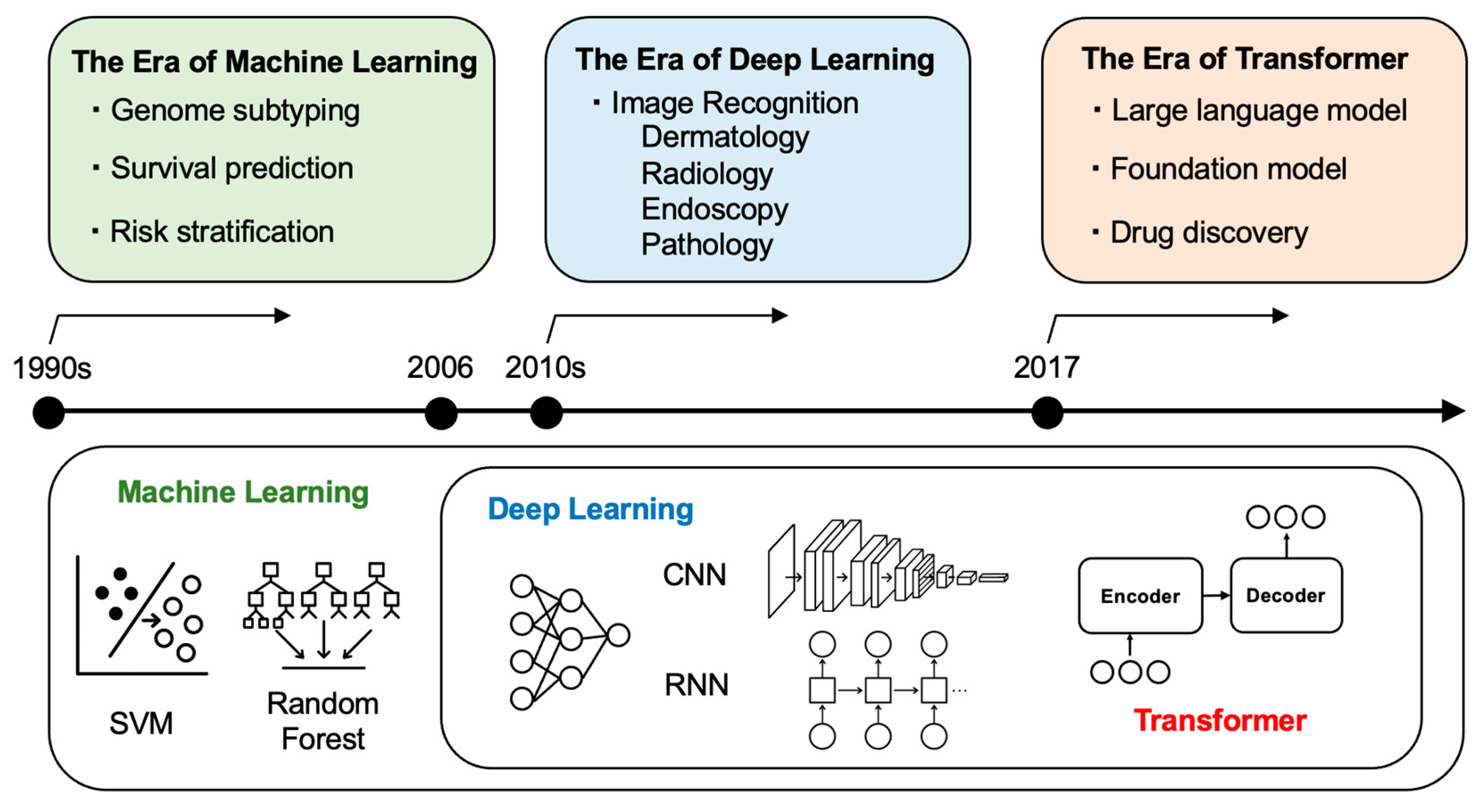
2. An Overview of AI Technologies and Their Relevance to Oncology
3. Productivity Enhancement in Clinical Oncology
3.1. AI for Image-Based Oncologic Diagnosis: Radiology, Digital Pathology, Endoscopy, and Dermatology
3.2. NLP for Data Structuring and Report Generation
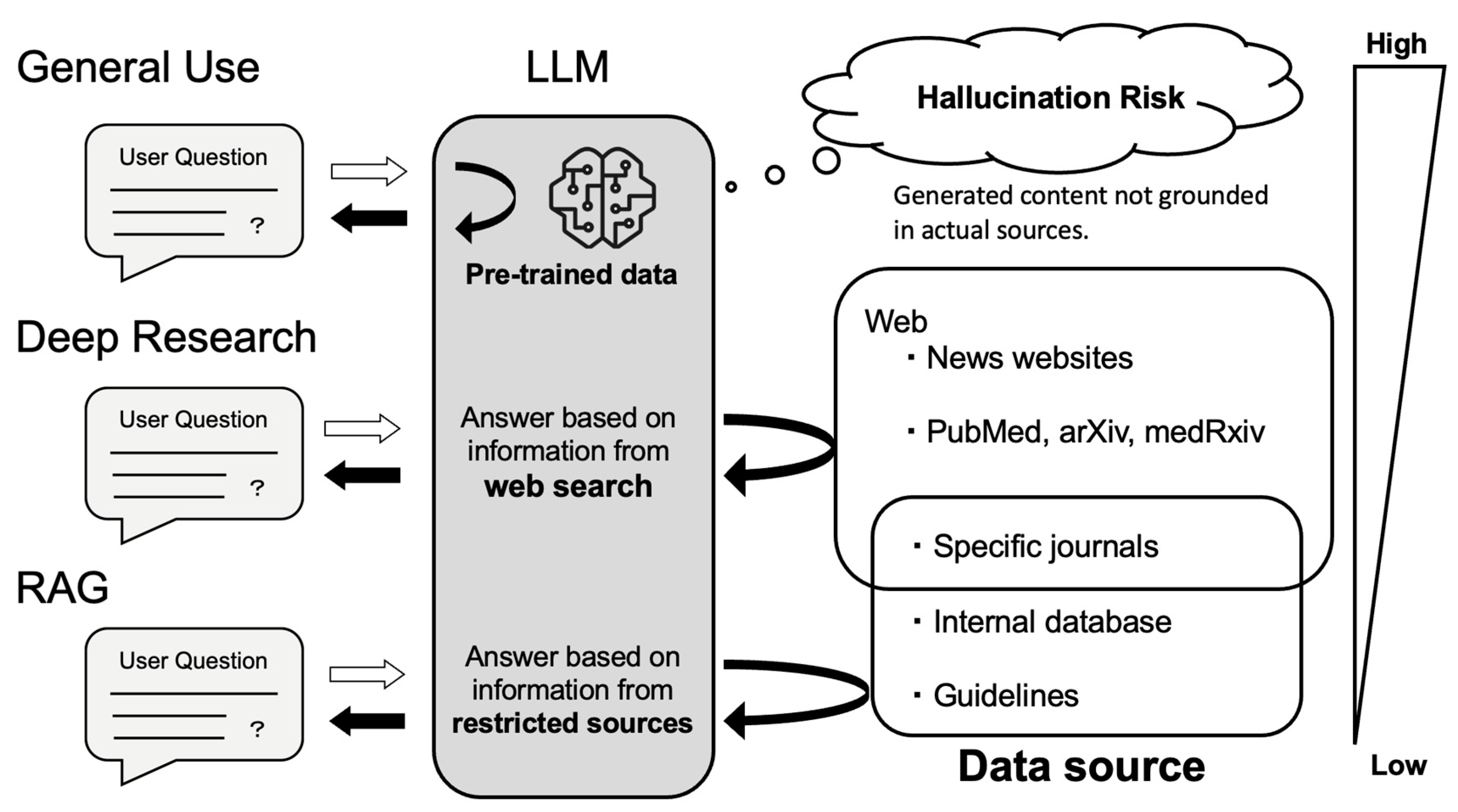
3.3. AI-Powered Research Support Tools
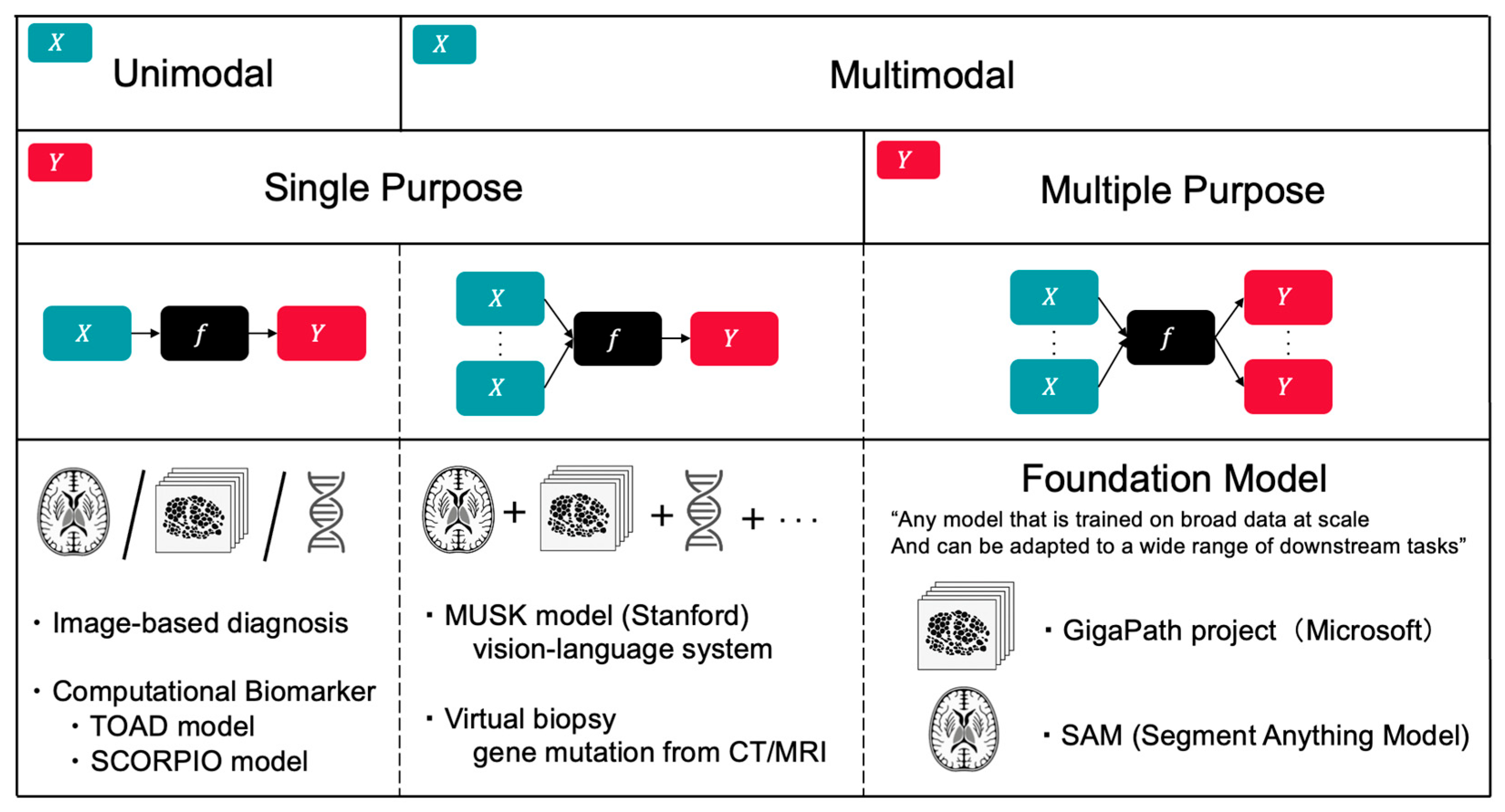
4. Creative Discovery in Clinical Oncology
4.1. Computational Biomarkers for Precision Oncology
4.2. Unsupervised Discovery and Deep Clinical Phenotyping
4.3. Drug Discovery
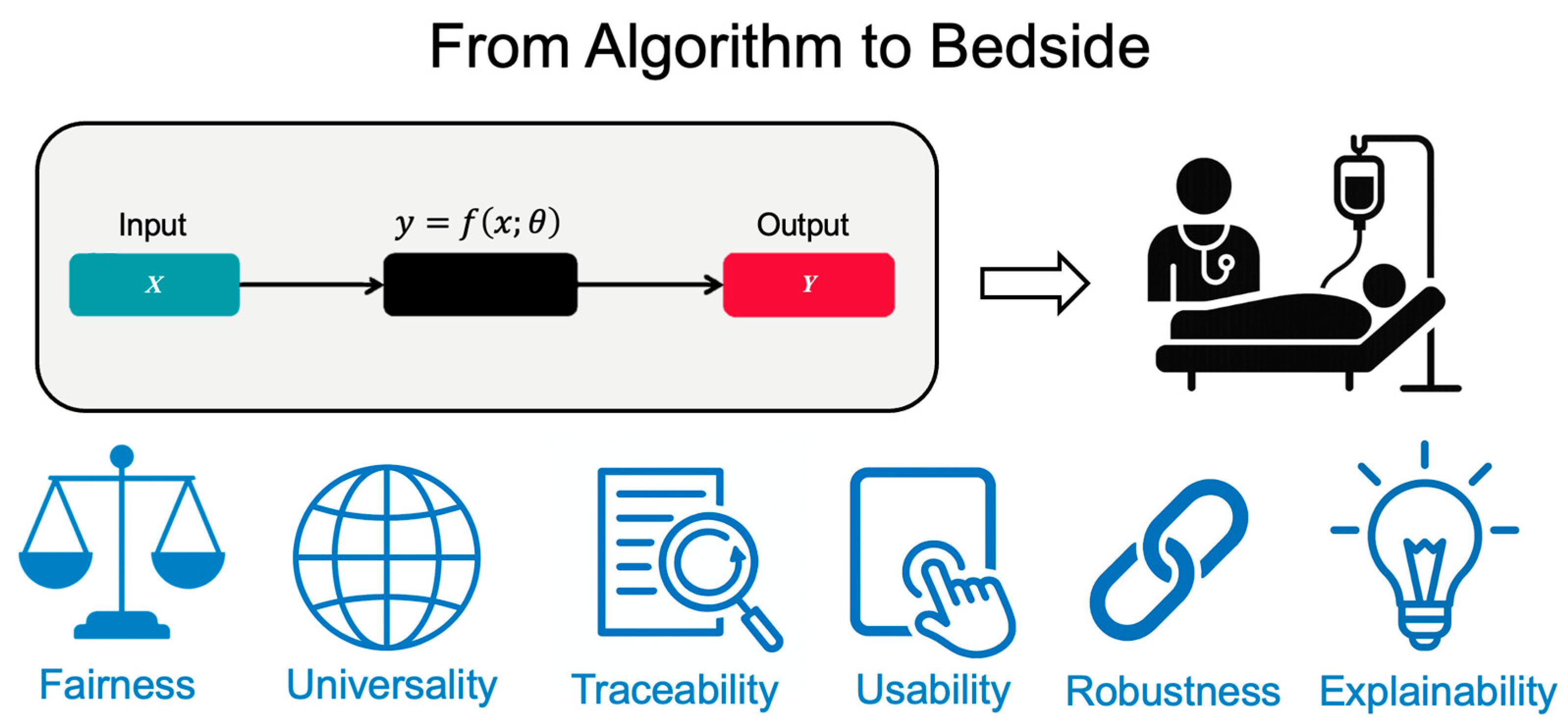
5. From Algorithm to Bedside: A FUTURE-AI–Informed Guide for Oncologists
5.1. Fairness: Ensuring Equitable AI Performance
5.2. Universality: Building Models for Diverse Clinical Settings
5.3. Traceability: Creating Transparent AI Systems
5.4. Usability: Integrating AI into Clinical Workflows
5.5. Robustness: Sustained Performance Under Real-World Conditions
5.6. Explainability: Making AI Decisions Understandable
5.7. Future Directions
6. Conclusions: Toward Collaborative Intelligence in Oncology
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADR | Adenoma Detection Rate |
| AI | Artificial Intelligence |
| ASCO | American Society of Clinical Oncology |
| AUROC: | Area Under the Receiver Operating Characteristic Curve |
| CDK | Cyclin-Dependent Kinase |
| CNN | Convolutional Neural Network |
| CT | Computed Tomography |
| CTNNB1 | Catenin Beta 1 |
| CUP | Cancer of Unknown Primary |
| DL | Deep Learning |
| EGFR | Epidermal Growth Factor Receptor |
| EHR | Electronic Health Record |
| FIGO: | International Federation of Gynecology and Obstetrics |
| FDA | Food and Drug Administration |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| H&E | Hematoxylin and Eosin |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| ICI | Immune Checkpoint Inhibitor |
| IDH | Isocitrate Dehydrogenase |
| LLM | Large Language Model |
| MASAI | Mammography Screening with Artificial Intelligence |
| MDL | Multimodal Deep Learning |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| MSI | Microsatellite Instability |
| mTOR | mammalian target of rapamycin |
| MUSK | Multimodal transformer with Unified maSKed modeling |
| NLP | Natural Language Processing |
| P4 | Predictive, Preventive, Personalized, and Participatory |
| PACS | Picture Archiving and Communication System |
| PD-L1 | Programmed Cell Death Ligand 1 |
| RAG | Retrieval-Augmented Generation |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| RNA | Ribonucleic Acid |
| RNN | Recurrent Neural Network |
| SCORPIO | Standard Clinical and labOratory featuRes for Prognostication of Immunotherapy Outcomes |
| SHAP | SHapley Additive exPlanations |
| SVM | Support Vector Machine |
| TMB | Tumor Mutational Burden |
| TNM | Tumor, Node, Metastasis |
| TOAD | Tumor Origin Assessment via Deep learning |
| TP53 | Tumor Protein p53 |
| UCSF | University of California, San Francisco |
| WSB | WD repeat and suppressor of cytokine signaling box containing |
| WSI | Whole-Slide Image |
References
- Lotter, W.; Hassett, M.J.; Schultz, N.; Kehl, K.L.; Van Allen, E.M.; Cerami, E. Artificial Intelligence in Oncology: Current Landscape, Challenges, and Future Directions. Cancer Discov. 2024, 14, 711–726. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Tiwari, A.; Mishra, S.; Kuo, T.-R. Current AI Technologies in Cancer Diagnostics and Treatment. Mol. Cancer 2025, 24, 159. [Google Scholar] [CrossRef]
- El Naqa, I.; Karolak, A.; Luo, Y.; Folio, L.; Tarhini, A.A.; Rollison, D.; Parodi, K. Translation of AI into Oncology Clinical Practice. Oncogene 2023, 42, 3089–3097. [Google Scholar] [CrossRef]
- Marra, A.; Morganti, S.; Pareja, F.; Campanella, G.; Bibeau, F.; Fuchs, T.; Loda, M.; Parwani, A.; Scarpa, A.; Reis-Filho, J.S.; et al. Artificial Intelligence Entering the Pathology Arena in Oncology: Current Applications and Future Perspectives. Ann. Oncol. 2025, 36, 712–725. [Google Scholar] [CrossRef]
- Beam, A.L.; Drazen, J.M.; Kohane, I.S.; Leong, T.-Y.; Manrai, A.K.; Rubin, E.J. Artificial Intelligence in Medicine. N. Engl. J. Med. 2023, 388, 1220–1221. [Google Scholar] [CrossRef]
- Howell, M.D.; Corrado, G.S.; DeSalvo, K.B. Three Epochs of Artificial Intelligence in Health Care. JAMA J. Am. Med. Assoc. 2024, 331, 242–244. [Google Scholar]
- Topol, E.J. High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep Learning in Cancer Diagnosis, Prognosis and Treatment Selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Merico, D.; Delong, A.; Frey, B.J. Deep Learning in Biomedicine. Nat. Biotechnol. 2018, 36, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Stancanello, J.; Pirrone, G.; Drigo, A.; Retico, A. The Evolution of Artificial Intelligence in Medical Imaging: From Computer Science to Machine and Deep Learning. Cancers 2024, 16, 3702. [Google Scholar] [CrossRef]
- Cruz, G.A.; Wishart, D.S. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform. 2007, 2, 59–77. [Google Scholar]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine Learning Applications in Cancer Prognosis and Prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Tseng, H.-H.; Wei, L.; Cui, S.; Luo, Y.; Haken, R.K.T.; El Naqa, I. Machine Learning and Imaging Informatics in Oncology. Oncology 2020, 98, 344–362. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Alex, K.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Andre, E.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and Mutation Prediction from Non-Small Cell Lung Cancer Histopathology Images Using Deep Learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Elman, J.L. Finding Structure in Time. Cogn. Sci. 1990, 14, 179–211. [Google Scholar] [CrossRef]
- Werbos, P.J. Backpropagation through time: What it does and how to do it. In Proceedings of the IEEE, Skokie, IL, USA, 4–6 October 1990; Volume 78, pp. 1550–1560. [Google Scholar]
- Azizi, S.; Bayat, S.; Yan, P.; Tahmasebi, A.; Kwak, J.T.; Xu, S.; Turkbey, B.; Choyke, P.; Pinto, P.; Wood, B.; et al. Deep Recurrent Neural Networks for Prostate Cancer Detection: Analysis of Temporal Enhanced Ultrasound. IEEE Trans. Med. Imaging 2018, 37, 2695–2703. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017. [Google Scholar] [CrossRef]
- Pai, S.; Bontempi, D.; Hadzic, I.; Prudente, V.; Sokač, M.; Chaunzwa, T.L.; Bernatz, S.; Hosny, A.; Mak, R.H.; Birkbak, N.J.; et al. Foundation Model for Cancer Imaging Biomarkers. Nat. Mach. Intell. 2024, 6, 354–367. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020. [Google Scholar] [CrossRef]
- Yan, S.; Yu, Z.; Primiero, C.; Vico-Alonso, C.; Wang, Z.; Yang, L.; Tschandl, P.; Hu, M.; Ju, L.; Tan, G.; et al. A Multimodal Vision Foundation Model for Clinical Dermatology. Nat. Med. 2025, 31, 2691–2702. [Google Scholar] [CrossRef]
- Cui, H.; Wang, C.; Maan, H.; Pang, K.; Luo, F.; Duan, N.; Wang, B. scGPT: Toward Building a Foundation Model for Single-Cell Multi-Omics Using Generative AI. Nat. Methods 2024, 21, 1470–1480. [Google Scholar]
- Bommasani, R.; Hudson, D.A.; Adeli, E.; Altman, R.; Arora, S.; von Arx, S.; Bernstein, M.S.; Bohg, J.; Bosselut, A.; Brunskill, E.; et al. On the Opportunities and Risks of Foundation Models. arXiv 2021. [Google Scholar] [CrossRef]
- Panagiotis, K.; Kline, T.L.; Meyer, H.M.; Khalid, S.; Leiner, T.; Loufek, B.T.; Blezek, D.; Vidal, D.E.; Hartman, R.P.; Joppa, L.J.; et al. Implementing Artificial Intelligence Algorithms in the Radiology Workflow: Challenges and Considerations. Mayo Clin. Proc. Digit. Health 2025, 3, 100188. [Google Scholar]
- Kristina, L.; Josefsson, V.; Larsson, A.-M.; Larsson, S.; Högberg, C.; Sartor, H.; Hofvind, S.; Andersson, I.; Rosso, A. Artificial Intelligence-Supported Screen Reading versus Standard Double Reading in the Mammography Screening with Artificial Intelligence Trial (MASAI): A Clinical Safety Analysis of a Randomised, Controlled, Non-Inferiority, Single-Blinded, Screening Accuracy Study. Lancet Oncol. 2023, 24, 936–944. [Google Scholar]
- Veronica, H.; Josefsson, V.; Sartor, H.; Schmidt, D.; Larsson, A.-M.; Hofvind, S.; Andersson, I.; Rosso, A.; Hagberg, O.; Lång, K. Screening Performance and Characteristics of Breast Cancer Detected in the Mammography Screening with Artificial Intelligence Trial (MASAI): A Randomised, Controlled, Parallel-Group, Non-Inferiority, Single-Blinded, Screening Accuracy Study. Lancet Digit. Health 2025, 7, e175–e183. [Google Scholar]
- Xiang, J.; Wang, X.; Zhang, X.; Xi, Y.; Eweje, F.; Chen, Y.; Li, Y.; Bergstrom, C.; Gopaulchan, M.; Kim, T.; et al. A Vision-Language Foundation Model for Precision Oncology. Nature 2025, 638, 769–778. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial Intelligence in Digital Pathology—New Tools for Diagnosis and Precision Oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital Pathology and Artificial Intelligence in Translational Medicine and Clinical Practice. Mod. Pathol. 2022, 35, 23–32. [Google Scholar]
- McGenity, C.; Clarke, E.L.; Jennings, C.; Matthews, G.; Cartlidge, C.; Freduah-Agyemang, H.; Stocken, D.D.; Treanor, D. Artificial Intelligence in Digital Pathology: A Systematic Review and Meta-Analysis of Diagnostic Test Accuracy. npj Digit. Med. 2024, 7, 114. [Google Scholar] [CrossRef]
- Xu, H.; Usuyama, N.; Bagga, J.; Zhang, S.; Rao, R.; Naumann, T.; Wong, C.; Gero, Z.; González, J.; Gu, Y.; et al. A Whole-Slide Foundation Model for Digital Pathology from Real-World Data. Nature 2024, 630, 181–188. [Google Scholar] [PubMed]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-Time Differentiation of Adenomatous and Hyperplastic Diminutive Colorectal Polyps during Analysis of Unaltered Videos of Standard Colonoscopy Using a Deep Learning Model. Gut 2019, 68, 94–100. [Google Scholar] [PubMed]
- Mori, Y.; Bretthauer, M.; Kalager, M. Hopes and Hypes for Artificial Intelligence in Colorectal Cancer Screening. Gastroenterology 2021, 161, 774–777. [Google Scholar] [CrossRef]
- Hassan, C.; Spadaccini, M.; Iannone, A.; Maselli, R.; Jovani, M.; Chandrasekar, V.T.; Antonelli, G.; Yu, H.; Areia, M.; Dinis-Ribeiro, M.; et al. Performance of Artificial Intelligence in Colonoscopy for Adenoma and Polyp Detection: A Systematic Review and Meta-Analysis. Gastrointest. Endosc. 2021, 93, 77–85.e6. [Google Scholar] [CrossRef]
- Hassan, C.; Spadaccini, M.; Mori, Y.; Foroutan, F.; Facciorusso, A.; Gkolfakis, P.; Tziatzios, G.; Triantafyllou, K.; Antonelli, G.; Khalaf, K.; et al. Real-Time Computer-Aided Detection of Colorectal Neoplasia during Colonoscopy: A Systematic Review and Meta-Analysis. Ann. Intern. Med. 2023, 176, 1209–1220. [Google Scholar] [CrossRef]
- Ahmad, A.; Wilson, A.; Haycock, A.; Humphries, A.; Monahan, K.; Suzuki, N.; Thomas-Gibson, S.; Vance, M.; Bassett, P.; Thiruvilangam, K.; et al. Evaluation of a Real-Time Computer-Aided Polyp Detection System during Screening Colonoscopy: AI-DETECT Study. Endoscopy 2023, 55, 313–319. [Google Scholar] [PubMed]
- Karsenti, D.; Tharsis, G.; Perrot, B.; Cattan, P.; Sert, A.P.D.; Venezia, F.; Zrihen, E.; Gillet, A.; Lab, J.-P.; Tordjman, G.; et al. Effect of Real-Time Computer-Aided Detection of Colorectal Adenoma in Routine Colonoscopy (COLO-GENIUS): A Single-Centre Randomised Controlled Trial. Lancet Gastroenterol. Hepatol. 2023, 8, 726–734. [Google Scholar]
- Yim, W.-W.; Yetisgen, M.; Harris, W.P.; Kwan, S.W. Natural Language Processing in Oncology: A Review: A Review. JAMA Oncol. 2016, 2, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Kehl, K.L.; Xu, W.; Lepisto, E.; Elmarakeby, H.; Hassett, M.J.; Van Allen, E.M.; Johnson, B.E.; Schrag, D. Natural Language Processing to Ascertain Cancer Outcomes from Medical Oncologist Notes. JCO Clin. Cancer Inform. 2020, 4, 680–690. [Google Scholar] [CrossRef]
- Savova, G.K.; Danciu, I.; Alamudun, F.; Miller, T.; Lin, C.; Bitterman, D.S.; Tourassi, G.; Warner, J.L. Use of Natural Language Processing to Extract Clinical Cancer Phenotypes from Electronic Medical Records. Cancer Res. 2019, 79, 5463–5470. [Google Scholar] [CrossRef]
- Jee, J.; Fong, C.; Pichotta, K.; Tran, T.N.; Luthra, A.; Waters, M.; Fu, C.; Altoe, M.; Liu, S.-Y.; Maron, S.B.; et al. Automated Real-World Data Integration Improves Cancer Outcome Prediction. Nature 2024, 636, 728–736. [Google Scholar] [CrossRef]
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large Language Models Encode Clinical Knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef]
- Chen, D.; Alnassar, S.A.; Avison, K.E.; Huang, R.S.; Raman, S. Large Language Model Applications for Health Information Extraction in Oncology: Scoping Review. JMIR Cancer 2025, 11, e65984. [Google Scholar] [CrossRef]
- Shah, N.H.; Entwistle 2025, D.; Pfeffer, M.A. Creation and Adoption of Large Language Models in Medicine. JAMA J. Am. Med. Assoc. 2023, 330, 866–869. [Google Scholar] [CrossRef]
- Zhu, M.; Lin, H.; Jiang, J.; Jinia, A.J.; Jee, J.; Pichotta, K.; Waters, M.; Rose, D.; Schultz, N.; Chalise, S.; et al. Large Language Model Trained on Clinical Oncology Data Predicts Cancer Progression. npj Digit. Med. 2025, 8, 397. [Google Scholar] [CrossRef]
- Rao, V.M.; Hla, M.; Moor, M.; Adithan, S.; Kwak, S.; Topol, E.J.; Rajpurkar, P. Multimodal Generative AI for Medical Image Interpretation. Nature 2025, 639, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Izhar, A.; Idris, N.; Japar, N. Medical Radiology Report Generation: A Systematic Review of Current Deep Learning Methods, Trends, and Future Directions. Artif. Intell. Med. 2025, 168, 103220. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Figueredo, G.; Li, R.; Zhang, W.E.; Chen, W.; Chen, X. A Survey of Deep-Learning-Based Radiology Report Generation Using Multimodal Inputs. Med. Image Anal. 2025, 103, 103627. [Google Scholar]
- Hong, K.E.; Ham, J.; Roh, B.; Gu, J.; Park, B.; Kang, S.; You, K.; Eom, J.; Bae, B.; Jo, J.-B.; et al. Diagnostic Accuracy and Clinical Value of a Domain-Specific Multimodal Generative AI Model for Chest Radiograph Report Generation. Radiology 2025, 314, e241476. [Google Scholar] [CrossRef]
- Huang, J.; Wittbrodt, M.T.; Teague, C.N.; Karl, E.; Galal, G.; Thompson, M.; Chapa, A.; Chiu, M.-L.; Herynk, B.; Linchangco, R.; et al. Efficiency and Quality of Generative AI-Assisted Radiograph Reporting. JAMA Netw. Open 2025, 8, e2513921. [Google Scholar] [CrossRef] [PubMed]
- Wornow, M.; Lozano, A.; Dash, D.; Jindal, J.; Mahaffey, K.W.; Shah, N.H. Zero-Shot Clinical Trial Patient Matching with LLMs. NEJM AI 2025, 2, AIcs2400360. [Google Scholar] [CrossRef]
- Von Itzstein, M.S.; Hullings, M.; Mayo, H.; Beg, M.S.; Williams, E.L.; Gerber, D.E. Application of Information Technology to Clinical Trial Evaluation and Enrollment: A Review: A Review. JAMA Oncol. 2021, 7, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Chow, R.; Midroni, J.; Kaur, J.; Boldt, G.; Liu, G.; Eng, L.; Liu, F.-F.; Haibe-Kains, B.; Lock, M.; Raman, S. Use of Artificial Intelligence for Cancer Clinical Trial Enrollment: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2023, 115, 365–374. [Google Scholar] [CrossRef]
- Beck, T.J.; Rammage, M.; Jackson, G.P.; Preininger, A.M.; Dankwa-Mullan, I.; Roebuck, M.C.; Torres, A.; Holtzen, H.; Coverdill, S.E.; Williamson, M.P.; et al. Artificial Intelligence Tool for Optimizing Eligibility Screening for Clinical Trials in a Large Community Cancer Center. JCO Clin. Cancer Inform. 2020, 4, 50–59. [Google Scholar] [CrossRef]
- Lu, X.; Yang, C.; Liang, L.; Hu, G.; Zhong, Z.; Jiang, Z. Artificial Intelligence for Optimizing Recruitment and Retention in Clinical Trials: A Scoping Review. J. Am. Med. Inform. Assoc. JAMIA 2024, 31, 2749–2759. [Google Scholar] [CrossRef]
- Saady, M.; Eissa, M.; Yacoub, A.S.; Hamed, A.B.; Azzazy, H.M.E.-S. Implementation of Artificial Intelligence Approaches in Oncology Clinical Trials: A Systematic Review. Artif. Intell. Med. 2025, 161, 103066. [Google Scholar] [CrossRef]
- Homolak, J. Opportunities and Risks of ChatGPT in Medicine, Science, and Academic Publishing: A Modern Promethean Dilemma. Croat. Med. J. 2023, 64, 1–3. [Google Scholar]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; Yih, W.-T.; Rocktäschel, T.; et al. Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. arXiv 2020. [Google Scholar] [CrossRef]
- Low, S.Y.; Jackson, M.L.; Hyde, R.J.; Brown, R.E.; Sanghavi, N.M.; Baldwin, J.D.; Pike, C.W.; Muralidharan, J.; Hui, G.; Alexander, N.; et al. Answering Real-World Clinical Questions Using Large Language Model, Retrieval-Augmented Generation, and Agentic Systems. Digit. Health 2025, 11, 20552076251348850. Available online: https://www.asco.org/practice-patients/guidelines/assistant (accessed on 1 September 2025). [CrossRef]
- Ahn, S. Data Science through Natural Language with ChatGPT’s Code Interpreter. Transl. Clin. Pharmacol. 2024, 32, 73–82. [Google Scholar] [CrossRef]
- Sapkota, R.; Roumeliotis, K.I.; Karkee, M. Vibe Coding vs. Agentic Coding: Fundamentals and Practical Implications of Agentic AI. arXiv 2025. [Google Scholar] [CrossRef]
- Rezaeikhonakdar, D. AI Chatbots and Challenges of HIPAA Compliance for AI Developers and Vendors. J. Am. Soc. Law Med. Ethics 2023, 51, 988–995. [Google Scholar] [CrossRef]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing Multimodal Data Integration to Advance Precision Oncology. Nat. Rev. Cancer 2022, 22, 114–126. [Google Scholar] [CrossRef]
- Lipkova, J.; Chen, R.J.; Chen, B.; Lu, M.Y.; Barbieri, M.; Shao, D.; Vaidya, A.J.; Chen, C.; Zhuang, L.; Williamson, D.F.; et al. Artificial Intelligence for Multimodal Data Integration in Oncology. Cancer Cell 2022, 40, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, P.; Fuchs, T.J.; Ho, D.J. Artificial Intelligence-Driven Cancer Diagnostics: Enhancing Radiology and Pathology through Reproducibility, Explainability, and Multimodality. Cancer Res. 2025, 85, 2356–2367. [Google Scholar] [CrossRef]
- Prelaj, M.V.; Zanitti, M.; Trovo, F.; Genova, C.; Viscardi, G.; Rebuzzi, S.E.; Mazzeo, L.; Provenzano, L.; Kosta, S.; Favali, M.; et al. Artificial Intelligence for Predictive Biomarker Discovery in Immuno-Oncology: A Systematic Review. Ann. Oncol. 2024, 35, 29–65. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.W.; van Amstel, P.; Martens, R.M.; Kooi, I.E.; Wesseling, P.; de Langen, A.J.; der Houven van Oordt, C.W.M.-V.; Jansen, B.H.E.; Moll, A.C.; Dorsman, J.C.; et al. Non-Invasive Tumor Genotyping Using Radiogenomic Biomarkers, a Systematic Review and Oncology-Wide Pathway Analysis. Oncotarget 2018, 9, 20134–20155. [Google Scholar] [CrossRef]
- Awais, M.; Rehman, A.; Bukhari, S.S. Advances in Liquid Biopsy and Virtual Biopsy for Care of Patients with Glioma: A Narrative Review. Expert Rev. Anticancer Ther. 2025, 25, 529–550. [Google Scholar] [CrossRef]
- Arthur, A.; Johnston, E.W.; Winfield, J.M.; Blackledge, M.D.; Jones, R.L.; Huang, P.H.; Messiou, C. Virtual Biopsy in Soft Tissue Sarcoma. How Close Are We? Front. Oncol. 2023, 12, 892620. [Google Scholar] [CrossRef]
- Cifci, D.; Foersch, S.; Kather, J.N. Artificial Intelligence to Identify Genetic Alterations in Conventional Histopathology. J. Pathol. 2022, 257, 430–444. [Google Scholar] [CrossRef]
- Campanella, G.; Kumar, N.; Nanda, S.; Singi, S.; Fluder, E.; Kwan, R.; Muehlstedt, S.; Pfarr, N.; Schüffler, P.J.; Häggström, I.; et al. Real-World Deployment of a Fine-Tuned Pathology Foundation Model for Lung Cancer Biomarker Detection. Nat. Med. 2025, 12, 892620. [Google Scholar] [CrossRef] [PubMed]
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.A.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J.; et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020, 159, 1406–1416.e11. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Lyu, T.; Liu, S.; Zhang, W.; Zhou, Y.; Zeng, C.; Wu, G. Learn to Estimate Genetic Mutation and Microsatellite Instability with Histopathology H&E Slides in Colon Carcinoma. Cancers 2022, 14, 4144. [Google Scholar] [CrossRef]
- Boehm, K.M.; El Nahhas, O.S.M.; Marra, A.; Waters, M.; Jee, J.; Braunstein, L.; Schultz, N.; Selenica, P.; Wen, H.Y.; Weigelt, B.; et al. Multimodal Histopathologic Models Stratify Hormone Receptor-Positive Early Breast Cancer. Nat. Commun. 2025, 16, 2106. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Chen, T.Y.; Williamson, D.F.K.; Zhao, M.; Shady, M.; Lipkova, J.; Mahmood, F. AI-Based Pathology Predicts Origins for Cancers of Unknown Primary. Nature 2021, 594, 106–110. [Google Scholar] [CrossRef]
- Yoo, S.-K.; Fitzgerald, C.W.; Cho, B.A.; Fitzgerald, B.G.; Han, C.; Koh, E.S.; Pandey, A.; Sfreddo, H.; Crowley, F.; Korostin, M.R.; et al. Prediction of Checkpoint Inhibitor Immunotherapy Efficacy for Cancer Using Routine Blood Tests and Clinical Data. Nat. Med. 2025, 31, 869–880. [Google Scholar] [CrossRef]
- Nangalia, J.; Campbell, P.J. Genome Sequencing during a Patient’s Journey through Cancer. N. Engl. J. Med. 2019, 381, 2145–2156. [Google Scholar] [CrossRef]
- Aleksakhina, S.N.; Imyanitov, E.N. Cancer Therapy Guided by Mutation Tests: Current Status and Perspectives. Int. J. Mol. Sci. 2021, 22, 10931. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical Management of Breast Cancer Heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of Artificial Intelligence for Preoperative Diagnostic and Prognostic Prediction in Epithelial Ovarian Cancer Based on Blood Biomarkers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 1248–1259. [Google Scholar]
- Fujita, M.; Chen, M.-J.M.; Siwak, D.R.; Sasagawa, S.; Oosawa-Tatsuguchi, A.; Arihiro, K.; Ono, A.; Miura, R.; Maejima, K.; Aikata, H.; et al. Proteo-Genomic Characterization of Virus-Associated Liver Cancers Reveals Potential Subtypes and Therapeutic Targets. Nat. Commun. 2022, 13, 6481. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yan, X.; Dong, Y.; Li, X.; Gao, B. Machine Learning and Multi-Omics Data Reveal Driver Gene-Based Molecular Subtypes in Hepatocellular Carcinoma for Precision Treatment. PLoS Comput. Biol. 2024, 20, e1012113. [Google Scholar] [CrossRef]
- Karin, J.; Mintz, R.; Raveh, B.; Nitzan, M. Interpreting Single-Cell and Spatial Omics Data Using Deep Neural Network Training Dynamics. Nat. Comput. Sci. 2024, 4, 941–954. [Google Scholar] [CrossRef]
- An, H.; Fang, W.; Chen, H.; Huang, W.; Liu, H.; Zhang, Z.; Zhao, H.; Zhang, Y.; Zhao, M.; Qiu, J.; et al. Spatial Transcriptomics Unveils Regional Heterogeneity and Subclonal Dynamics in the Lung Adenocarcinoma Microenvironment. Comput. Methods Programs Biomed. 2025, 270, 108929. [Google Scholar] [CrossRef]
- Takano, Y.; Suzuki, J.; Nomura, K.; Fujii, G.; Zenkoh, J.; Kawai, H.; Kuze, Y.; Kashima, Y.; Nagasawa, S.; Nakamura, Y.; et al. Spatially Resolved Gene Expression Profiling of Tumor Microenvironment Reveals Key Steps of Lung Adenocarcinoma Development. Nat. Commun. 2024, 15, 10637. [Google Scholar] [CrossRef]
- BBischoff, P.; Trinks, A.; Obermayer, B.; Pett, J.P.; Wiederspahn, J.; Uhlitz, F.; Liang, X.; Lehmann, A.; Jurmeister, P.; Elsner, A.; et al. Single-Cell RNA Sequencing Reveals Distinct Tumor Microenvironmental Patterns in Lung Adenocarcinoma. Oncogene 2021, 40, 6748–6758. [Google Scholar] [CrossRef]
- Yurkovich, J.T.; Tian, Q.; Price, N.D.; Hood, L. A Systems Approach to Clinical Oncology Uses Deep Phenotyping to Deliver Personalized Care. Nature Reviews. Clin. Oncol. 2020, 17, 183–194. [Google Scholar]
- Reicher, L.; Shilo, S.; Godneva, A.; Lutsker, G.; Zahavi, L.; Shoer, S.; Krongauz, D.; Rein, M.; Kohn, S.; Segev, T.; et al. Deep Phenotyping of Health-Disease Continuum in the Human Phenotype Project. Nat. Med. 2025, 31, 3191–3203. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.M.S.-F.; Contrepois, K.; Moneghetti, K.J.; Zhou, W.; Mishra, T.; Mataraso, S.; Dagan-Rosenfeld, O.; Ganz, A.B.; Dunn, J.; Hornburg, D.; et al. A Longitudinal Big Data Approach for Precision Health. Nat. Med. 2019, 25, 792–804. [Google Scholar] [CrossRef]
- Yurkovich, J.T.; Evans, S.J.; Rappaport, N.; Boore, J.L.; Lovejoy, J.C.; Price, N.D.; Hood, L.E. The Transition from Genomics to Phenomics in Personalized Population Health. Nature Reviews. Genetics 2024, 25, 286–302. [Google Scholar] [PubMed]
- Tian, P.N.D.; Hood, L. Systems Cancer Medicine: Towards Realization of Predictive, Preventive, Personalized and Participatory (P4) Medicine: Key Symposium: Systems Cancer Medicine. J. Intern. Med. 2012, 271, 111–121. [Google Scholar] [CrossRef]
- Sakurada, K.; Ishikawa, T.; Oba, J.; Kuno, M.; Okano, Y.; Sakamaki, T.; Tamura, T. Medical AI and AI for Medical Sciences. JMA J. 2025, 8, 26–37. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ren, F.; Ding, X.; Zheng, M.; Korzinkin, M.; Cai, X.; Zhu, W.; Mantsyzov, A.; Aliper, A.; Aladinskiy, V.; Cao, Z.; et al. AlphaFold Accelerates Artificial Intelligence Powered Drug Discovery: Efficient Discovery of a Novel CDK20 Small Molecule Inhibitor. Chem. Sci. 2023, 14, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.N.; Nguyen, P.K.; Nguyen, T.P.T.; Nguyen, H.Q.; Tam, D.N.H.; Huynh, H.H.; Huynh, P.K.; Le, N.Q.K. An In-Depth Review of AI-Powered Advancements in Cancer Drug Discovery. Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167680. [Google Scholar]
- Weng, Y.; Pan, C.; Shen, Z.; Chen, S.; Xu, L.; Dong, X.; Chen, J. Identification of Potential WSB1 Inhibitors by AlphaFold Modeling, Virtual Screening, and Molecular Dynamics Simulation Studies. Evid. Based Complement. Altern. Med. 2022, 2022, 4629392. [Google Scholar]
- Schauperl, M.; Denny, R.A. AI-Based Protein Structure Prediction in Drug Discovery: Impacts and Challenges. J. Chem. Inf. Model. 2022, 62, 3142–3156. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; McShan, A.C. The Power and Pitfalls of AlphaFold2 for Structure Prediction beyond Rigid Globular Proteins. Nat. Chem. Biol. 2024, 20, 950–959. [Google Scholar] [CrossRef]
- Nussinov, R.; Zhang, M.; Liu, Y.; Jang, H. AlphaFold, Allosteric, and Orthosteric Drug Discovery: Ways Forward. Drug Discov. Today 2023, 28, 103551. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold Predictions Are Valuable Hypotheses and Accelerate but Do Not Replace Experimental Structure Determination. Nat. Methods 2024, 21, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Lekadir, K.; Frangi, A.F.; Porras, A.R.; Glocker, B.; Cintas, C.; Langlotz, C.P.; Weicken, E.; Asselbergs, F.W.; Prior, F.; Collins, G.S.; et al. FUTURE-AI: International Consensus Guideline for Trustworthy and Deployable Artificial Intelligence in Healthcare. BMJ 2025, 388, e081554. [Google Scholar] [CrossRef] [PubMed]
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.A.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.M.; Bailey, E.E.; Gevaert, O.; et al. Disparities in Dermatology AI Performance on a Diverse, Curated Clinical Image Set. Sci. Adv. 2022, 8, eabq6147. [Google Scholar] [CrossRef]
- Adamson, A.S.; Smith, A. Machine Learning and Health Care Disparities in Dermatology. JAMA Dermatol. 2018, 154, 1247–1248. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting Racial Bias in an Algorithm Used to Manage the Health of Populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing Machine Learning in Health Care—Addressing Ethical Challenges. N. Engl. J. Med. 2018, 378, 981–983. [Google Scholar]
- Seyyed-Kalantari, L.; Zhang, H.; McDermott, M.B.A.; Chen, I.Y.; Ghassemi, M. Underdiagnosis Bias of Artificial Intelligence Algorithms Applied to Chest Radiographs in Under-Served Patient Populations. Nat. Med. 2021, 27, 2176–2182. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Lungren, M.P. The Current and Future State of AI Interpretation of Medical Images. N. Engl. J. Med. 2023, 388, 1981–1990. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, H.; Gichoya, J.W.; Katabi, D.; Ghassemi, M. The Limits of Fair Medical Imaging AI in Real-World Generalization. Nat. Med. 2024, 30, 2838–2848. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International Evaluation of an AI System for Breast Cancer Screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Ng, A.Y.; James, J.J.; Khara, G.; Ambrózay, É.; Austin, C.C.; Forrai, G.; Fox, G.; Glocker, B.; Heindl, A.; et al. Multi-Vendor Evaluation of Artificial Intelligence as an Independent Reader for Double Reading in Breast Cancer Screening on, 275,900 Mammograms. BMC Cancer 2023, 23, 460. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Hardt, M.; Howell, M.D.; Corrado, G.; Chin, M.H. Ensuring Fairness in Machine Learning to Advance Health Equity. Ann. Intern. Med. 2018, 169, 866–872. [Google Scholar] [CrossRef]
- Seyyed-Kalantari, L.; Liu, G.; McDermott, M.; Chen, I.Y.; Ghassemi, M. CheXclusion: Fairness Gaps in Deep Chest X-Ray Classifiers. arXiv 2020. [Google Scholar] [CrossRef]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated Learning in Medicine: Facilitating Multi-Institutional Collaborations without Sharing Patient Data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. npj Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Zhang, H.B.; Lemoine, B.; Mitchell, M. Mitigating Unwanted Biases with Adversarial Learning. arXiv 2018. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key Challenges for Delivering Clinical Impact with Artificial Intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef]
- Schömig-Markiefka, B.; Pryalukhin, A.; Hulla, W.; Bychkov, A.; Fukuoka, J.; Madabhushi, A.; Achter, V.; Nieroda, L.; Büttner, R.; Quaas, A.; et al. Quality Control Stress Test for Deep Learning-Based Diagnostic Model in Digital Pathology. Mod. Pathol. 2021, 34, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the Effects of Data Augmentation and Stain Color Normalization in Convolutional Neural Networks for Computational Pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef]
- Napravnik, M.; Hržić, F.; Urschler, M.; Miletić, D.; Štajduhar, I. Lessons Learned from Radiology NET Foundation Models for Transfer Learning in Medical Radiology. Sci. Rep. 2025, 15, 21622. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding Transfer Learning for Medical Imaging. arXiv 2019. [Google Scholar] [CrossRef]
- Ming, Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-Efficient and Weakly Supervised Computational Pathology on Whole-Slide Images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar]
- Fedorov, A.; Longabaugh, W.J.R.; Pot, D.; Clunie, D.A.; Pieper, S.D.; Gibbs, D.L.; Bridge, C.; Herrmann, M.D.; Homeyer, A.; Lewis, R.; et al. National Cancer Institute Imaging Data Commons: Toward Transparency, Reproducibility, and Scalability in Imaging Artificial Intelligence. RadioGraphics 2023, 43, e230180. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Ghassemi, M.; Oakden-Rayner, L.; Beam, A.L. The False Hope of Current Approaches to Explainable Artificial Intelligence in Health Care. Lancet Digit. Health 2021, 3, e745–e750. [Google Scholar]
- Schelter, L.; Klein, T. Automatically Tracking Metadata and Provenance of Machine Learning Experiments. In Machine Learning Systems Workshop at NIPS. 2017. Available online: https://neurips.cc/virtual/2017/workshop/8774 (accessed on 20 October 2025).
- Namli, T.; Sınacı, A.A.; Gönül, S.; Herguido, C.R.; Garcia-Canadilla, P.; Muñoz, A.M.; Esteve, A.V.; Ertürkmen, G.B.L. A Scalable and Transparent Data Pipeline for AI-Enabled Health Data Ecosystems. Front. Med. 2024, 11, 1393123. [Google Scholar] [CrossRef]
- Ratwani, R.M.; Bates, D.W.; Classen, D.C. Patient Safety and Artificial Intelligence in Clinical Care. JAMA Health Forum 2024, 5, e235514. [Google Scholar] [CrossRef]
- Kwong, J.C.C.; Khondker, A.; Lajkosz, K.; McDermott, M.B.A.; Frigola, X.B.; McCradden, M.D.; Mamdani, M.; Kulkarni, G.S.; Johnson, A.E.W. APPRAISE-AI Tool for Quantitative Evaluation of AI Studies for Clinical Decision Support. JAMA Netw. Open 2023, 6, e2335377. [Google Scholar] [CrossRef]
- Shah, N.H.; Halamka, J.D.; Saria, S.; Pencina, M.; Tazbaz, T.; Tripathi, M.; Callahan, A.; Hildahl, H.; Anderson, B. A Nationwide Network of Health AI Assurance Laboratories. JAMA J. Am. Med. Assoc. 2024, 331, 245–249. [Google Scholar] [CrossRef]
- Kolla, L.; Parikh, R.B. Uses and Limitations of Artificial Intelligence for Oncology. Cancer 2024, 130, 2101–2107. [Google Scholar] [CrossRef]
- Sujan, M.; Furniss, D.; Grundy, K.; Grundy, H.; Nelson, D.; Elliott, M.; White, S.; Habli, I.; Reynolds, N. Human Factors Challenges for the Safe Use of Artificial Intelligence in Patient Care. BMJ Health Care Inform. 2019, 26, e100081. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, Z.-H.; Cao, Y.-H.; Peng, S. Clinical Impact and Quality of Randomized Controlled Trials Involving Interventions Evaluating Artificial Intelligence Prediction Tools: A Systematic Review. npj Digit. Med. 2021, 4, 154. [Google Scholar] [CrossRef] [PubMed]
- Balendran, A.; Beji, C.; Bouvier, F.; Khalifa, O.; Evgeniou, T.; Ravaud, P.; Porcher, R. A Scoping Review of Robustness Concepts for Machine Learning in Healthcare. npj Digit. Med. 2025, 8, 38. [Google Scholar] [CrossRef]
- Joel, M.Z.; Umrao, S.; Chang, E.; Choi, R.; Yang, D.X.; Duncan, J.S.; Omuro, A.; Herbst, R.; Krumholz, H.M.; Aneja, S. Using Adversarial Images to Assess the Robustness of Deep Learning Models Trained on Diagnostic Images in Oncology. JCO Clin. Cancer Inform. 2022, 6, e2100170. [Google Scholar] [CrossRef] [PubMed]
- Subasri, V.; Krishnan, A.; Kore, A.; Dhalla, A.; Pandya, D.; Wang, B.; Malkin, D.; Razak, F.; Verma, A.A.; Goldenberg, A.; et al. Detecting and Remediating Harmful Data Shifts for the Responsible Deployment of Clinical AI Models. JAMA Netw. Open 2025, 8, e2513685. [Google Scholar] [CrossRef]
- Goceri, E. Medical Image Data Augmentation: Techniques, Comparisons and Interpretations. Artif. Intell. Rev. 2023, 56, 12561–12605. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, S.G.; Subbaswamy, A.; Singh, K.; Bowers, J.; Kupke, A.; Zittrain, J.; Kohane, I.S.; Saria, S. The Clinician and Dataset Shift in Artificial Intelligence. N. Engl. J. Med. 2021, 385, 283–286. [Google Scholar]
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual Lifelong Learning with Neural Networks: A Review. Neural Netw. 2019, 113, 54–71. [Google Scholar] [CrossRef]
- Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial Intelligence versus Clinicians: Systematic Review of Design, Reporting Standards, and Claims of Deep Learning Studies. BMJ 2020, 368, m689. [Google Scholar] [CrossRef]
- Guan, H.; Bates, D.; Zhou, L. Keeping Medical AI Healthy: A Review of Detection and Correction Methods for System Degradation. arXiv 2020. [Google Scholar] [CrossRef]
- Qazi, M.A.; Hashmi, A.U.R.; Sanjeev, S.; Almakky, I.; Saeed, N.; Gonzalez, C.; Yaqub, M. Continual Learning in Medical Imaging: A Survey and Practical Analysis. arXiv 2025. [Google Scholar] [CrossRef]
- Perkonigg, M.; Hofmanninger, J.; Herold, C.J.; Brink, J.A.; Pianykh, O.; Prosch, H.; Langs, G. Dynamic Memory to Alleviate Catastrophic Forgetting in Continual Learning with Medical Imaging. Nat. Commun. 2021, 12, 5678. [Google Scholar] [CrossRef]
- Qayyum, A.; Qadir, J.; Bilal, M.; Al-Fuqaha, A. Secure and Robust Machine Learning for Healthcare: A Survey. IEEE Rev. Biomed. Eng. 2021, 14, 156–180. [Google Scholar] [PubMed]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Salahuddin, Z.; Woodruff, H.C.; Chatterjee, A.; Lambin, P. Transparency of Deep Neural Networks for Medical Image Analysis: A Review of Interpretability Methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef]
- Nazir, S.; Dickson, D.M.; Akram, M.U. Survey of Explainable Artificial Intelligence Techniques for Biomedical Imaging with Deep Neural Networks. Comput. Biol. Med. 2023, 156, 106668. [Google Scholar] [CrossRef]
- E Ihongbe, I.; Fouad, S.; Mahmoud, T.F.; Rajasekaran, A.; Bhatia, B. Evaluating Explainable Artificial Intelligence (XAI) Techniques in Chest Radiology Imaging through a Human-Centered Lens. PLoS ONE 2024, 19, e0308758. [Google Scholar]
- Abas Mohamed, Y.; Khoo, B.E.; Asaari, M.S.M.; Aziz, M.E.; Ghazali, F.R. Decoding the Black Box: Explainable AI (XAI) for Cancer Diagnosis, Prognosis, and Treatment Planning-A State-of-the Art Systematic Review. Int. J. Med. Inform. 2025, 193, 105689. [Google Scholar]
- Arravalli, T.; Chadaga, K.; Muralikrishna, H.; Sampathila, N.; Cenitta, D.; Chadaga, R.; Swathi, K.S. Detection of Breast Cancer Using Machine Learning and Explainable Artificial Intelligence. Sci. Rep. 2025, 15, 26931. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Vanitha, K.; Mahesh, T.R.; Sree, S.S.; Guluwadi, S. Deep Learning Ensemble Approach with Explainable AI for Lung and Colon Cancer Classification Using Advanced Hyperparameter Tuning. BMC Med. Inform. Decis. Mak. 2024, 24, 222. [Google Scholar]
- Talaat, F.M.; Gamel, S.A.; El-Balka, R.M.; Shehata, M.; ZainEldin, H. Grad-CAM Enabled Breast Cancer Classification with a 3D Inception-ResNet V2: Empowering Radiologists with Explainable Insights. Cancers 2024, 16, 3668. [Google Scholar]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017. [Google Scholar] [CrossRef]
- Caruana, R.; Lou, Y.; Gehrke, J.; Koch, P.; Sturm, M.; Elhadad, N. Intelligible Models for HealthCare: Predicting Pneumonia Risk and Hospital 30-Day Readmission. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 10–13 August 2015. [Google Scholar]
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Precise4Q consortium Explainability for Artificial Intelligence in Healthcare: A Multidisciplinary Perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar]
- Spaanderman, D.J.; Marzetti, M.; Wan, X.; Scarsbrook, A.F.; Robinson, P.; Oei, E.H.G.; Visser, J.J.; Hemke, R.; van Langevelde, K.; Hanff, D.F.; et al. AI in Radiological Imaging of Soft-Tissue and Bone Tumours: A Systematic Review Evaluating against CLAIM and FUTURE-AI Guidelines. eBioMedicine 2025, 114, 105642. [Google Scholar]
- Fiske, A.; Radhuber, I.M.; Willem, T.; Buyx, A.; Celi, L.A.; McLennan, S. Climate Change and Health: The next Challenge of Ethical AI. Lancet Glob. Health 2025, 13, e1314–20. [Google Scholar] [PubMed]
- Hao, K. Training a Single AI Model Can Emit as Much Carbon as Five Cars in Their Lifetimes. Available online: https://www.technologyreview.com/2019/06/06/239031/training-a-single-ai-model-can-emit-as-much-carbon-as-five-cars-in-their-lifetimes/ (accessed on 1 October 2025).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuno, M.; Osumi, H.; Udagawa, S.; Yoshikawa, K.; Ooki, A.; Shinozaki, E.; Ishikawa, T.; Oba, J.; Yamaguchi, K.; Sakurada, K. Artificial Intelligence in Clinical Oncology: From Productivity Enhancement to Creative Discovery. Curr. Oncol. 2025, 32, 588. https://doi.org/10.3390/curroncol32110588
Kuno M, Osumi H, Udagawa S, Yoshikawa K, Ooki A, Shinozaki E, Ishikawa T, Oba J, Yamaguchi K, Sakurada K. Artificial Intelligence in Clinical Oncology: From Productivity Enhancement to Creative Discovery. Current Oncology. 2025; 32(11):588. https://doi.org/10.3390/curroncol32110588
Chicago/Turabian StyleKuno, Masahiro, Hiroki Osumi, Shohei Udagawa, Kaoru Yoshikawa, Akira Ooki, Eiji Shinozaki, Tetsuo Ishikawa, Junna Oba, Kensei Yamaguchi, and Kazuhiro Sakurada. 2025. "Artificial Intelligence in Clinical Oncology: From Productivity Enhancement to Creative Discovery" Current Oncology 32, no. 11: 588. https://doi.org/10.3390/curroncol32110588
APA StyleKuno, M., Osumi, H., Udagawa, S., Yoshikawa, K., Ooki, A., Shinozaki, E., Ishikawa, T., Oba, J., Yamaguchi, K., & Sakurada, K. (2025). Artificial Intelligence in Clinical Oncology: From Productivity Enhancement to Creative Discovery. Current Oncology, 32(11), 588. https://doi.org/10.3390/curroncol32110588




