Knockdown of Rab9 Recovers Defective Morphological Differentiation Induced by Chemical ER Stress Inducer or PMD-Associated PLP1 Mutant Protein in FBD-102b Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture and Differentiation
- (1)
- The FBD-102b cell line (kindly provided by Dr. Yasuhiro Tomo-oka [Riken, Saitama, Japan/Tokyo University of Science, Chiba, Japan]), a mouse oligodendroglial precursor cell line, was cultured in Dulbecco’s modified Eagle medium (DMEM)/Ham’s F-12 nutrient mixed medium (Nacalai Tesque, Kyoto, Japan; Fujifilm, Tokyo, Japan) containing 10% heat-inactivated fetal bovine serum (FBS) and PenStrep mixed antibiotics (Thermo Fisher Scientific, Waltham, MA, USA).
- (2)
- The cells were seeded at 2 × 106 per 10 cm diameter of Nunc cell and tissue culture dishes (Thermo Fisher Scientific, Waltham, MA, USA) and grown until 8 × 106 per 10 cm diameter of culture dishes.
- (3)
- The dishes were cultured in 5% carbon dioxide at 37 °C until 20 passages.
- (4)
- To induce differentiation, the cells were plated at 4 × 106 per 10 cm diameter polylysine (Nacalai Tesque)-coated cell culture dishes in culture medium containing 1% FBS and maintained in 5% carbon dioxide at 37 °C.
- (5)
- The cells were allowed to be differentiated for several days.
- (6)
- The cells displaying secondary branches from primary ones or exhibiting myelin membrane-like widespread membranes (cells large enough to contain a circle with a diameter exceeding 0.03 mm) were considered to have differentiated phenotypes [39].
- (7)
- Cell morphologies were captured using a microscopic system equipped with i-NTER LENS (Micronet, Saitama, Japan) and i-NTER SHOT ver.2 (Micronet).
- (8)
- The images in the figures are representative of multiple captures and were analyzed with Image J software (ver. Java 8, https://imagej.nih.gov/ on 1 July 2024).
2.3. Reverse Transcription and Polymerase Chain Reaction (RT-PCR) and Routine PCR
- (1)
- Total cellular RNA was prepared from cells grown on a 10 cm diameter culture dish using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions.
- (2)
- Single-strand cDNAs were generated from their RNAs (1 mg of RNAs per one reaction) using the PrimeScript RT Master Mix kit (Takara Bio, Kyoto, Japan) in accordance with the manufacturer’s instructions.
- (3)
- PCR amplification was performed using 1:40 of the RT product with Gflex DNA polymerase (Takara Bio) for 35 cycles.
- (4)
- Each cycle consisted of a denaturation reaction at 98 °C (0.2 min), an annealing reaction at 56 to 65 °C (0.25 min), depending on the annealing temperature, and an extension reaction at 68 °C (0.5 min).
- (5)
- The resultant PCR products were loaded onto premade agarose gels (Nacalai Tesque; Fujifilm).
2.4. siRNA and/or Plasmid Transfection
- (1)
- The cells were transfected with the respective siRNAs using the ScreenFect siRNA transfection kit (Fujifilm) in accordance with the manufacturer’s instructions. Alternatively, the cells were cotransfected with the respective siRNAs and plasmids using the ScreenFect A transfection kit (Fujifilm).
- (2)
- The medium was replaced with fresh medium 4 h after transfection.
- (3)
- The cells were treated with chemicals or their vehicle controls at 24 h after transfection.
- (4)
- The cells were generally used for biochemical experiments more than 48 h after transfection.
2.5. Cell Lysis and Polyacrylamide Gel Electrophoresis
- (1)
- The cells were lysed in cell and tissue extraction buffer (50 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 3 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/mL leupeptin, 1 mM EDTA, 1 mM Na3VO4, and 10 mM NaF) with a mild detergent (0.5% NP-40) [39]. Generally, 0.1 mL of extraction buffer was used per 1 × 106 cells.
- (2)
- For denaturing conditions, cell lysates were mixed with premade sample buffer (Fujifilm) in accordance with the manufacturer’s instructions.
- (3)
- Denatured samples (0.025 mg of proteins per lane) were separated on premade sodium dodecyl sulfate-polyacrylamide gel (Nacalai Tesque).
2.6. Blotting and Detection of Immunoreactive Bands
- (1)
- Electrophoretically separated proteins were transferred to a polyvinylidene fluoride membrane (Fujifilm).
- (2)
- The membrane was blocked using the Skim Blocker kit (Fujifilm).
- (3)
- The blocked membrane was immunoblotted with primary antibodies.
- (4)
- This was followed by incubation with peroxidase enzyme-conjugated secondary antibodies.
- (5)
- The bands were chemically reacted with the Chemi-Lumi One Super or Ultra kit (Nacalai Tesque).
- (6)
- Peroxidase-reactive bands were detected on X-ray film (Fujifilm) at the same exposure time (within 0.5 h) for each sample being compared.
- (7)
- The reactive bands on the film were captured using the CanoScan LiDE 400 (Canon, Tokyo, Japan) and CanoScan LiDE 400 Scanner Driver Ver.1.01.
- (8)
- Multiple sets of experiments were conducted for immunoblotting studies, and the quantification of immunoreactive bands was performed using Image J software (ver. Java 8), with the immunoreactive band of another sample set as the 100% reference.
2.7. Statistical Analyses
- (1)
- For all analyses, the investigator was blinded to the sample conditions. The values are expressed as means ± standard deviation (SD) from separate experiments.
- (2)
- Intergroup comparisons were performed using the unpaired t-test with Student’s correction or Welch’s correction, conducted with Excel software (ver. 2024, Microsoft, Redmond, WA, USA).
- (3)
- Differences were considered significant at p < 0.05.
2.8. Ethics Statement
3. Results
3.1. Rab9 Negatively Regulates Cell Morphogenesis
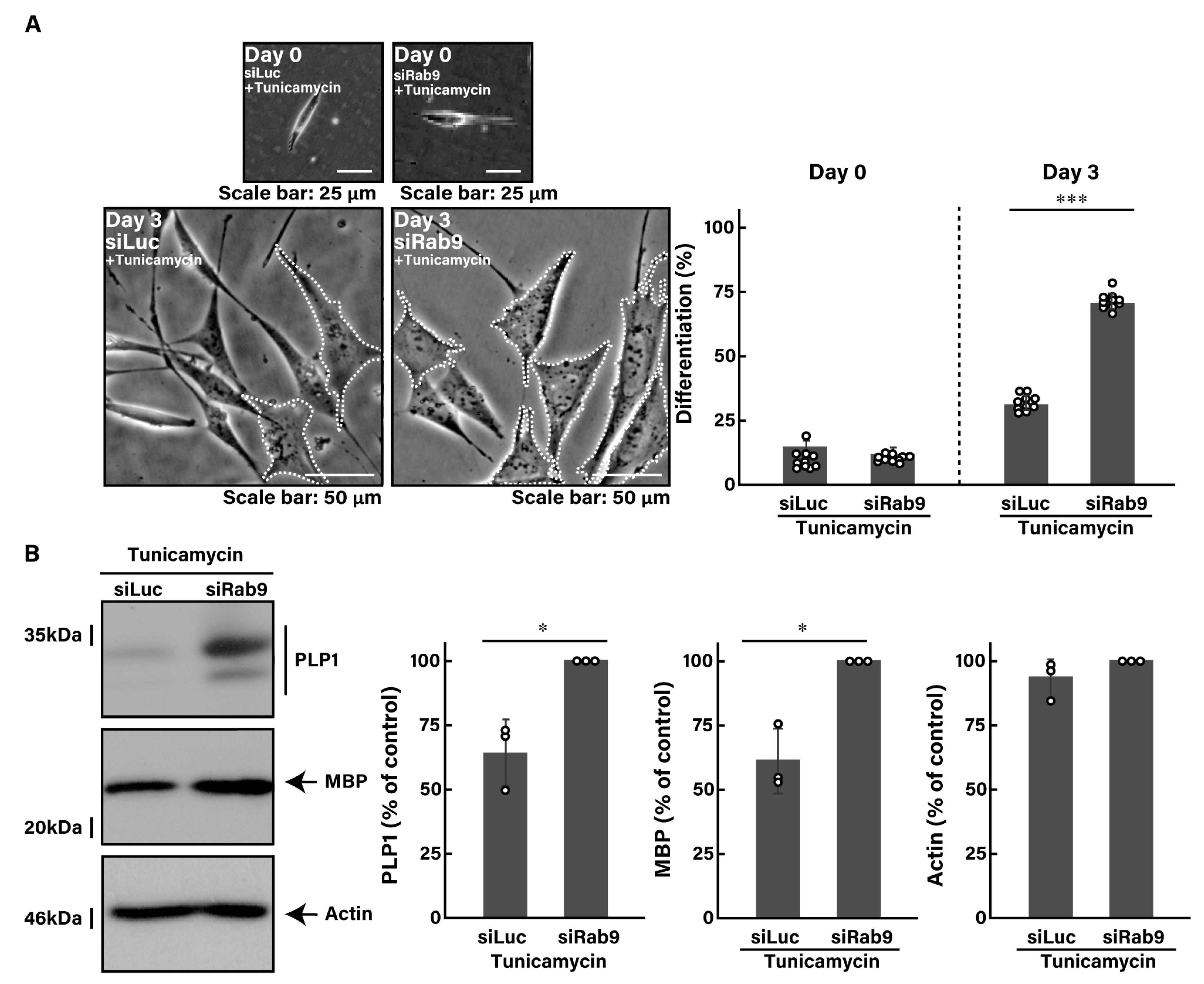
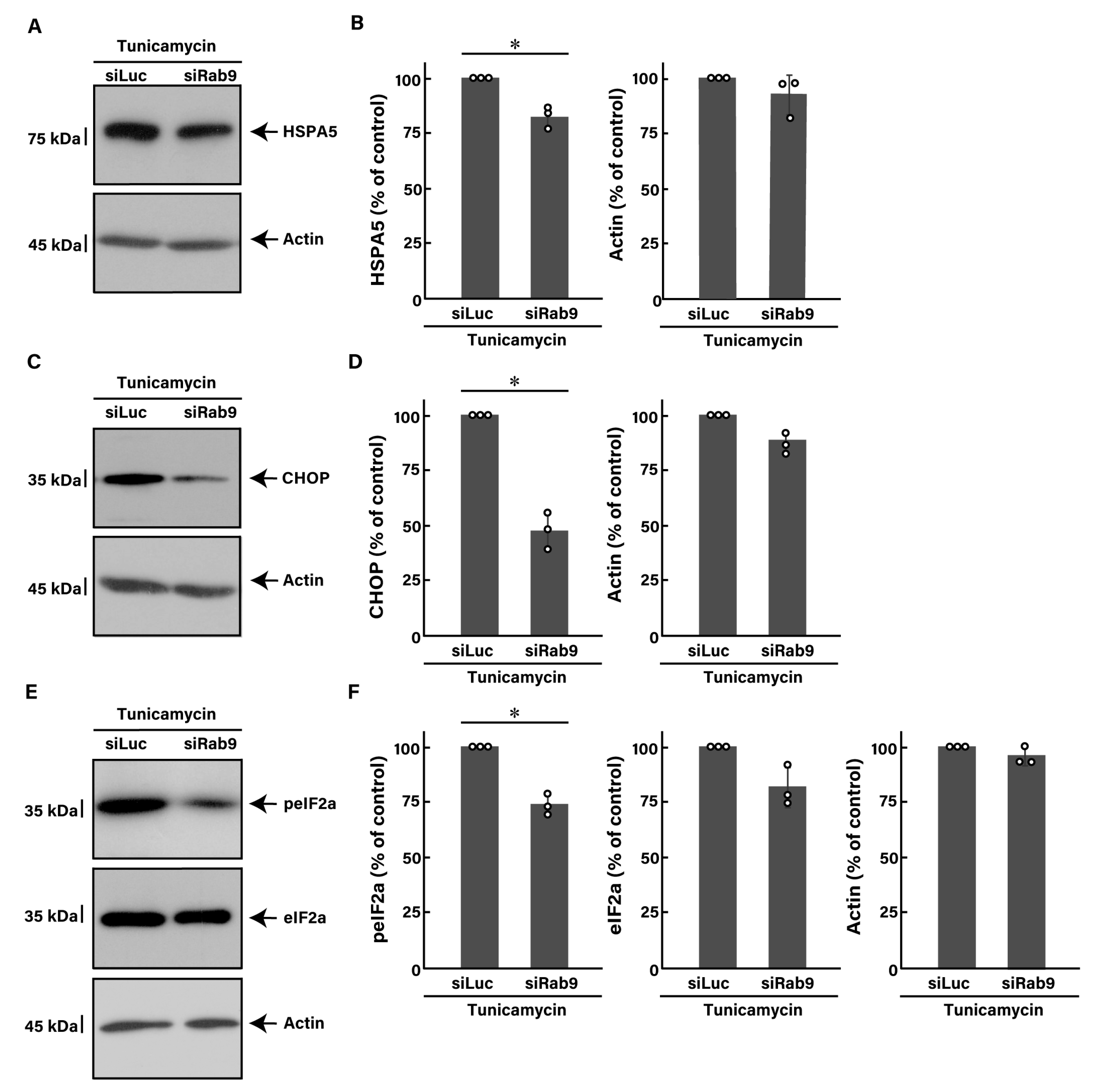
3.2. Knockdown of Rab9 Leads to Recovering Tunicamycin-Induced Cell Phenotypes
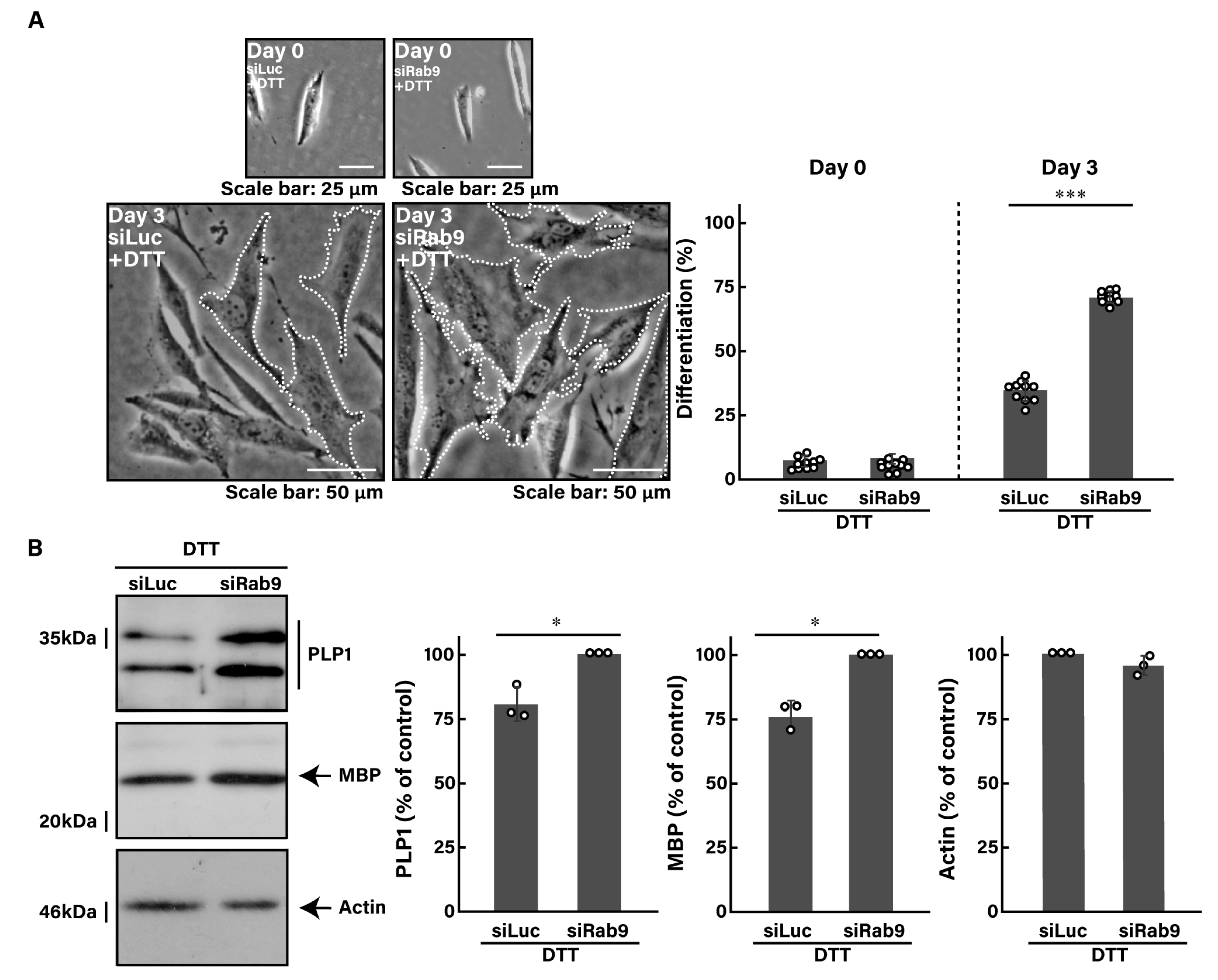
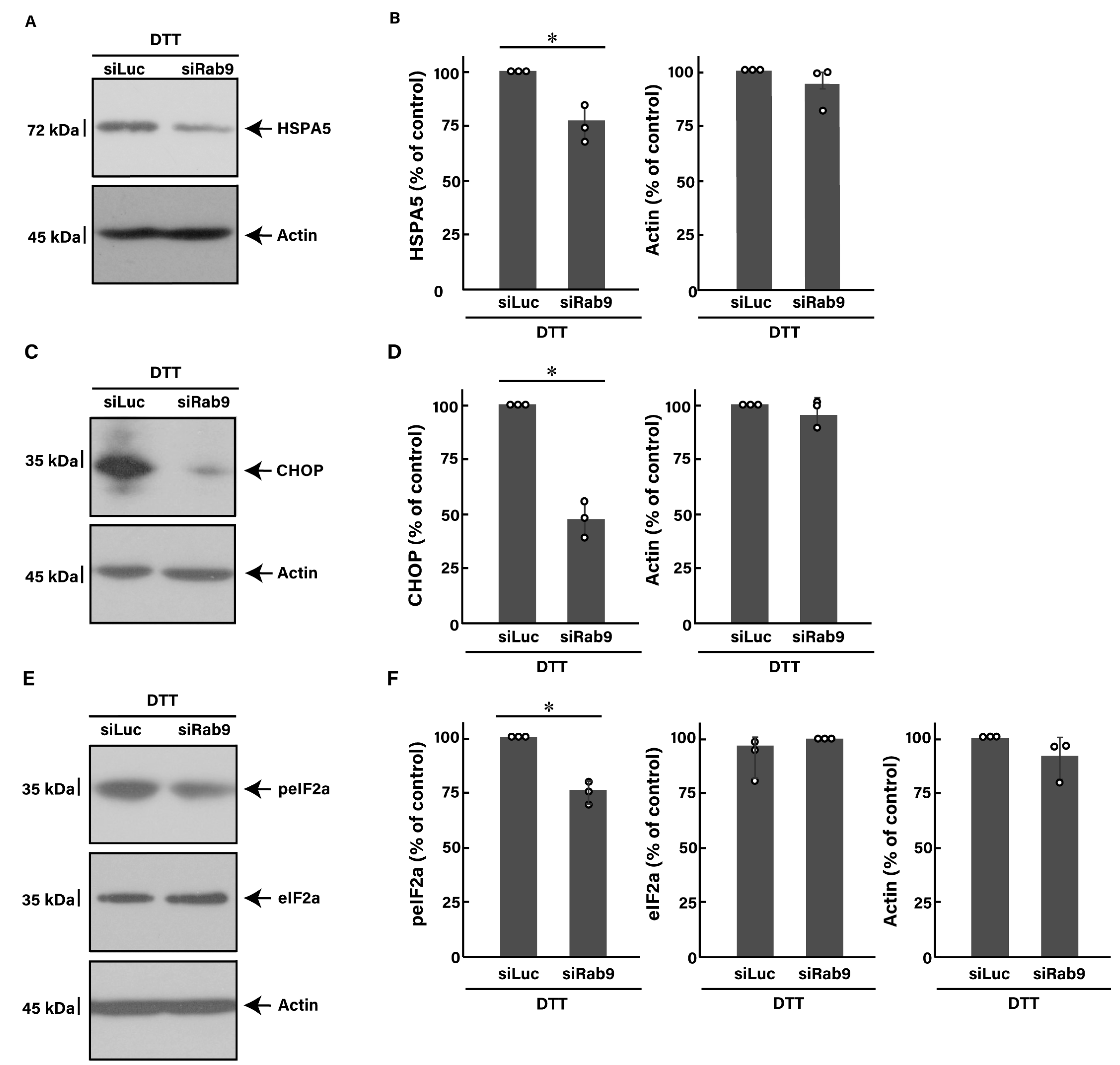
3.3. Knockdown of Rab9 Leads to Recovering Dithiothreitol (DTT)-Induced Cell Phenotypes
3.4. Knockdown of Rab9 Leads to Recovering Cell Phenotypes Induced by PLP1 with the A243V Mutation
3.5. Effects of ER Stress Inducers and Rab9 Knockdown on Akt Signaling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simons, N.; Pham-Dinh, D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001, 81, 871–927. [Google Scholar]
- Simons, M.; Lyons, D.A. Axonal selection and myelin sheath generation in the central nervous system. Curr. Opin. Cell Biol. 2013, 25, 512–519. [Google Scholar] [CrossRef]
- Saab, A.S.; Nave, K.A. Myelin dynamics: Protecting and shaping neuronal functions. Curr. Opin. Neurobiol. 2017, 47, 104–112. [Google Scholar] [CrossRef]
- Abu-Rub, M.; Miller, R.H. Emerging cellular and molecular strategies for enhancing central nervous system (CNS) remyelination. Brain Sci. 2018, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, S.; Gritti, L.; Crooks, D.; Dombrowski, Y. Oligodendrocytes in development, myelin generation and beyond. Cells 2019, 8, 1424. [Google Scholar] [CrossRef] [PubMed]
- Stadelmann, C.; Timmler, S.; Barrantes-Freer, A.; Saab, M. Myelin in the central nervous system: Structure, function, and pathology. Physiol. Rev. 2019, 99, 1381–1431. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Guan, T.; Guo, Y.; Kong, J. The initial myelination in the central nervous system. ASN Neuro 2023, 15, 17590914231163039. [Google Scholar] [CrossRef]
- Barnes-Vélez, J.A.; Aksoy Yasar, F.B.; Hu, J. Myelin lipid metabolism and its role in myelination and myelin maintenance. Innovation 2023, 4, 100360. [Google Scholar] [CrossRef]
- Dobson, R.; Giovannoni, G. Multiple sclerosis-a review Eur. J. Neurol. 2019, 26, 27–40. [Google Scholar]
- Shirai, R.; Yamauchi, J. New insights into risk genes and their candidates in multiple sclerosis. Neurol. Int. 2022, 15, 24–39. [Google Scholar] [CrossRef]
- Garbern, J.; Cambi, F.; Shy, M.; Kamholz, J. The molecular pathogenesis of Pelizaeus-Merzbacher disease. Arch. Neurol. 1999, 56, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Wolf, W.; Vanderver, A.; Bernard, G.; Wolf, I.; Dreha-Kulczewksi, F.; Deoni, L.; Bertini, E.; Kohlschütter, A.; Richardson, W.; Ffrench-Constant, C.; et al. Hypomyelinating leukodystrophies: Translational research progress and prospects. Ann. Neurol. 2014, 76, 5–19. [Google Scholar]
- Inoue, K. Pelizaeus-Merzbacher disease: Molecular and cellular pathologies and associated phenotypes. Adv. Exp. Med. Biol. 2019, 1190, 201–216. [Google Scholar] [PubMed]
- Wolf, N.I.; Ffrench-Constant, C.; van der Knaap, M.S. Hypomyelinating leukodystrophies-unravelling myelin biology. Nat. Rev. Neurol. 2021, 17, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Torii, T.; Yamauchi, J. Molecular pathogenic mechanisms of hypomyelinating leukodystrophies (HLDs). Neurol. Int. 2023, 15, 1155–1173. [Google Scholar] [CrossRef]
- Dhaunchak, A.S.; Colman, D.R.; Nave, K.A. Misalignment of PLP/DM20 transmembrane domains determines protein misfolding in Pelizaeus-Merzbacher disease. J. Neurosci. 2011, 31, 14961–14971. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef]
- Langemeyer, L.; Fröhlich, F.; Ungermann, C. Rab GTPase function in endosome and lysosome biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Guadagno, N.A.; Progida, C. Rab GTPases: Switching to human diseases. Cells 2019, 8, 909. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, S.; Kümmel, D. Insights into the role of the membranes in Rab GTPase regulation. Curr. Opin. Cell Biol. 2023, 83, 102177. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, D.; Soldati, T.; Riederer, M.A.; Goda, Y.; Zerial, M.; Pfeffer, S.R. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993, 12, 677–682. [Google Scholar] [CrossRef]
- Barbero, P.; Bittova, L.; Pfeffer, S.R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002, 156, 511–518. [Google Scholar] [CrossRef]
- Ganley, I.G.; Carroll, K.; Bittova, L.; Pfeffer, S. Rab9 GTPase regulates late endosome size and requires effector interaction for its stability. Mol. Biol. Cell 2004, 15, 5420–5430. [Google Scholar] [CrossRef]
- Ao, X.; Zou, L.; Wu, Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014, 21, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Kucera, A.; Bakke, O.; Progida, C. The multiple roles of Rab9 in the endolysosomal system. Commun. Integr. Biol. 2016, 9, e1204498. [Google Scholar] [CrossRef]
- Kucera, A.; Borg Distefano, M.; Berg-Larsen, A.; Skjeldal, F.; Repnik, U.; Bakke, O.; Progida, C. Spatiotemporal resolution of Rab9 and CI-MPR dynamics in the endocytic pathway. Traffic 2016, 17, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Oakes, S.A.; Papa, F.R. The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 2015, 10, 173–194. [Google Scholar] [CrossRef]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, S.J.; Chambers, J.E.; Ron, D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 2022, 21, 115–140. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Harding, H.P.; Ron, D.; Popko, B. Endoplasmic reticulum stress modulates the response of myelinating oligodendrocytes to the immune cytokine interferon-gamma. J. Cell Biol. 2005, 169, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Potter, E. CHOP and the endoplasmic reticulum stress response in myelinating glia. Bone 2008, 23, 1–7. [Google Scholar]
- D’Antonio, M.; Feltri, M.L.; Wrabetz, L. Myelin under stress. J. Neurosci. Res. 2009, 87, 3241. [Google Scholar] [CrossRef]
- Tong, B.C.K. Endoplasmic reticulum stress and the unfolded protein response in disorders of myelinating glia. Physiol. Behav. 2017, 176, 139–148. [Google Scholar]
- Bouverat, B.P.; Krueger, W.H.; Coetzee, T.; Bansal, R.; Pfeiffer, S.E. Expression of Rab GTP-binding proteins during oligodendrocyte differentiation in culture. J. Neurosci. Res. 2000, 59, 446–453. [Google Scholar] [CrossRef]
- Anitei, M.; Cowan, A.E.; Pfeiffer, S.E.; Bansal, R. Role for Rab3a in oligodendrocyte morphological differentiation. J. Neurosci. Res. 2009, 87, 342–352. [Google Scholar] [CrossRef]
- Fukushima, N.; Shirai, R.; Sato, T.; Nakamura, S.; Ochiai, A.; Miyamoto, Y.; Yamauchi, J. Knockdown of Rab7B, but not of Rab7A, which antagonistically regulates oligodendroglial cell morphological differentiation, recovers tunicamycin-induced defective differentiation in FBD-102b cells. J. Mol. Neurosci. 2023, 73, 363–374. [Google Scholar] [CrossRef]
- Singh, V.; Menard, M.A.; Serrano, G.E.; Beach, T.G.; Zhao, H.T.; Riley-DiPaolo, A.; Subrahmanian, N.; LaVoie, M.J.; Volpicelli-Daley, L.A. Cellular and subcellular localization of Rab10 and phospho-T73 Rab10 in the mouse and human brain. Acta Neuropathol. Commun. 2023, 11, 201. [Google Scholar]
- Horiuchi, M.; Tomooka, Y. An oligodendroglial progenitor cell line FBD-102b possibly secretes a radial glia-inducing factor. Neurosci. Res. 2006, 56, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Tomooka, Y. A role of Sema6A expressed in oligodendrocyte precursor cells. Neurosci. Lett. 2013, 539, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Morimura, T.; Numata, Y.; Nakamura, S.; Hirano, E.; Gotoh, L.; Goto, Y.I.; Urushitani, M.; Inoue, K. Attenuation of endoplasmic reticulum stress in Pelizaeus-Merzbacher disease by an anti-malaria drug, chloroquine. Exp. Biol. Med. 2014, 239, 489–501. [Google Scholar] [CrossRef]
- Higa, A.; Taouji, S.; Lhomond, S.; Jensen, D.; Fernandez-Zapico, M.E.; Simpson, J.C.; Pasquet, J.-M.; Schekman, R.; Chevet, E. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol. Cell. Biol. 2014, 34, 1839–1849. [Google Scholar] [CrossRef]
- Healy, S.; McMahon, J.; FitzGerald, U. UPR induction prevents iron accumulation and oligodendrocyte loss in ex vivo cultured hippocampal slices. Front. Neurosci. 2018, 12, 969. [Google Scholar] [CrossRef]
- Kuchitsu, Y.; Homma, Y.; Fujita, N.; Fukuda, M. Rab7 knockout unveils regulated autolysosome maturation induced by glutamine starvation. J. Cell Sci. 2018, 131, jcs215442. [Google Scholar] [CrossRef] [PubMed]
- Kuchitsu, Y.; Fukuda, M. Revisiting Rab7 functions in mammalian autophagy: Rab7 knockout studies. Cells 2018, 7, 215. [Google Scholar] [CrossRef]
- Xing, R.; Zhou, H.; Jian, Y.; Li, L.; Wang, M.; Liu, N.; Yin, Q.; Liang, Z.; Guo, W.; Yang, C. The Rab7 effector WDR91 promotes autophagy-lysosome degradation in neurons by regulating lysosome fusion. J. Cell Biol. 2021, 220, e202007061. [Google Scholar] [CrossRef] [PubMed]
- Torisu, H.; Iwaki, A.; Takeshita, K.; Hiwatashi, A.; Sanefuji, M.; Fukumaki, Y.; Hara, T. Clinical and genetic characterization of a 2-year-old boy with complete PLP1 deletion. Brain Dev. 2012, 34, 852–856. [Google Scholar] [CrossRef]
- Tsvetanova, N.G.; Riordan, D.P.; Brown, P.O. The yeast Rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 RNA. PLoS Genet. 2012, 8, e1002862. [Google Scholar] [CrossRef]
- Paira, S.; Chakraborty, A.; Das, B. The Sequential recruitments of Rab-GTPase Ypt1p and the NNS Complex onto pre-HAC1 mRNA promote its nuclear degradation in baker’s yeast. Mol. Cell. Biol. 2023, 43, 371–400. [Google Scholar] [CrossRef] [PubMed]
- Haile, Y.; Deng, X.; Ortiz-Sandoval, C.; Tahbaz, N.; Janowicz, A.; Lu, J.Q.; Kerr, B.J.; Gutowski, N.J.; Holley, J.E.; Eggleton, P.; et al. Rab32 connects ER stress to mitochondrial defects in multiple sclerosis. J. Neuroinflamm. 2017, 14, 19. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Zhang, J. Bridges between mitochondrial oxidative stress, ER stress and mTOR signaling in pancreatic beta cells. Cell. Signal. 2016, 28, 1099–1104. [Google Scholar]
- Kim, S.; Woo, C.H. Laminar flow inhibits ER stress-induced endothelial apoptosis through PI3K/Akt-dependent signaling Pathway. Mol. Cells 2018, 41, 964–970. [Google Scholar] [PubMed]
- Zhu, Y.; Shi, F.; Wang, M.; Ding, J. Knockdown of Rab9 suppresses the progression of gastric cancer through regulation of Akt signaling pathway. Technol. Cancer Res. Treat. 2020, 19, 153303. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Davis, R.J. Cell signaling and stress responses. Cold Spring Harb. Perspect. Biol. 2016, 8, a006072. [Google Scholar]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef]
- Schinzel, R.T.; Higuchi-Sanabria, R.; Shalem, O.; Moehle, E.A.; Webster, B.M.; Joe, L.; Bar-Ziv, R.; Frankino, P.A.; Durieux, J.; Pender, C.; et al. The hyaluronidase, TMEM2, promotes ER homeostasis and longevity independent of the UPR. Cell 2019, 179, 1306–1318.e18. [Google Scholar] [CrossRef] [PubMed]

| Reagents or Sources | Company or Source | Cat. No. | Lot. No. | Concentration Used |
|---|---|---|---|---|
| Antibodies | ||||
| Anti-proteolipid protein 1 (PLP1) | Atlas Antibodies (Stockholm, Sweden) | HPA004128 | 8115828 | Immunoblotting (IB), 1:500 |
| Anti-myelin basic protein (MBP) | BioLegend (San Diego, CA, USA) | 836504 | B225469 | IB, 1:500 |
| Anti-GSTpi | MBL (Tokyo, Japan) | 312 | 67 | IB, 1:500 |
| Anti-actin (also called pan-beta type actin) | MBL | M177-3 | 007 | IB, 1:5000 |
| Anti-eukaryotic initiation factor 2 alpha (eIF2a) | Santa Cruz Biotechnology (San Diego, CA, USA) | sc-13312 | J1922 | IB, 1:1000 |
| Anti-phosphorylated eukaryotic initiation factor 2 alpha kinase (peIF2a) | PGI Proteintech Group Inc. (Rosemont, IL, USA) | 28740-1-AP | 00089246 | IB, 1:1000 |
| Anti-C/EBP homologous protein (CHOP) | PGI Proteintech Group, Inc. | 15204-AP | 00117318 | IB, 1:1000 |
| Anti-heat shock protein family A member 5 (HSPA5) | PGI Proteintech Group, Inc. | 11587-AP | 00085813 | IB, 1:1000 |
| Anti-phosphorylated Akt kinase (pS473) | CST (Danvers, MA, USA) | 4060S | 27 | IB, 1:1000 |
| Anti-Akt kinase | CST | 4691T | 28 | IB, 1:1000 |
| Anti-IgG (H+L chain) (Mouse) pAb-HRP | MBL | 330 | 365 | IB, 1:5000 |
| Anti-IgG (H+L chain) (Rabbit) pAb-HRP | MBL | 458 | 353 | IB, 1:5000 |
| Key chemicals | ||||
| Tunicamycin (TMC) | Cayman chemical campany (Arbor, MI, USA) | 11445 | 0637439-4 | 100 ng/mL for typical experiments |
| Dimethyl sulfoxide (DMSO) | FUJIFILM Wako Pure Chemical Corporation (Tokyo, Japan) | 047-29353 | CDN0170 | Less than 0.1% |
| Dithiothreitol (DTT) | Nacalai Tesque (Kyoto, Japan) | 14128-46 | not described | 1 mM for typical experiments |
| Key reagents | ||||
| ScreenFect TM siRNA Transfection Reagent | FUJIFILM Wako Pure Chemical Corporation | 292-75013 | CAM0357 | According to manufacturer’s instructions |
| ScreenFect TM Dilution Buffer | FUJIFILM Wako Pure Chemical Corporation | 194-18181 | SKF5794 | According to manufacturer’s instructions |
| ImmunoStar TM Zeta | FUJIFILM Wako Pure Chemical Corporation | 295-72404 | WTL5319 | According to manufacturer’s instructions |
| Chemi-Lumi TM One Ultra | Nacalai Tesque | 11644-40 | L2P1314 | According to manufacturer’s instructions |
| Skim milk | FUJIFILM Wako Pure Chemical Corporation | 190-12865 | SKG4901 | According to manufacturer’s instructions |
| Western blotting stripping solution | Nacalai Tesque | 05364-55 | L5M5218 | According to manufacturer’s instructions |
| Fujifilm TM Sample buffer | FUJIFILM Wako Pure Chemical Corporation | 191-13282 | WDP4995 | According to manufacturer’s instructions |
| Isogen | Nippon Gene (Tokyo, Japan) | 311-02501 | 75009K | According to manufacturer’s instructions |
| 5×PrimeScript master mix | TaKaRa Bio (Shiga, Japan) | RR036A | AIE0440A | According to manufacturer’s instructions |
| Gflex DNA polymerase | TaKaRa Bio | R060A | AL80564A | According to manufacturer’s instructions |
| 2×Gflex PCR buffer (Mg2+, dNTP plus; with or without dye) | TaKaRa Bio | R060A | AL80564A | According to manufacturer’s instructions |
| TaKaRa Bio TM loading buffer | TaKaRa Bio | 9157 | A7201A | According to manufacturer’s instructions |
| Pre-stained Protein Markers (Broad Range) for SDS-PAGE | Nacalai Tesque | 02525-35 | L9M9989 | According to manufacturer’s instructions |
| ExcelBand All Blue Regular Range Protein Marker | Cosmo Bio (Toyko, Japan) | PM1500 | PM1500211500-5 | According to manufacturer’s instructions |
| ExcelBrand 3 Color Regular Range Protein Marker | Smo Bio (New Taipei City, Taipei) | PM2500-2 | PM25002112601-2 | According to manufacturer’s instructions |
| Cells used | ||||
| FBD-102b cells (mouse oligodendrocyte progenitor cells) | Dr. Yasuhiro Tomo-oka (Riken, Saitama, Japan and Tokyo University of Science, Chiba, Japan) | N/A | N/A | 1,000,000 cells per 6 cm dish culture |
| siRNA sequences (5′ to 3′) | ||||
| Sense chain for siLuciferase-105th (control siRNA) GCCAUUCUAUCCUCUAGAG-dTdT Antisense chain for siLuciferase-105th CUCUAGAGGAUAGAAUGGC-dTdT | This manuscript | N/A | N/A | 50 nM per one transfection |
| Sense chain for siRab9-142th GAUCUGGAGGUGGACGGAC-dTdT Antisense chain for siRab9b-142th GUCCGUCCACCUCCAGAUC-dTdT | This manuscript | N/A | N/A | 50 nM per one transfection |
| Sense chain for siRab9-199th GAACGCUUCCGAAGCCUGA-dTdT Antisense chain for siRab9b-199th UCAGGCUUCGGAAGCGUUC-dTdT | This manuscript | N/A | N/A | 50 nM per one transfection |
| Sense chain for siRab9-212th GCCUGAGGACGCCAUUUUA-dTdT Antisense chain for siRab9b-212th UAAAAUGGCGUCCUCAGGC-dTdT | This manuscript | N/A | N/A | 50 nM per one transfection |
| PCR primers (5′ to 3′) | ||||
| Sense primer for actin (internal control) ATGGATGACGATATCGCTGCGCTGGTC Antisense primer for actin CTAGAAGCACTTGCGGTGCACGATGGAG | This manuscript | N/A | N/A | 250 nM per one reaction |
| Sense primer for Rab9 ATGGCAGGAAAATCGTCTCTTTTTAAAATAATTCTTCTTG Antisense primer for Rab9b TCAACAGCAAGATGAGTTTGGCTTGG | This manuscript | N/A | N/A | 250 nM per one reaction |
| Plasmids | ||||
| pcDNA3.1(+)-c-eGFP-human PLP1/wild type PLP1 | GenScript Japan (Tokyo, Japan) | TK797353 | J908Z095G0-2 | 1 microgram per one transfection for 6 cm diameter-culture dish |
| pcDNA3.1(+)-c-eGFP-human PLP1 (A243V)/PLP1 p.Ala243Val | Genscript | TM203178 | J357RPHAG0-1 | 1 microgram per one transfection for 6 cm diameter-culture dish |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukushima, N.; Miyamoto, Y.; Yamauchi, J. Knockdown of Rab9 Recovers Defective Morphological Differentiation Induced by Chemical ER Stress Inducer or PMD-Associated PLP1 Mutant Protein in FBD-102b Cells. Pathophysiology 2024, 31, 420-435. https://doi.org/10.3390/pathophysiology31030032
Fukushima N, Miyamoto Y, Yamauchi J. Knockdown of Rab9 Recovers Defective Morphological Differentiation Induced by Chemical ER Stress Inducer or PMD-Associated PLP1 Mutant Protein in FBD-102b Cells. Pathophysiology. 2024; 31(3):420-435. https://doi.org/10.3390/pathophysiology31030032
Chicago/Turabian StyleFukushima, Nana, Yuki Miyamoto, and Junji Yamauchi. 2024. "Knockdown of Rab9 Recovers Defective Morphological Differentiation Induced by Chemical ER Stress Inducer or PMD-Associated PLP1 Mutant Protein in FBD-102b Cells" Pathophysiology 31, no. 3: 420-435. https://doi.org/10.3390/pathophysiology31030032







