Enhancing Annotation Efficiency with Machine Learning: Automated Partitioning of a Lung Ultrasound Dataset by View
Abstract
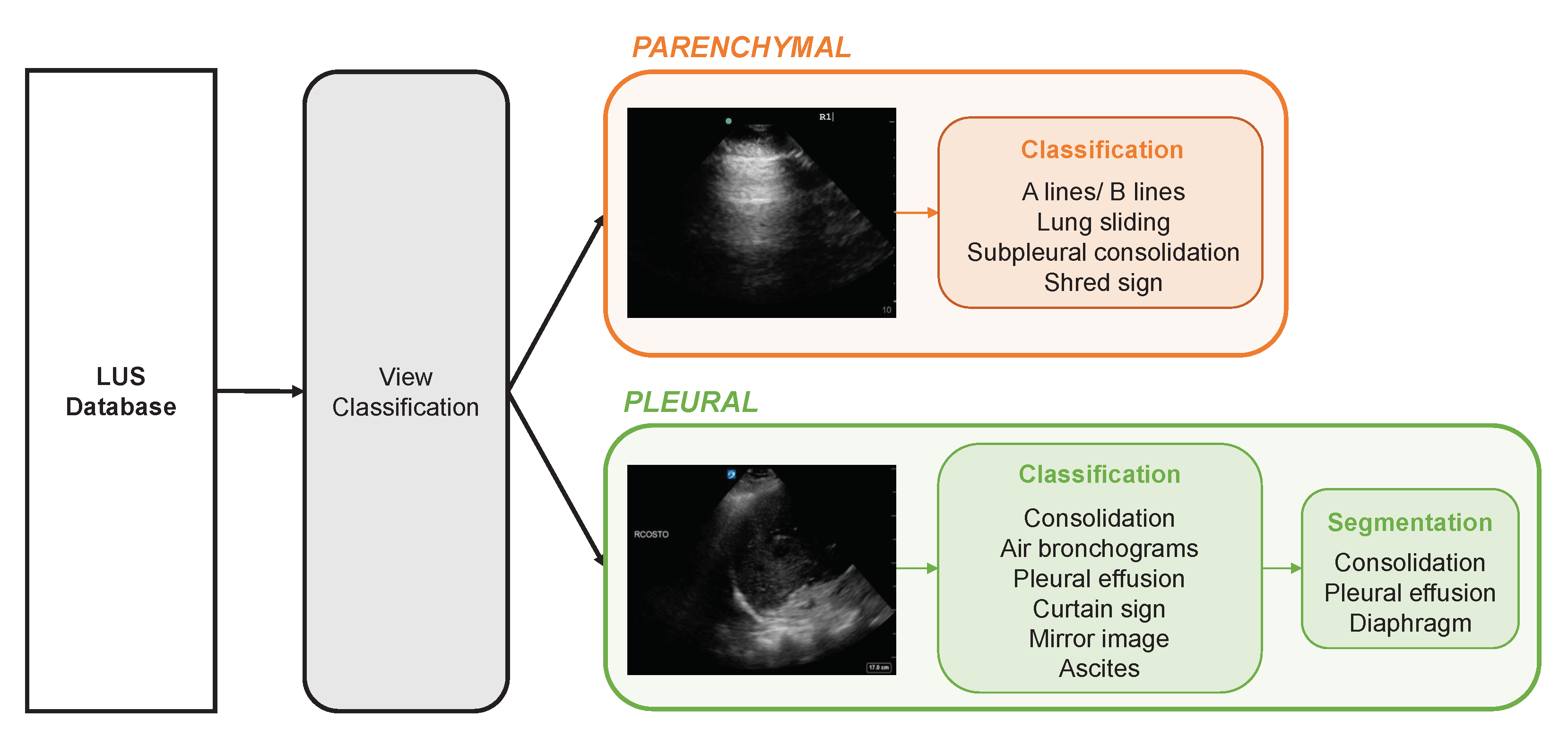
:1. Introduction
2. Materials and Methods
2.1. Data Curation and Annotation
2.2. View (Parenchymal vs. Pleural) Classifier
2.2.1. Clip-Level Data
2.2.2. Frame-Based Data
2.2.3. Dataset Pre-Processing
2.2.4. Model Architecture
2.2.5. Clip Predictions
2.2.6. Validation Strategy
2.3. Automating the View Annotation Task
2.3.1. Partitioning a LUS Dataset by View
2.3.2. The Annotation Task
2.3.3. Statistical Analysis
3. Results
3.1. View Classifier Validation
3.1.1. Frame-Based Performance
3.1.2. Clip-Based Performance
3.1.3. Frame-Based Explainability
3.1.4. Clip-Based Explainability
3.2. Automating the View Annotation Task
3.2.1. Performance on an Auto-Partitioned Dataset
3.2.2. Annotation Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area under the receiver operating curve |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| LUS | Lung ultrasound |
| ReLU | Rectified linear activation function |
| ROC | Receiver operator curve |
Appendix A. Alternative Model Architectures
| Base | Accuracy | AUC | Parameters | ||
|---|---|---|---|---|---|
| Train | Validation | Train | Validation | ||
| Inceptionv3 [31] | 2.18 × 107 | ||||
| ResNet14v2 [32] | 1.45 × 106 | ||||
| ResNet50v2 [32] | 2.36 × 107 | ||||
| EfficientNetB0 [26] | 4.21 × 106 | ||||
| EfficientNetB7 [26] | 6.44 × 107 | ||||
Appendix B. Training Details
Appendix C. Explainability


References
- Long, L.; Zhao, H.T.; Zhang, Z.Y.; Wang, G.Y.; Zhao, H.L. Lung ultrasound for the diagnosis of pneumonia in adults: A meta-analysis. Medicine 2017, 96, e5713. [Google Scholar] [CrossRef] [PubMed]
- Ma, O.J.; Mateer, J.R. Trauma ultrasound examination versus chest radiography in the detection of hemothorax. Ann. Emerg. Med. 1997, 29, 312–316. [Google Scholar] [CrossRef]
- Lichenstein, D.; Meziere, G. Relevance of lung ultrasound in the diagnosis of acute respiratory failure. Chest 2008, 134, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Chiumello, D.; Umbrello, M.; Papa, G.F.S.; Angileri, A.; Gurgitano, M.; Formenti, P.; Coppola, S.; Froio, S.; Cammaroto, A.; Carrafiello, G. Global and regional diagnostic accuracy of lung ultrasound compared to CT in patients with acute respiratory distress syndrome. Crit. Care Med. 2019, 47, 1599–1606. [Google Scholar] [CrossRef] [PubMed]
- Nazerian, P.; Volpicelli, G.; Vanni, S.; Gigli, C.; Betti, L.; Bartolucci, M.; Zanobetti, M.; Ermini, F.R.; Iannello, C.; Grifoni, S. Accuracy of lung ultrasound for the diagnosis of consolidations when compared to chest computed tomography. Am. J. Emerg. Med. 2015, 33, 620–625. [Google Scholar] [CrossRef]
- Stassen, J.; Bax, J.J. How to do lung ultrasound. Eur. Heart J.-Cardiovasc. Imaging 2022, 23, 447–449. [Google Scholar] [CrossRef]
- Ruaro, B.; Baratella, E.; Confalonieri, P.; Confalonieri, M.; Vassallo, F.G.; Wade, B.; Geri, P.; Pozzan, R.; Caforio, G.; Marrocchio, C.; et al. High-Resolution Computed Tomography and Lung Ultrasound in Patients with Systemic Sclerosis: Which One to Choose? Diagnostics 2021, 11, 2293. [Google Scholar] [CrossRef]
- Ginsburg, A.S.; Lenahan, J.L.; Jehan, F.; Bila, R.; Lamorte, A.; Hwang, J.; Madrid, L.; Nisar, M.I.; Vitorino, P.; Kanth, N.; et al. Performance of lung ultrasound in the diagnosis of pediatric pneumonia in Mozambique and Pakistan. Pediatr. Pulmonol. 2021, 56, 551–560. [Google Scholar] [CrossRef]
- Alsup, C.; Lipman, G.S.; Pomeranz, D.; Huang, R.W.; Burns, P.; Juul, N.; Phillips, C.; Jurkiewicz, C.; Cheffers, M.; Evans, K.; et al. Interstitial pulmonary edema assessed by lung ultrasound on ascent to high altitude and slight association with acute mountain sickness: A prospective observational study. High Alt. Med. Biol. 2019, 20, 150–156. [Google Scholar] [CrossRef]
- Lichtenstein, D.A. Lung ultrasound in the critically ill. Ann. Intensive Care 2014, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lichtenstein, D.A. BLUE-protocol and FALLS-protocol: Two applications of lung ultrasound in the critically ill. Chest 2015, 147, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Gargani, L.; Volpicelli, G. How I do it: Lung ultrasound. Cardiovasc. Ultrasound 2014, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, D.A.; Mezière, G.A.; Lagoueyte, J.F.; Biderman, P.; Goldstein, I.; Gepner, A. A-lines and B-lines: Lung ultrasound as a bedside tool for predicting pulmonary artery occlusion pressure in the critically ill. Chest 2009, 136, 1014–1020. [Google Scholar] [CrossRef]
- Lichtenstein, D.; Meziere, G.; Biderman, P.; Gepner, A.; Barre, O. The comet-tail artifact: An ultrasound sign of alveolar-interstitial syndrome. Am. J. Respir. Crit. Care Med. 1997, 156, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.C.Y. The Curtain Sign in Lung Ultrasound. J. Med. Ultrasound 2017, 25, 101–104. [Google Scholar] [CrossRef]
- Eisen, L.A.; Leung, S.; Gallagher, A.E.; Kvetan, V. Barriers to ultrasound training in critical care medicine fellowships: A survey of program directors. Crit. Care Med. 2010, 38, 1978–1983. [Google Scholar] [CrossRef]
- Wong, J.; Montague, S.; Wallace, P.; Negishi, K.; Liteplo, A.; Ringrose, J.; Dversdal, R.; Buchanan, B.; Desy, J.; Ma, I.W. Barriers to learning and using point-of-care ultrasound: A survey of practicing internists in six North American institutions. Ultrasound J. 2020, 12, 1–7. [Google Scholar] [CrossRef]
- Azar, A.T.; El-Metwally, S.M. Decision tree classifiers for automated medical diagnosis. Neural Comput. Appl. 2013, 23, 2387–2403. [Google Scholar] [CrossRef]
- Wang, F.; Kaushal, R.; Khullar, D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann. Intern. Med. 2020, 172, 59–60. [Google Scholar] [CrossRef]
- Rahimi, S.; Oktay, O.; Alvarez-Valle, J.; Bharadwaj, S. Addressing the Exorbitant Cost of Labeling Medical Images with Active Learning. In Proceedings of the International Conference on Machine Learning in Medical Imaging and Analysis, Singapore, 24–25 May 2021. [Google Scholar]
- Zhang, L.; Tong, Y.; Ji, Q. Active Image Labeling and Its Application to Facial Action Labeling. In Proceedings of the Computer Vision—ECCV 2008, Marseille, France, 12–18 October 2008; Forsyth, D., Torr, P., Zisserman, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 706–719. [Google Scholar]
- Gu, Y.; Leroy, G. Mechanisms for Automatic Training Data Labeling for Machine Learning. In Proceedings of the ICIS, Munich, Germany, 15–18 December 2019. [Google Scholar]
- Gong, T.; Li, S.; Wang, J.; Tan, C.L.; Pang, B.C.; Lim, C.C.T.; Lee, C.K.; Tian, Q.; Zhang, Z. Automatic labeling and classification of brain CT images. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 1581–1584. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019. [Google Scholar] [CrossRef]
- Smit, A.; Jain, S.; Rajpurkar, P.; Pareek, A.; Ng, A.; Lungren, M.P. CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. In Proceedings of the EMNLP, Online, 16–20 November 2020. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning. PMLR, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision And Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Arntfield, R.; Wu, D.; Tschirhart, J.; VanBerlo, B.; Ford, A.; Ho, J.; McCauley, J.; Wu, B.; Deglint, J.; Chaudhary, R.; et al. Automation of Lung Ultrasound Interpretation via Deep Learning for the Classification of Normal versus Abnormal Lung Parenchyma: A Multicenter Study. Diagnostics 2021, 11, 2049. [Google Scholar] [CrossRef]
- Heinisch, O.; Cochran, W.G. Sampling Techniques, 2. Aufl. John Wiley and Sons, New York, London 1963. Preis s. Biom. Z. 1965, 7, 203. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized Gradient-based Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Louppe, G.; Kumar, M. Bayesian Optimization with Skopt. 2016. Available online: https://scikit-optimize.github.io/stable/auto_examples/bayesian-optimization.html (accessed on 25 July 2022).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]







| Training Data | Holdout Data | |||
|---|---|---|---|---|
| Clip label | Parenchymal | Pleural | Parenchymal | Pleural |
| Patients | 611 | 342 | 441 | 466 |
| Number of clips | 1454 | 1454 | 457 | 488 |
| Frames | 369,832 | 330,191 | 107,205 | 100,616 |
| Average clips/patient | 2.38 | 4.25 | 1.04 | 1.05 |
| Class-patient overlap | 303/650 | 32/875 | ||
| Age (std) | 64.0 (17.2) | 64.5 (16.2) | 64.1 (18.0) | 64.4 (17.4) |
| Sex | Female: 238 (39%) | Female: 134 (39%) | Female: 156 (35%) | Female: 205 (44%) |
| Male: 347 (57%) | Male: 193 (56%) | Male: 269 (61%) | Male: 235 (50%) | |
| Unknown: 26 (4%) | Unknown: 15 (4%) | Unknown: 16 (4%) | Unknown: 26 (6%) | |
| Control Data | Auto-Partitioned Data | |||
|---|---|---|---|---|
| Clip Label | Parenchymal | Pleural | Parenchymal | Pleural |
| Patients | 339 | 371 | 660 | 34 |
| Number of clips | 351 | 383 | 701 | 35 |
| Average clips per patient | 1.04 | 1.03 | 1.06 | 1.03 |
| Patient overlap across classes | 25/685 | 5/689 | ||
| Mean age (std) | 63.7 (18.1) | 64.0 (16.1) | 64.0 (16.6) | 63.7 (18.3) |
| Sex | Female: 117 (35%) | Female: 156 (42%) | Female: 259 (39%) | Female: 12 (35%) |
| Male: 193 (57%) | Male: 201 (54%) | Male: 374 (57%) | Male: 21 (62%) | |
| Unknown: 29 (9%) | Unknown: 14 (4%) | Unknown: 27 (4%) | Unknown: 1 (3%) | |
| Accuracy | Negative Predictive Value | Positive Predictive Value | AUC | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Fold | Frames | Clips | Frames | Clips | Frames | Clips | Frames |
| Training | 1 | |||||||
| 2 | ||||||||
| 3 | ||||||||
| 4 | ||||||||
| 5 | ||||||||
| 6 | ||||||||
| 7 | ||||||||
| 8 | ||||||||
| 9 | ||||||||
| 10 | ||||||||
| Mean | ||||||||
| (STD) | ||||||||
| Holdout | − | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
VanBerlo, B.; Smith, D.; Tschirhart, J.; VanBerlo, B.; Wu, D.; Ford, A.; McCauley, J.; Wu, B.; Chaudhary, R.; Dave, C.; et al. Enhancing Annotation Efficiency with Machine Learning: Automated Partitioning of a Lung Ultrasound Dataset by View. Diagnostics 2022, 12, 2351. https://doi.org/10.3390/diagnostics12102351
VanBerlo B, Smith D, Tschirhart J, VanBerlo B, Wu D, Ford A, McCauley J, Wu B, Chaudhary R, Dave C, et al. Enhancing Annotation Efficiency with Machine Learning: Automated Partitioning of a Lung Ultrasound Dataset by View. Diagnostics. 2022; 12(10):2351. https://doi.org/10.3390/diagnostics12102351
Chicago/Turabian StyleVanBerlo, Bennett, Delaney Smith, Jared Tschirhart, Blake VanBerlo, Derek Wu, Alex Ford, Joseph McCauley, Benjamin Wu, Rushil Chaudhary, Chintan Dave, and et al. 2022. "Enhancing Annotation Efficiency with Machine Learning: Automated Partitioning of a Lung Ultrasound Dataset by View" Diagnostics 12, no. 10: 2351. https://doi.org/10.3390/diagnostics12102351
APA StyleVanBerlo, B., Smith, D., Tschirhart, J., VanBerlo, B., Wu, D., Ford, A., McCauley, J., Wu, B., Chaudhary, R., Dave, C., Ho, J., Deglint, J., Li, B., & Arntfield, R. (2022). Enhancing Annotation Efficiency with Machine Learning: Automated Partitioning of a Lung Ultrasound Dataset by View. Diagnostics, 12(10), 2351. https://doi.org/10.3390/diagnostics12102351






