Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus
Abstract
:1. Introduction
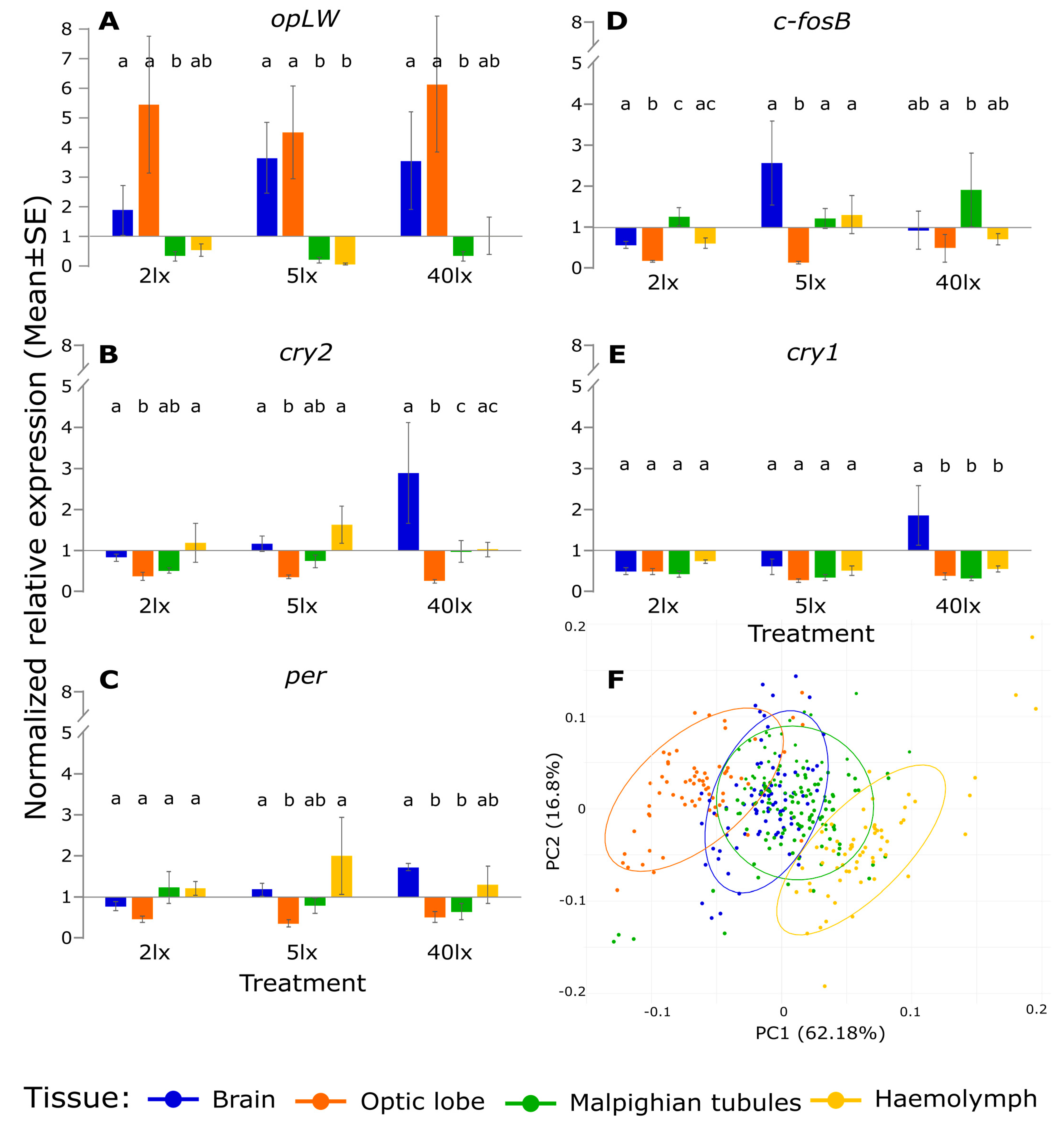
2. Results
3. Discussion
4. Materials and Methods
4.1. Insect Rearing Conditions
4.2. Light-Pulse Experiments and Sample Preparation
4.3. Primers and qPCR
4.4. Data-Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borges, R.M. Impacts of Artificial Light at Night on Biological Timings. Indian J. Entomol. 2022, 84, 483–492. [Google Scholar] [CrossRef]
- Saunders, D.S. Insect Photoperiodism: Seeing the Light. Physiol. Entomol. 2012, 37, 207–218. [Google Scholar] [CrossRef]
- Pittendrigh, C.S. Circadian Rhythms and the Circadian Organization of Living Systems. Cold Spring Harb. Symp. Quant. Biol. 1960, 25, 159–184. [Google Scholar] [CrossRef]
- Kronfeld-Schor, N.; Visser, M.E.; Salis, L.; van Gils, J.A. Chronobiology of Interspecific Interactions in a Changing World. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160248. [Google Scholar] [CrossRef]
- Ampleford, E.J.; Steel, C.G.H. Circadian Control of Ecdysis in Rhodnius prloixus (Hempitera). J. Comp. Physiol. A 1982, 147, 281–286. [Google Scholar] [CrossRef]
- Helm, B.; Visser, M.E.; Schwartz, W.; Kronfeld-Schor, N.; Gerkema, M.; Piersma, T.; Bloch, G. Two Sides of a Coin: Ecological and Chronobiological Perspectives of Timing in the Wild. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160246. [Google Scholar] [CrossRef]
- Saunders, D.S.; Steel, C.G.H.; Vafopoulou, X.; Lewis, R.D. Circadian Rhythms of Activity in Individual Insects. In Insect Clocks; Elsevier: Amsterdam, The Netherlands, 2002; pp. 7–42. [Google Scholar]
- Merrow, M.; Spoelstra, K.; Roenneberg, T. The Circadian Cycle: Daily Rhythms from Behaviour to Genes. EMBO Rep. 2005, 6, 930–935. [Google Scholar] [CrossRef]
- Horch, H.W.; Mito, T.; Popadi, A.; Ohuchi, H.; Noji, S. The Cricket as a Model Organism; Springer: Tokyo, Japan, 2017; ISBN 978-4-431-56476-8. [Google Scholar]
- Moaraf, S.; Vistoropsky, Y.; Pozner, T.; Heiblum, R.; Okuliarová, M.; Zeman, M.; Barnea, A. Artificial Light at Night Affects Brain Plasticity and Melatonin in Birds. Neurosci. Lett. 2020, 716, 134639. [Google Scholar] [CrossRef]
- Falchi, F.; Furgoni, R.; Gallaway, T.A.; Rybnikova, N.A.; Portnov, B.A.; Baugh, K.; Cinzano, P.; Elvidge, C.D. Light Pollution in USA and Europe: The Good, the Bad and the Ugly. J. Environ. Manag. 2019, 248, 109227. [Google Scholar] [CrossRef]
- Hölker, F.; Wolter, C.; Perkin, E.K.; Tockner, K. Light Pollution as a Biodiversity Threat. Trends Ecol. Evol. 2010, 25, 681–682. [Google Scholar] [CrossRef]
- Dominoni, D.; Quetting, M.; Partecke, J. Artificial Light at Night Advances Avian Reproductive Physiology. Proc. R. Soc. B Biol. Sci. 2013, 280, 20123017. [Google Scholar] [CrossRef]
- Amichai, E.; Kronfeld-Schor, N. Artificial Light at Night Promotes Activity Throughout the Night in Nesting Common Swifts (Apus apus). Sci. Rep. 2019, 9, 11052. [Google Scholar] [CrossRef]
- Kumar, D.; Soni, S.K.; Kronfeld-Schor, N.; Singaravel, M. Wheel-Running Activity Rhythms and Masking Responses in the Diurnal Palm Squirrel, Funambulus pennantii. Chronobiol. Int. 2020, 37, 1693–1708. [Google Scholar] [CrossRef]
- Buchanan, B.W. Observed and Potential Effects of Artificial Night Lighting on Anuran Amphibians. Ecol. Conseq. Artif. Night Light. 2006, 09, 192–220. [Google Scholar]
- Levy, K.; Wegrzyn, Y.; Efronny, R.; Barnea, A.; Ayali, A. Lifelong Exposure to Artificial Light at Night Impacts Stridulation and Locomotion Activity Patterns in the Cricket Gryllus bimaculatus. Proc. R. Soc. B Biol. Sci. 2021, 288, 20211626. [Google Scholar] [CrossRef]
- Durrant, J.; Michaelides, E.B.; Rupasinghe, T.; Tull, D.; Green, M.P.; Jones, T.M. Constant Illumination Reduces Circulating Melatonin and Impairs Immune Function in the Cricket Teleogryllus commodus. PeerJ 2015, 3, e1075. [Google Scholar] [CrossRef]
- Raap, T.; Pinxten, R.; Eens, M. Light Pollution Disrupts Sleep in Free-Living Animals. Sci. Rep. 2015, 5, 13557. [Google Scholar] [CrossRef]
- Gaston, K.J.; Davies, T.W.; Nedelec, S.L.; Holt, L.A. Impacts of Artificial Light at Night on Biological Timings. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 49–68. [Google Scholar] [CrossRef]
- Owens, A.C.S.; Lewis, S.M. The Impact of Artificial Light at Night on Nocturnal Insects: A Review and Synthesis. Ecol. Evol. 2018, 8, 11337–11358. [Google Scholar] [CrossRef]
- Sanders, D.; Kehoe, R.; Cruse, D.; van Veen, F.J.F.; Gaston, K.J. Low Levels of Artificial Light at Night Strengthen Top-Down Control in Insect Food Web. Curr. Biol. 2018, 28, 2474–2478.e3. [Google Scholar] [CrossRef]
- Bolliger, J.; Hennet, T.; Wermelinger, B.; Bösch, R.; Pazur, R.; Blum, S.; Haller, J.; Obrist, M.K. Effects of Traffic-Regulated Street Lighting on Nocturnal Insect Abundance and Bat Activity. Basic Appl. Ecol. 2020, 47, 44–56. [Google Scholar] [CrossRef]
- Foster, J.J.; Tocco, C.; Smolka, J.; Khaldy, L.; Baird, E.; Byrne, M.J.; Nilsson, D.E.; Dacke, M. Light Pollution Forces a Change in Dung Beetle Orientation Behavior. Curr. Biol. 2021, 31, 3935–3942.e3. [Google Scholar] [CrossRef]
- Giavi, S.; Blösch, S.; Schuster, G.; Knop, E. Artificial Light at Night Can Modify Ecosystem Functioning beyond the Lit Area. Sci. Rep. 2020, 10, 11870. [Google Scholar] [CrossRef]
- Knop, E.; Zoller, L.; Ryser, R.; Gerpe, C.; Hörler, M.; Fontaine, C. Artificial Light at Night as a New Threat to Pollination. Nature 2017, 548, 206–209. [Google Scholar] [CrossRef]
- Rich, C.; Longcore, T. Ecological Consequences of Artificial Night Lighting; Island Press: Washington, DC, USA, 2006; ISBN 9781559631297. [Google Scholar]
- Eisenbeis, G.; Hänel, A. Light Pollution and the Impact of Artificial Night Lighting on Insects. In Ecology of Cities and Towns: A Comparative Approach; McDonnell, M.J., Hahs, A.K., Breuste, J.H., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 243–263. ISBN 9780511609763. [Google Scholar]
- Perkin, E.K.; Hölker, F.; Tockner, K. The Effects of Artificial Lighting on Adult Aquatic and Terrestrial Insects. Freshw. Biol. 2014, 59, 368–377. [Google Scholar] [CrossRef]
- Davies, T.W.; Bennie, J.; Gaston, K.J. Street Lighting Changes the Composition of Invertebrate Communities. Biol. Lett. 2012, 8, 764–767. [Google Scholar] [CrossRef]
- Sanders, D.; Frago, E.; Kehoe, R.; Patterson, C.; Gaston, K.J. A Meta-Analysis of Biological Impacts of Artificial Light at Night. Nat. Ecol. Evol. 2021, 5, 74–81. [Google Scholar] [CrossRef]
- Sanders, D.; Gaston, K.J. How Ecological Communities Respond to Artificial Light at Night. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2018, 329, 394–400. [Google Scholar] [CrossRef]
- Owens, A.C.S.; Cochard, P.; Durrant, J.; Farnworth, B.; Perkin, E.K.; Seymoure, B. Light Pollution Is a Driver of Insect Declines. Biol. Conserv. 2020, 241, 108259. [Google Scholar] [CrossRef]
- Bachleitner, W.; Kempinger, L.; Wülbeck, C.; Rieger, D.; Helfrich-Förster, C. Moonlight Shifts the Endogenous Clock of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2007, 104, 3538–3543. [Google Scholar] [CrossRef]
- Kempinger, L.; Dittmann, R.; Rieger, D.; Helfrich-Förster, C. The Nocturnal Activity of Fruit Flies Exposed to Artificial Moonlight Is Partly Caused by Direct Light Effects on the Activity Level That Bypass the Endogenous Clock. Chronobiol. Int. 2009, 26, 151–166. [Google Scholar] [CrossRef]
- Kronfeld-Schor, N.; Dominoni, D.; la Iglesia, H.D.; Levy, O.; Herzog, E.D.; Dayan, T.; Helfrich-Forster, C. Chronobiology by Moonlight. Proc. R. Soc. B Biol. Sci. 2013, 1765, 20123088. [Google Scholar] [CrossRef]
- Helfrich-Förster, C. The Circadian Clock in the Brain: A Structural and Functional Comparison between Mammals and Insects. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2004, 190, 601–613. [Google Scholar] [CrossRef]
- Tomioka, K.; Matsumoto, A. The Circadian System in Insects: Cellular, Molecular, and Functional Organization. In Advances in Insect Physiology; Academic Press Inc.: Cambridge, MA, USA, 2019; Volume 56, pp. 73–115. ISBN 9780081028421. [Google Scholar]
- Helfrich-Förster, C. Light Input Pathways to the Circadian Clock of Insects with an Emphasis on the Fruit Fly Drosophila melanogaster. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2020, 206, 259–272. [Google Scholar] [CrossRef]
- Tomioka, K.; Chiba, Y. Post-Embryonic Development of Circadian Rhythm in the Cricket, Gryllus bimaculatus: A Rhythm Reversal. J. Comp. Physiol. A 1982, 147, 299–304. [Google Scholar] [CrossRef]
- Tomioka, K.; Chiba, Y. Circadian Rhythm in the Neurally Isolated Lamina-Medulla-Complex of the Cricket, Gryllus bimaculatus. J. Insect Physiol. 1986, 32, 747–755. [Google Scholar] [CrossRef]
- Abe, Y.; Ushirogawa, H.; Tomioka, K. Circadian Locomotor Rhythms in the Cricket, Gryllodes sigillatus I. Localization of the Pacemaker and the Photoreceptor. Zool. Sci. 1997, 14, 719–727. [Google Scholar] [CrossRef]
- Komada, S.; Kamae, Y.; Koyanagi, M.; Tatewaki, K.; Hassaneen, E.; Saifullah, A.; Yoshii, T.; Terakita, A.; Tomioka, K. Green-Sensitive Opsin Is the Photoreceptor for Photic Entrainment of an Insect Circadian Clock. Zool. Lett. 2015, 1, 11. [Google Scholar] [CrossRef]
- Tomioka, K. Chronobiology of Crickets: A Review. Zool. Sci. 2014, 31, 624–632. [Google Scholar] [CrossRef]
- Loher, W. Temporal Organization of Reproductive Behavior. In Cricket Behavior and Neurobiology; Huber, F., Moore, T.E., Loher, W., Eds.; Cornell University Press: Ithaca, NY, USA; London, UK, 1989; pp. 83–113. [Google Scholar]
- Moriyama, Y.; Kamae, Y.; Uryu, O.; Tomioka, K. Gb’clock Is Expressed in the Optic Lobe and Is Required for the Circadian Clock in the Cricket Gryllus bimaculatus. J. Biol. Rhythm. 2012, 27, 467–477. [Google Scholar] [CrossRef]
- Masayuki, I.; Kenji, T. Temperature Dependency of the Circadian Locomotor Rhythm in the Cricket Gryllus bimaculatus. Zool. Sci. 1993, 10, 597–604. [Google Scholar] [CrossRef]
- Tomioka, K. Analysis of Coupling between Optic Lobe Circadian Pacemakers in the Cricket Gryllus bimaculatus. J. Comp. Physiol. A 1993, 172, 401–408. [Google Scholar] [CrossRef]
- Tokuoka, A.; Itoh, T.Q.; Hori, S.; Uryu, O.; Danbara, Y.; Nose, M.; Bando, T.; Tanimura, T.; Tomioka, K. Cryptochrome Genes Form an Oscillatory Loop Independent of the per/Tim Loop in the Circadian Clockwork of the Cricket Gryllus bimaculatus. Zool. Lett. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Kutaragi, Y.; Tokuoka, A.; Tomiyama, Y.; Nose, M.; Watanabe, T.; Bando, T.; Moriyama, Y.; Tomioka, K. A Novel Photic Entrainment Mechanism for the Circadian Clock in an Insect: Involvement of c-Fos and Cryptochromes. Zool. Lett. 2018, 4, 26. [Google Scholar] [CrossRef]
- Moriyama, Y.; Takeuchi, K.; Shinohara, T.; Miyagawa, K.; Matsuka, M.; Yoshii, T.; Tomioka, K. Timeless Plays an Important Role in Compound Eye-Dependent Photic Entrainment of the Circadian Rhythm in the Cricket Gryllus bimaculatus. Zool. Sci. 2022, 39, 397–405. [Google Scholar] [CrossRef]
- Alaasam, V.J.; Liu, X.; Niu, Y.; Habibian, J.S.; Pieraut, S.; Ferguson, B.S.; Zhang, Y.; Ouyang, J.Q. Effects of Dim Artificial Light at Night on Locomotor Activity, Cardiovascular Physiology, and Circadian Clock Genes in a Diurnal Songbird. Environ. Pollut. 2021, 282, 117036. [Google Scholar] [CrossRef]
- Dominoni, D.M.; de Jong, M.; van Oers, K.; O’Shaughnessy, P.; Blackburn, G.J.; Atema, E.; Mateman, A.C.; D’Amelio, P.B.; Trost, L.; Bellingham, M.; et al. Integrated Molecular and Behavioural Data Reveal Deep Circadian Disruption in Response to Artificial Light at Night in Male Great Tits (Parus major). Sci. Rep. 2022, 12, 1553. [Google Scholar] [CrossRef]
- Touzot, M.; Lefebure, T.; Lengagne, T.; Secondi, J.; Dumet, A.; Konecny-Dupre, L.; Veber, P.; Navratil, V.; Duchamp, C.; Mondy, N. Transcriptome-Wide Deregulation of Gene Expression by Artificial Light at Night in Tadpoles of Common Toads. Sci. Total Environ. 2022, 818, 151734. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, W.; Tzeng, D.T.W.; Owens, A.C.S.; Tang, H.; Wu, C.; Lin, S.; Zhong, S.; Yang, E. Effects of Artificial Light at Night (ALAN) on Gene Expression of Aquatica ficta Fire Fly Larvae. Environ. Pollut. 2021, 281, 116944. [Google Scholar] [CrossRef]
- Uryu, O.; Tomioka, K. Circadian Oscillations Outside the Optic Lobe in the Cricket Gryllus bimaculatus. J. Insect Physiol. 2010, 56, 1284–1290. [Google Scholar] [CrossRef]
- Tomioka, K.; Abdelsalam, S. Circadian Organization in Hemimetabolous Insects. Zool. Sci. 2004, 21, 1153–1162. [Google Scholar] [CrossRef] [Green Version]
- Tomioka, K.; Chiba, Y. Photoperiodic Entrainment of Locomotor Activity in Crickets (Gryllus bimaculatus) Lacking the Optic Lobe Pacemaker. J. Insect Physiol. 1989, 35, 827–835. [Google Scholar] [CrossRef]
- Hege, D.M.; Stanewsky, R.; Hall, J.C.; Giebultowicz, J.M. Rhythmic Expression of a PER-Reporter in the Malpighian Tubules of Decapitated Drosophila: Evidence for a Brain-Independent Circadian Clock. J. Biol. Rhythm. 1997, 12, 300–308. [Google Scholar] [CrossRef]
- Sadik, N.; Cruz, L.; Gurtner, A.; Rodosthenous, R.S.; Dusoswa, S.A.; Ziegler, O.; Van Solinge, T.S.; Wei, Z.; Salvador-Garicano, A.M.; Gyorgy, B.; et al. Extracellular RNAs: A New Awareness of Old Perspectives. In Extracellular RNA. Methods in Molecular Biology; Patel, T., Ed.; Springer: New York, NY, USA, 2018; Volume 1740, pp. 1–15. ISBN 978-1-4939-7652-2. [Google Scholar]
- Adewoye, A.B.; Kyriacou, C.P.; Tauber, E. Identification and Functional Analysis of Early Gene Expression Induced by Circadian Light-Resetting in Drosophila. BMC Genom. 2015, 16, 570. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Parikh, V.; Itsukaichi, T.; Bae, K.; Edery, I. Resetting the Drosophila Clock by Photic Regulation of PER and a PER-TIM Complex. Science 1996, 271, 1740–1744. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Lindenberg, A.; Albert, S.; Grübel, K.; Spaethe, J.; Rössler, W.; Groh, C. Age-Related and Light-Induced Plasticity in Opsin Gene Expression and in Primary and Secondary Visual Centers of the Nectar-Feeding Ant Camponotus rufipes. Dev. Neurobiol. 2016, 76, 1041–1057. [Google Scholar] [CrossRef]
- Yan, S.; Zhu, J.; Zhu, W.; Zhang, X.; Li, Z.; Liu, X.; Zhang, Q. The Expression of Three Opsin Genes from the Compound Eye of Helicoverpa armigera (Lepidoptera: Noctuidae) Is Regulated by a Circadian Clock, Light Conditions and Nutritional Status. PLoS ONE 2014, 9, e111683. [Google Scholar] [CrossRef]
- Tomioka, K. Light and Serotonin Phase-Shift the Circadian Clock in the Cricket Optic Lobe in Vitro. J. Comp. Physiol.—A Sens. Neural Behav. Physiol. 1999, 185, 437–444. [Google Scholar] [CrossRef]
- Okada, Y.; Tomioka, K.; Chiba, Y. Circadian Phase-Response Curves for Light in Nymphal and Adult Crickets, Gryllus bimaculatus. J. Insect Physiol. 1991, 37, 583–590. [Google Scholar] [CrossRef]
- Hölker, F.; Bolliger, J.; Davies, T.W.; Giavi, S.; Jechow, A.; Kalinkat, G.; Longcore, T.; Spoelstra, K.; Tidau, S.; Visser, M.E.; et al. 11 Pressing Research Questions on How Light Pollution Affects Biodiversity. Front. Ecol. Evol. 2021, 9, 767177. [Google Scholar] [CrossRef]
- Grubisic, M.; van Grunsven, R.H. Artificial Light at Night Disrupts Species Interactions and Changes Insect Communities. Curr. Opin. Insect Sci. 2021, 47, 136–141. [Google Scholar] [CrossRef]
- Shortall, R.C.; Moore, A.; Smith, E.; Hall, J.M.; Woiwod, P.I.; Harrington, R. Long-Term Changes in the Abundance of Flying Insects. Insect Conserv. Divers. 2009, 2, 251–260. [Google Scholar] [CrossRef]
- Hallmann, C.A.; Sorg, M.; Jongejans, E.; Siepel, H.; Hofland, N.; Schwan, H.; Stenmans, W.; Müller, A.; Sumser, H.; Hörren, T.; et al. More than 75 Percent Decline over 27 Years in Total Flying Insect Biomass in Protected Areas. PLoS ONE 2017, 12, e0185809. [Google Scholar] [CrossRef]
- Altermatt, F.; Ebert, D. Reduced Flight-to-Light Behaviour of Moth Populations Exposed to Long-Term Urban Light Pollution. Biol. Lett. 2016, 12, 2016–2019. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate QPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Pinheiro, J.B. Nlme: Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-157; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Levy, K.; Fishman, B.; Barnea, A.; Ayali, A.; Tauber, E. Supplementary material and data from: Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus. Figshare 2022. [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levy, K.; Fishman, B.; Barnea, A.; Ayali, A.; Tauber, E. Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus. Int. J. Mol. Sci. 2022, 23, 11358. https://doi.org/10.3390/ijms231911358
Levy K, Fishman B, Barnea A, Ayali A, Tauber E. Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus. International Journal of Molecular Sciences. 2022; 23(19):11358. https://doi.org/10.3390/ijms231911358
Chicago/Turabian StyleLevy, Keren, Bettina Fishman, Anat Barnea, Amir Ayali, and Eran Tauber. 2022. "Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus" International Journal of Molecular Sciences 23, no. 19: 11358. https://doi.org/10.3390/ijms231911358
APA StyleLevy, K., Fishman, B., Barnea, A., Ayali, A., & Tauber, E. (2022). Transcriptional Response of Circadian Clock Genes to an ‘Artificial Light at Night’ Pulse in the Cricket Gryllus bimaculatus. International Journal of Molecular Sciences, 23(19), 11358. https://doi.org/10.3390/ijms231911358








