NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging
Abstract
:1. Introduction
2. Dietary Interventions to Increase Lifespan and Promote Healthy Aging
3. Nutrient Sensing Pathways: Their Role in Molecular Aging
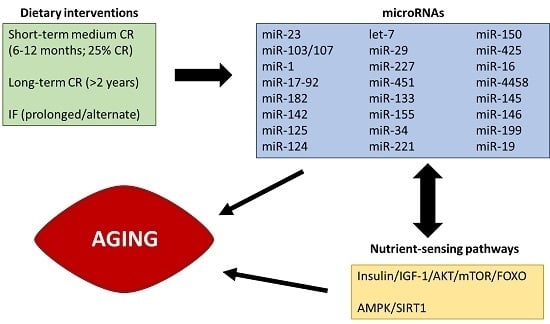
4. MiRagings: What and Which Are They? How Can They Be Modulated by Diet?
5. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| T2DM | Type 2 diabetes mellitus |
| CVD | Cardiovascular disease |
| CR | Caloric restriction |
| IF | Intermittent fasting |
| PA | Physical activity |
| RISC | RNA-induced silencing complex |
| PBMCs | Peripheral blood mononuclear cells |
| BAT | Brown adipose tissue |
| HFD | High-fat diet |
References
- Eurostat. Yearbook; Eurostat: Luxembourg, 2015. [Google Scholar]
- Willcox, D.C.; Scapagnini, G.; Willcox, B.J. Healthy aging diets other than the mediterranean: A focus on the okinawan diet. Mech. Ageing Dev. 2014, 136–137, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Antebi, A.; Bartke, A.; Barzilai, N.; Brown-Borg, H.M.; Caruso, C.; Curiel, T.J.; de Cabo, R.; Franceschi, C.; Gems, D.; et al. Interventions to slow aging in humans: Are we ready? Aging Cell 2015, 14, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Geller, A.M.; Zenick, H. Aging and the environment: A research framework. Environ. Health Perspect. 2005, 113, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Most, J.; Tosti, V.; Redman, L.M.; Fontana, L. Calorie restriction in humans: An update. Ageing Res. Rev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Willeit, P.; Drozdov, I.; Kiechl, S.; Mayr, M. Profiling of circulating micrornas: From single biomarkers to re-wired networks. Cardiovasc. Res. 2012, 93, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhou, B.; Ross, S.A.; Zempleni, J. Nutrition, micrornas, and human health. Adv. Nutr. 2017, 8, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, A.P.; Espanol, Y.; Lopez-Otin, C. Micromanaging aging with mirnas: New messages from the nuclear envelope. Nucleus 2011, 2, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Marin, T.; Gongol, B.; Chen, Z.; Woo, B.; Subramaniam, S.; Chien, S.; Shyy, J.Y. Mechanosensitive micrornas-role in endothelial responses to shear stress and redox state. Free Radic. Biol. Med. 2013, 64, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Suh, Y. Regulation of IGF-1 signaling by micrornas. Front. Genet. 2014, 5, 472. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.P.; Masoro, E.J.; Murata, I.; Bertrand, H.A.; Lynd, F.T. Life span study of SPF Fischer 344 male rats fed ad libitum or restricted diets: Longevity, growth, lean body mass and disease. J. Gerontol. 1982, 37, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, S.E.; Seo, A.Y.; Marzetti, E.; Lees, H.A.; Leeuwenburgh, C. Skeletal muscle autophagy and apoptosis during aging: Effects of calorie restriction and life-long exercise. Exp. Gerontol. 2010, 45, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Donati, A.; Recchia, G.; Cavallini, G.; Bergamini, E. Effect of aging and anti-aging caloric restriction on the endocrine regulation of rat liver autophagy. J. Geront. A Biol. Sci. Med. Sci. 2008, 63, 550–555. [Google Scholar] [CrossRef]
- Anson, R.M.; Guo, Z.; de Cabo, R.; Iyun, T.; Rios, M.; Hagepanos, A.; Ingram, D.K.; Lane, M.A.; Mattson, M.P. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc. Natl. Acad. Sci. USA 2003, 100, 6216–6220. [Google Scholar] [CrossRef] [PubMed]
- Varady, K.A.; Roohk, D.J.; Hellerstein, M.K. Dose effects of modified alternate-day fasting regimens on in vivo cell proliferation and plasma insulin-like growth factor-1 in mice. J. Appl. Physiol. 2007, 103, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Varady, K.A.; Roohk, D.J.; Loe, Y.C.; McEvoy-Hein, B.K.; Hellerstein, M.K. Effects of modified alternate-day fasting regimens on adipocyte size, triglyceride metabolism, and plasma adiponectin levels in mice. J. Lipid Res. 2007, 48, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R. Dietary restriction in rats and mice: A meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res. Rev. 2012, 11, 254–270. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.L.; Smith, E.D.; Tsuchiya, M.; Welton, K.L.; Thomas, J.H.; Fields, S.; Kennedy, B.K.; Kaeberlein, M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 2006, 5, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.D.; Wilson, M.A.; Zhu, M.; Wolkow, C.A.; de Cabo, R.; Ingram, D.K.; Zou, S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 2006, 5, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Colman, R.J.; Beasley, T.M.; Kemnitz, J.W.; Johnson, S.C.; Weindruch, R.; Anderson, R.M. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat. Commun. 2014, 5, 3557. [Google Scholar] [CrossRef] [PubMed]
- Mattison, J.A.; Roth, G.S.; Beasley, T.M.; Tilmont, E.M.; Handy, A.M.; Herbert, R.L.; Longo, D.L.; Allison, D.B.; Young, J.E.; Bryant, M.; et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 2012, 489, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Maxmen, A. Calorie restriction falters in the long run. Nature 2012, 488, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Wilcox, B.J.; Wilcox, C.D. Implications from and for food cultures for cardiovascular disease: Longevity. Asia Pac. J. Clin. Nutr. 2001, 10, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Chatzianagnostou, K.; Del Turco, S.; Pingitore, A.; Sabatino, L.; Vassalle, C. The Mediterranean Lifestyle as a Non-Pharmacological and Natural Antioxidant for Healthy Aging. Antioxidants 2015, 4, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Ros, E.; Salas-Salvado, J.; Covas, M.I.; Corella, D.; Aros, F.; Gomez-Gracia, E.; Ruiz-Gutierrez, V.; Fiol, M.; Lapetra, J.; et al. Primary prevention of cardiovascular disease with a mediterranean diet. N. Engl. J. Med. 2013, 368, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Calzon, S.; Martinez-Gonzalez, M.A.; Razquin, C.; Aros, F.; Lapetra, J.; Martinez, J.A.; Zalba, G.; Marti, A. Mediterranean diet and telomere length in high cardiovascular risk subjects from the PREDIMED-NAVARRA study. Clin. Nutr. 2016, 35, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Calzon, S.; Gea, A.; Razquin, C.; Corella, D.; Lamuela-Raventos, R.M.; Martinez, J.A.; Martinez-Gonzalez, M.A.; Zalba, G.; Marti, A. Longitudinal association of telomere length and obesity indices in an intervention study with a Mediterranean diet: The PREDIMED-NAVARRA trial. Int. J. Obes. 2014, 38, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Masana, M.F.; Koyanagi, A.; Haro, J.M.; Tyrovolas, S. n-3 Fatty acids, mediterranean diet and cognitive function in normal aging: A systematic review. Exp. Gerontol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Nishihira, J.; Tokashiki, T.; Higashiuesato, Y.; Willcox, D.C.; Mattek, N.; Shinto, L.; Ohya, Y.; Dodge, H.H. Associations between serum ω-3 fatty acid levels and cognitive functions among community-dwelling octogenarians in Okinawa, Japan: The KOCOA Study. J. Alzheimer Dis. 2016, 51, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Heilbronn, L.K.; de Jonge, L.; Frisard, M.I.; DeLany, J.P.; Larson-Meyer, D.E.; Rood, J.; Nguyen, T.; Martin, C.K.; Volaufova, J.; Most, M.M.; et al. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: A randomized controlled trial. JAMA 2006, 295, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Ravussin, E.; Redman, L.M.; Rochon, J.; Das, S.K.; Fontana, L.; Kraus, W.E.; Romashkan, S.; Williamson, D.A.; Meydani, S.N.; Villareal, D.T.; et al. A 2-year randomized controlled trial of human caloric restriction: Feasibility and effects on predictors of health span and longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Larson-Meyer, D.E.; Newcomer, B.R.; Heilbronn, L.K.; Volaufova, J.; Smith, S.R.; Alfonso, A.J.; Lefevre, M.; Rood, J.C.; Williamson, D.A.; Ravussin, E. Effect of 6-month calorie restriction and exercise on serum and liver lipids and markers of liver function. Obesity 2008, 16, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, M.; Redman, L.M.; Heilbronn, L.K.; Smith, J.V.; Martin, C.K.; Rood, J.C.; Greenway, F.L.; Williamson, D.A.; Smith, S.R.; Ravussin, E. Caloric restriction alone and with exercise improves CVD risk in healthy non-obese individuals. Atherosclerosis 2009, 203, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Redman, L.M.; Veldhuis, J.D.; Rood, J.; Smith, S.R.; Williamson, D.; Ravussin, E. The effect of caloric restriction interventions on growth hormone secretion in nonobese men and women. Aging Cell 2010, 9, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.S.; Frost, E.A.; Xie, W.; Rood, J.; Ravussin, E.; Redman, L.M. No effect of caloric restriction on salivary cortisol levels in overweight men and women. Metabolism 2014, 63, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Redman, L.M.; Heilbronn, L.K.; Martin, C.K.; de Jonge, L.; Williamson, D.A.; Delany, J.P.; Ravussin, E. Metabolic and behavioral compensations in response to caloric restriction: Implications for the maintenance of weight loss. PLoS ONE 2009, 4, e4377. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.A.; Martin, C.K.; Anton, S.D.; York-Crowe, E.; Han, H.; Redman, L.; Ravussin, E. Is caloric restriction associated with development of eating-disorder symptoms? Results from the CALERIE trial. Health Psychol. 2008, 27, S32–S42. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Meyer, T.E.; Klein, S.; Holloszy, J.O. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl. Acad. Sci. USA 2004, 101, 6659–6663. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Klein, S.; Holloszy, J.O. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age 2010, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Soare, A.; Cangemi, R.; Omodei, D.; Holloszy, J.O.; Fontana, L. Long-term calorie restriction, but not endurance exercise, lowers core body temperature in humans. Aging 2011, 3, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Heilbronn, L.K.; Smith, S.R.; Martin, C.K.; Anton, S.D.; Ravussin, E. Alternate-day fasting in nonobese subjects: Effects on body weight, body composition, and energy metabolism. Am. J. Clin. Nutr. 2005, 81, 69–73. [Google Scholar] [PubMed]
- Heilbronn, L.K.; Civitarese, A.E.; Bogacka, I.; Smith, S.R.; Hulver, M.; Ravussin, E. Glucose tolerance and skeletal muscle gene expression in response to alternate day fasting. Obes. Res. 2005, 13, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Hoddy, K.K.; Bhutani, S.; Phillips, S.A.; Varady, K.A. Effects of different degrees of insulin resistance on endothelial function in obese adults undergoing alternate day fasting. Nutr. Healthy Aging 2016, 4, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Harder-Lauridsen, N.M.; Nielsen, S.T.; Mann, S.P.; Lyngbaek, M.P.; Benatti, F.B.; Langkilde, A.R.; Law, I.; Wedell-Neergaard, A.S.; Thomsen, C.; Moller, K.; et al. The effect of alternate-day caloric restriction on the metabolic consequences of 8 days of bed rest in healthy lean men: A randomized trial. J. Appl. Physiol. 2017, 122, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Harvie, M.N.; Pegington, M.; Mattson, M.P.; Frystyk, J.; Dillon, B.; Evans, G.; Cuzick, J.; Jebb, S.A.; Martin, B.; Cutler, R.G.; et al. The effects of intermittent or continuous energy restriction on weight loss and metabolic disease risk markers: A randomized trial in young overweight women. Int. J. Obes. 2011, 35, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Lettieri-Barbato, D.; Giovannetti, E.; Aquilano, K. Effects of dietary restriction on adipose mass and biomarkers of healthy aging in human. Aging 2016, 8, 3341–3355. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.P.; Racette, S.B.; Villareal, D.T.; Fontana, L.; Steger-May, K.; Schechtman, K.B.; Klein, S.; Ehsani, A.A.; Holloszy, J.O. Lower extremity muscle size and strength and aerobic capacity decrease with caloric restriction but not with exercise-induced weight loss. J. Appl. Physiol. 2007, 102, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Taormina, G.; Fabrizio, P.; Wei, M.; Hu, J.; Longo, V.D. Serine- and threonine/valine-dependent activation of PDK and Tor orthologs converge on Sch9 to promote aging. PLoS Genet. 2014, 10, e1004113. [Google Scholar] [CrossRef] [PubMed]
- Mair, W.; Piper, M.D.; Partridge, L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005, 3, e223. [Google Scholar] [CrossRef] [PubMed]
- Segall, P.E.; Timiras, P.S. Patho-physiologic findings after chronic tryptophan deficiency in rats: A model for delayed growth and aging. Mech. Ageing Dev. 1976, 5, 109–124. [Google Scholar] [CrossRef]
- Ooka, H.; Segall, P.E.; Timiras, P.S. Histology and survival in age-delayed low-tryptophan-fed rats. Mech. Ageing Dev. 1988, 43, 79–98. [Google Scholar] [CrossRef]
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [PubMed]
- Miller, R.A.; Buehner, G.; Chang, Y.; Harper, J.M.; Sigler, R.; Smith-Wheelock, M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 2005, 4, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Gallinetti, J.; Harputlugil, E.; Mitchell, J.R. Amino acid sensing in dietary-restriction-mediated longevity: Roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 2013, 449, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Altintas, O.; Park, S.; Lee, S.J. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep. 2016, 49, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Lithgow, G.J.; Link, W. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016, 15, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Fernandez, E.; Flurkey, K.; Han, M.; Javors, M.A.; Li, X.; Nadon, N.L.; Nelson, J.F.; et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 2014, 13, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Tullet, J.M.; Hertweck, M.; An, J.H.; Baker, J.; Hwang, J.Y.; Liu, S.; Oliveira, R.P.; Baumeister, R.; Blackwell, T.K. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 2008, 132, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Uno, M.; Honjoh, S.; Matsuda, M.; Hoshikawa, H.; Kishimoto, S.; Yamamoto, T.; Ebisuya, M.; Matsumoto, K.; Nishida, E. A fasting-responsive signaling pathway that extends life span in C. elegans. Cell Rep. 2013, 3, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Giannakou, M.E.; Goss, M.; Partridge, L. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: Not required, but its activity modulates the response. Aging Cell 2008, 7, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, I.; Komatsu, T.; Hayashi, N.; Kim, S.E.; Kawata, T.; Park, S.; Hayashi, H.; Yamaza, H.; Chiba, T.; Mori, R. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging Cell 2015, 14, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Warr, M.R.; Binnewies, M.; Flach, J.; Reynaud, D.; Garg, T.; Malhotra, R.; Debnath, J.; Passegue, E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 2013, 494, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.M.; Song, X.L.; Hong, Y.Q.; Zhu, H.L.; Li, C.; Zhang, T.; Chen, W.; Zhao, S.C.; Chen, Q. Association between FOXO3A gene polymorphisms and human longevity: A meta-analysis. Asian J. Androl. 2014, 16, 446–452. [Google Scholar] [PubMed]
- Willcox, B.J.; Tranah, G.J.; Chen, R.; Morris, B.J.; Masaki, K.H.; He, Q.; Willcox, D.C.; Allsopp, R.C.; Moisyadi, S.; Poon, L.W.; et al. The FoxO3 gene and cause-specific mortality. Aging Cell 2016, 15, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Mercken, E.M.; Crosby, S.D.; Lamming, D.W.; JeBailey, L.; Krzysik-Walker, S.; Villareal, D.T.; Capri, M.; Franceschi, C.; Zhang, Y.; Becker, K.; et al. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell 2013, 12, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Adams, G.B.; Perin, L.; Wei, M.; Zhou, X.; Lam, B.S.; da Sacco, S.; Mirisola, M.; Quinn, D.I.; Dorff, T.B.; et al. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell 2014, 14, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Chen, D.; Riddle, D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 2004, 131, 3897–3906. [Google Scholar] [CrossRef] [PubMed]
- Honjoh, S.; Yamamoto, T.; Uno, M.; Nishida, E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 2009, 457, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Adelaiye, R.M.; Rastelli, A.L.; Miles, K.M.; Ciamporcero, E.; Longo, V.D.; Nguyen, H.; Vessella, R.; Pili, R. Dietary protein restriction inhibits tumor growth in human xenograft models. Oncotarget 2013, 4, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Solon-Biet, S.M.; McMahon, A.C.; Ballard, J.W.; Ruohonen, K.; Wu, L.E.; Cogger, V.C.; Warren, A.; Huang, X.; Pichaud, N.; Melvin, R.G.; et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014, 19, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Weiss, E.P.; Villareal, D.T.; Klein, S.; Holloszy, J.O. Long-term effects of calorie or protein restriction on serum IGF-1 and IGFBP-3 concentration in humans. Aging Cell 2008, 7, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.; Atzmon, G.; Cho, M.O.; Hwang, D.; Liu, B.; Leahy, D.J.; Barzilai, N.; Cohen, P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. USA 2008, 105, 3438–3442. [Google Scholar] [CrossRef] [PubMed]
- Milman, S.; Atzmon, G.; Huffman, D.M.; Wan, J.; Crandall, J.P.; Cohen, P.; Barzilai, N. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 2014, 13, 769–771. [Google Scholar] [CrossRef] [PubMed]
- Allard, J.S.; Heilbronn, L.K.; Smith, C.; Hunt, N.D.; Ingram, D.K.; Ravussin, E.; de Cabo, R. In vitro cellular adaptations of indicators of longevity in response to treatment with serum collected from humans on calorie restricted diets. PLoS ONE 2008, 3, e3211. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Auwerx, J. Targeting sirtuin 1 to improve metabolism: All you need is NAD+? Pharmacol. Rev. 2012, 64, 166–187. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Defossez, P.A.; Guarente, L. Requirement of nad and sir2 for life-span extension by calorie restriction in saccharomyces cerevisiae. Science 2000, 289, 2126–2128. [Google Scholar] [CrossRef] [PubMed]
- Scheibye-Knudsen, M.; Mitchell, S.J.; Fang, E.F.; Iyama, T.; Ward, T.; Wang, J.; Dunn, C.A.; Singh, N.; Veith, S.; Hasan-Olive, M.M.; et al. A high-fat diet and NAD+ activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014, 20, 840–855. [Google Scholar] [CrossRef] [PubMed]
- Satoh, A.; Brace, C.S.; Ben-Josef, G.; West, T.; Wozniak, D.F.; Holtzman, D.M.; Herzog, E.D.; Imai, S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 2010, 30, 10220–10232. [Google Scholar] [CrossRef] [PubMed]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.D.; Yamada, K.A.; Imai, S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Mouchiroud, L.; Houtkooper, R.H.; Auwerx, J. NAD+ metabolism: A therapeutic target for age-related metabolic disease. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Novelle, M.G.; Ali, A.; Dieguez, C.; Bernier, M.; de Cabo, R. Metformin: A hopeful promise in aging research. Cold Spring Harb. Perspect. Med. 2016, 6, a025932. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Boudreau, D.M.; Park, Y.; Simonds, N.I.; Freedman, A.N. Commonly used diabetes and cardiovascular medications and cancer recurrence and cancer-specific mortality: A review of the literature. Expert Opin. Drug Saf. 2014, 13, 1071–1099. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.P.; Feng, L.; Yap, K.B.; Lee, T.S.; Tan, C.H.; Winblad, B. Long-term metformin usage and cognitive function among older adults with diabetes. J. Alzheimer Dis. 2014, 41, 61–68. [Google Scholar]
- Zhang, C.; Li, S.; Yang, L.; Huang, P.; Li, W.; Wang, S.; Zhao, G.; Zhang, M.; Pang, X.; Yan, Z.; et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 2013, 4, 2163. [Google Scholar] [CrossRef] [PubMed]
- Breen, D.M.; Rasmussen, B.A.; Cote, C.D.; Jackson, V.M.; Lam, T.K. Nutrient-sensing mechanisms in the gut as therapeutic targets for diabetes. Diabetes 2013, 62, 3005–3013. [Google Scholar] [CrossRef] [PubMed]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojarvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Segura, L.; Perez-Andrade, M.; Miranda-Rios, J. The emerging role of microRNAs in the regulation of gene expression by nutrients. J. Nutrigenet. Nutrigenom. 2013, 6, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Claycombe, K.J.; Martinez, J.A.; Friso, S.; Schalinske, K.L. Nutritional epigenomics: A portal to disease prevention. Adv. Nutr. 2013, 4, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Davalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Saeidimehr, S.; Ebrahimi, A.; Saki, N.; Goodarzi, P.; Rahim, F. MicroRNA-based linkage between aging and cancer: From epigenetics view point. Cell J. 2016, 18, 117–126. [Google Scholar] [PubMed]
- Frost, R.J.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shyh-Chang, N.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Pullen, T.J.; da Silva Xavier, G.; Kelsey, G.; Rutter, G.A. miR-29a and miR-29b contribute to pancreatic β-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell. Biol. 2011, 31, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Zhang, M.F.; Wen, H.Y.; Hu, C.L.; Liu, R.; Wei, H.Y.; Ai, C.M.; Wang, G.; Liao, X.X.; Li, X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics 2013, 68, 75–80. [Google Scholar] [CrossRef]
- Goren, Y.; Kushnir, M.; Zafrir, B.; Tabak, S.; Lewis, B.S.; Amir, O. Serum levels of microRNAs in patients with heart failure. Eur. J. Heart Fail. 2012, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Tijsen, A.J.; Creemers, E.E.; Moerland, P.D.; de Windt, L.J.; van der Wal, A.C.; Kok, W.E.; Pinto, Y.M. Mir423-5p as a circulating biomarker for heart failure. Circ. Res. 2010, 106, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-κB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Frasca, D.; Diaz, A.; Romero, M.; Ferracci, F.; Blomberg, B.B. MicroRNAs miR-155 and miR-16 decrease AID and E47 in B cells from elderly individuals. J. Immunol. 2015, 195, 2134–2140. [Google Scholar] [CrossRef] [PubMed]
- Serna, E.; Gambini, J.; Borras, C.; Abdelaziz, K.M.; Mohammed, K.; Belenguer, A.; Sanchis, P.; Avellana, J.A.; Rodriguez-Mañas, L.; Viña, J. Centenarians, but not octogenarians, up-regulate the expression of microRNAs. Sci. Rep. 2012, 2, 961. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Capri, M.; Bonafè, M.; Morsiani, C.; Jung, H.J.; Spazzafumo, L.; Viña, J.; Suh, Y. Circulating miRNAs and miRNA shuttles as biomarkers: Perspective trajectories of healthy and unhealthy aging. Mech. Ageing Dev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Xiong, H.; Yang, H.; Ou, Y.; Xu, Y.; Huang, Q.; Lai, L.; Chen, S.; Zhang, Z.; Cai, Y.; et al. Circulating miR-34a levels correlate with age-related hearing loss in mice and humans. Exp. Gerontol. 2016, 76, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Tugay, K.; Guay, C.; Marques, A.C.; Allagnat, F.; Locke, J.M.; Harries, L.W.; Rutter, G.A.; Regazzi, R. Role of microRNAs in the age-associated decline of pancreatic β cell function in rat islets. Diabetologia 2016, 59, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Po, A.; Miele, E.; Ventriglia, G.; Ceccarelli, E.; Bugliani, M.; Marselli, L.; Marchetti, P.; Gulino, A.; Ferretti, E.; et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol. 2015, 52, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Wang, H.; Ye, Y.; Shu, Y.; Deng, Y.; He, X.; Lu, G.; Zhang, S. miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson's disease. Am. J. Transl. Res. 2016, 8, 2127–2137. [Google Scholar] [PubMed]
- Harada, M.; Jinnin, M.; Wang, Z.; Hirano, A.; Tomizawa, Y.; Kira, T.; Igata, T.; Masuguchi, S.; Fukushima, S.; Ihn, H. The expression of miR-124 increases in aged skin to cause cell senescence and it decreases in squamous cell carcinoma. Biosci. Trends 2017, 10, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zheng, W.; Li, N.; Su, Z.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017, 7, 41942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.; Tan, F.; Yu, N.; Pei, H. STAT1 Inhibits miR-181a expression to suppress colorectal cancer cell proliferation through PTEN/Akt. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, C.; Qi, W.; Zhang, Y.; Zhang, F.; Wu, J.X.; Hu, Y.N.; Wu, D.M.; Liu, Y.; Yan, T.T.; et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012, 55, 2032–2043. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramirez-Chavez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Berber, P.; Grassmann, F.; Kiel, C.; Weber, B.H. An eye on age-related macular degeneration: The role of microRNAs in disease pathology. Mol. Diagn. Ther. 2017, 21, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Doss, C.G.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. WIREs RNA 2014, 5, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Nesca, V.; Guay, C.; Jacovetti, C.; Menoud, V.; Peyot, M.L.; Laybutt, D.R.; Prentki, M.; Regazzi, R. Identification of particular groups of microRNAs that positively or negatively impact on β cell function in obese models of type 2 diabetes. Diabetologia 2013, 56, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Pang, L.; Cai, X.; Liu, X.; Yuan, S.; Fan, X.; Jiang, B.; Zhang, X.; Dou, Y.; Gorospe, M.; et al. let-7-repressesed SHC translation delays replicative senescence. Aging Cell 2014, 13, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Hyun, S.; Lee, J.H.; Jin, H.; Nam, J.; Namkoong, B.; Lee, G.; Chung, J.; Kim, V.N. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009, 139, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, X.; Chen, X.; Cai, R.; Chen, F.; Dong, W.; Yang, G.; Pang, W. MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biol. 2017, 14, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shen, J.K.; Hornicek, F.J.; Kan, Q.; Duan, Z. Regulation of microRNA-1 (miR-1) expression in human cancer. Biochim. Biophys. Acta 2017, 1860, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Ugalde, A.P.; Fernández, A.F.; Osorio, F.G.; Fueyo, A.; Freije, J.M.; López-Otín, C. Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc. Natl. Acad. Sci. USA 2010, 107, 16268–16273. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Qin, Y.; Jia, J.; Lin, T.; Chen, L.; Zeng, H.; Han, Y.; Wu, L.; Huang, S.; Wang, M.; et al. MiR-155 enhances insulin sensitivity by coordinated regulation of multiple genes in mice. PLoS Genet. 2016, 12, e1006308. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Deng, H.; Xu, H.; Yang, Q.; Zhou, Y.; Zhang, J.; Zhao, D.; Liu, F. MicroRNA-223 promotes mast cell apoptosis by targeting the insulin-like growth factor 1 receptor. Exp. Ther. Med. 2016, 11, 2171–2176. [Google Scholar] [PubMed]
- Teteloshvili, N.; Kluiver, J.; van der Geest, K.S.; van der Lei, R.J.; Jellema, P.; Pawelec, G.; Brouwer, E.; Kroesen, B.J.; Boots, A.M.; van den Berg, A. Age-associated differences in miRNA signatures are restricted to CD45RO negative T cells and are associated with changes in the cellular composition, activation and cellular ageing. PLoS ONE 2015, 10, e0137556. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Lin, X.; Du, T.; Xu, K.; Shen, H.; Wei, F.; Hao, W.; Lin, T.; Qin, Y.; Wang, H.; et al. Targeted Disruption of miR-17-92 impairs mouse spermatogenesis by activating mTOR signaling pathway. Medicine 2016, 95, e2713. [Google Scholar] [CrossRef] [PubMed]
- Grillari, J.; Hackl, M.; Grillari-Voglauer, R. miR-17–92 cluster: Ups and downs in cancer and aging. Biogerontology 2010, 11, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.U.; Thirion, C.; Polesskaya, A.; Bauersachs, S.; Kaiser, S.; Krause, S.; Pfaffl, M.W. TNF-α and IGF1 modify the microRNA signature in skeletal muscle cell differentiation. Cell Commun. Signal. 2015, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Law, P.T.; Ching, A.K.; Chan, A.W.; Wong, Q.W.; Wong, C.K.; To, K.F.; Wong, N. MiR-145 modulates multiple components of the insulin-like growth factor pathway in hepatocellular carcinoma. Carcinogenesis 2012, 33, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Budzinska, M.; Owczarz, M.; Pawlik-Pachucka, E.; Roszkowska-Gancarz, M.; Slusarczyk, P.; Puzianowska-Kuznicka, M. MiR-96, miR-145 and miR-9 expression increases, and IGF-1R and FOXO1 expression decreases in peripheral blood mononuclear cells of aging humans. BMC Geriatr. 2016, 16, 200. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lei, N.; Gong, A.Y.; Chen, X.M.; Hu, G. Cryptosporidium parvum induces SIRT1 expression in host epithelial cells through downregulating let-7i. Hum. Immunol. 2014, 75, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Menghini, R.; Casagrande, V.; Cardellini, M.; Martelli, E.; Terrinoni, A.; Amati, F.; Vasa-Nicotera, M.; Ippoliti, A.; Novelli, G.; Melino, G.; et al. Microrna 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 2009, 120, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A.; Owczarz, M.; Polosak, J.; Jonas, M.I.; Lisik, W.; Jonas, M.; Chmura, A.; Puzianowska-Kuznicka, M. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int. J. Obes. 2016, 40, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Yamac, A.H.; Kucukbuzcu, S.; Ozansoy, M.; Gok, O.; Oz, K.; Erturk, M.; Yilmaz, E.; Ersoy, B.; Zeybek, R.; Goktekin, O.; et al. Altered expression of micro-RNA 199a and increased levels of cardiac SIRT1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc. Pathol. 2016, 25, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Wu, X.; Gimble, J.M.; Guan, X.; Freitas, M.A.; Bunnell, B.A. Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging Cell 2011, 10, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wei, Z.; Ding, H.; Wang, Q.; Zhou, Z.; Zheng, S.; Zhang, Y.; Hou, D.; Liu, Y.; Zen, K.; et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis 2015, 241, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guan, M.; Townsend, K.L.; Huang, T.L.; An, D.; Yan, X.; Xue, R.; Schulz, T.J.; Winnay, J.; Mori, M.; et al. MicroRNA-455 regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1α signaling network. EMBO Rep. 2015, 16, 1378–1393. [Google Scholar] [CrossRef] [PubMed]
- Nidadavolu, L.S.; Niedernhofer, L.J.; Khan, S.A. Identification of microRNAs dysregulated in cellular senescence driven by endogenous genotoxic stress. Aging 2013, 5, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Min, K.H.; Lee, W. MicroRNA expression analysis in the liver of high fat diet-induced obese mice. Data Brief 2016, 9, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.B.; Wei, M.X.; Han, J.Y.; Wu, X.Y.; Li, C.; Wang, J.; Shen, W.; Lu, P.H. MicroRNA-451 regulates AMPK/mTORC1 signaling and fascin1 expression in HT-29 colorectal cancer. Cell Signal. 2014, 26, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Mercken, E.M.; Majounie, E.; Ding, J.; Guo, R.; Kim, J.; Bernier, M.; Mattison, J.; Cookson, M.R.; Gorospe, M.; de Cabo, R.; et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging 2013, 5, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Hu, Y.; Ma, L.; Du, M.; Xia, L.; Hu, Z. miR-425 inhibits melanoma metastasis through repression of PI3K-Akt pathway by targeting IGF-1. Biomed. Pharmacother. 2015, 75, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ihle, M.A.; Trautmann, M.; Kuenstlinger, H.; Huss, S.; Heydt, C.; Fassunke, J.; Wardelmann, E.; Bauer, S.; Schildhaus, H.U.; Buettner, R.; et al. miRNA-221 and miRNA-222 induce apoptosis via the KIT/AKT signalling pathway in gastrointestinal stromal tumours. Mol. Oncol. 2015, 9, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Iekushi, K.; Seeger, F.; Assmus, B.; Zeiher, A.M.; Dimmeler, S. Regulation of cardiac microRNAs by bone marrow mononuclear cell therapy in myocardial infarction. Circulation 2012, 125, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Liu, X.; Chen, X.; Li, J.; Yang, X.; Fan, J.; Yang, Y.; Chen, N. Ampelopsin attenuates brain aging of D-gal-induced rats through miR-34a-mediated SIRT1/mTOR signal pathway. Oncotarget 2016, 7, 74484–74495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Q.; Wang, G.D.; Wang, H.S.; Huang, Y.; Liu, X.M.; Cai, X.H. miR-16 inhibits cell proliferation by targeting IGF1R and theRraf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013, 587, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Ho-Fun, L.V.; Wong, A.M.; Kwong, D.L.; Zhu, Y.H.; Dong, S.S.; Kong, K.L.; Chen, J.; Tsao, S.W.; Guan, X.Y.; et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis 2013, 34, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cui, C.L.; Chen, W.L.; Fu, Z.Y.; Cui, X.Y.; Gong, X. miR-144 suppresses the growth and metastasis of laryngeal squamous cell carcinoma by targeting IRS1. Am. J. Transl. Res. 2016, 8, 1–11. [Google Scholar] [PubMed]
- Xiang, C.; Cui, S.P.; Ke, Y. MiR-144 inhibits cell proliferation of renal cell carcinoma by targeting MTOR. J. Huazhong Univ. Sci. Technol. Med. Sci. 2016, 36, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Turczynska, K.M.; Bhattachariya, A.; Sall, J.; Goransson, O.; Sward, K.; Hellstrand, P.; Albinsson, S. Stretch-sensitive down-regulation of the miR-144/451 cluster in vascular smooth muscle and its role in AMP-activated protein kinase signaling. PLoS ONE 2013, 8, e65135. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Ahtiainen, M.; Lazzarini, R.; Pöllänen, E.; Capri, M.; Lorenzi, M.; Fulgenzi, G.; Albertini, M.C.; Salvioli, S.; Alen, M.J.; et al. Hormone replacement therapy enhances IGF-1 signaling in skeletal muscle by diminishing miR-182 and miR-223 expressions: A study on postmenopausal monozygotic twin pairs. Aging Cell 2014, 13, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Tessitore, A.; Cicciarelli, G.; Del Vecchio, F.; Gaggiano, A.; Verzella, D.; Fischietti, M.; Mastroiaco, V.; Vetuschi, A.; Sferra, R.; Barnabei, R.; et al. MicroRNA expression analysis in high fat diet-induced NAFLD-NASH-HCC progression: Study on C57BL/6J mice. BMC Cancer 2016, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Fu, W.Q.; Zhao, S.; Zhao, X. Regulation of insulin-like growth factor 1 receptor signaling by microRNA-4458 in the development of lumbar disc degeneration. Am. J. Transl. Res. 2016, 8, 2309–2316. [Google Scholar] [PubMed]
- Li, X.; Sun, X.; Wu, J.; Li, Z. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am. J. Cancer Res. 2016, 6, 2869–2879. [Google Scholar] [PubMed]
- Csiszar, A.; Gautam, T.; Sosnowska, D.; Tarantini, S.; Banki, E.; Tucsek, Z.; Toth, P.; Losonczy, G.; Koller, A.; Reglodi, D.; et al. Caloric restriction confers persistent anti-oxidative, pro-angiogenic, and anti-inflammatory effects and promotes anti-aging miRNA expression profile in cerebromicrovascular endothelial cells of aged rats. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H292–H306. [Google Scholar] [CrossRef] [PubMed]
- Olivo-Marston, S.E.; Hursting, S.D.; Perkins, S.N.; Schetter, A.; Khan, M.; Croce, C.; Harris, C.C.; Lavigne, J. Effects of calorie restriction and diet-induced obesity on murine colon carcinogenesis, growth and inflammatory factors, and microRNA expression. PLoS ONE 2014, 9, e94765. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lim, M.; Zhao, S.; Sano, Y.; Simone, B.A.; Savage, J.E.; Wickstrom, E.; Camphausen, K.; Pestell, R.G.; Simone, N.L. The metastatic potential of triple-negative breast cancer is decreased via caloric restriction-mediated reduction of the miR-17~92 cluster. Breast Cancer Res. Treat. 2014, 146, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Sangiao-Alvarellos, S.; Pena-Bello, L.; Manfredi-Lozano, M.; Tena-Sempere, M.; Cordido, F. Perturbation of hypothalamic microRNA expression patterns in male rats after metabolic distress: Impact of obesity and conditions of negative energy balance. Endocrinology 2014, 155, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- De Siqueira, K.C.; de Lima, F.M.; Lima, F.S.; Taki, M.S.; da Cunha, C.F.; de Lima Reis, S.R.; Camargo, R.L.; Batista, T.M.; Vanzela, E.C.; Nardelli, T.R.; et al. miR-124a expression contributes to the monophasic pattern of insulin secretion in islets from pregnant rats submitted to a low-protein diet. Eur. J. Nutr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Cuesta Torres, L.F.; Ong, K.L.; Shrestha, S.; Choteau, S.A.; Barter, P.J.; Clifton, P.; Rye, K.A. High-Density Lipoprotein-Associated miR-223 is Altered after Diet-Induced Weight loss in Overweight and Obese Males. PLoS ONE 2016, 11, e0151061. [Google Scholar] [CrossRef] [PubMed]

| MicroRNA | Relationship with Aging | Related Nutrient Sensing Pathway | Effect of Diet | References |
|---|---|---|---|---|
| let-7 | Associated with the regulation of cellular senescence | IGF-1/PI3K/AKT mTOR SIRT1 | It is differentially expressed because of CR | [11,125,139,153,164] |
| miR-1 | Upregulated in the liver of progeria models | IGF-1/PI3K/AKT mTOR | Downregulated in the adipose tissue of mice fed a high-fat diet | [128,129,130] |
| miR-155 | Increased in β-cells in the elderly | IGF1-1/PI3K/AKT | It is differentially expressed because of CR | [109,110,131,164] |
| miR-16 | Increased in β-cells in the elderly | IGF-1R | It is differentially expressed because of CR | [110,154,164] |
| miR-34a | Increased in aging. Circulating levels upregulated in age-related hearing loss and in age-related macular degeneration | SIRT1 mTOR | It is differentially expressed by CR | [113,114,152,153] |
| miR-124a | Increased in aging and in aged skin | AKT3/FOXA2 SIRT1 | A low-protein diet reduced its expression in pancreatic islets of pregnant rats | [114,115,116,117,167] |
| miR-383 | Increased in aging | IGF-1 and IGF-1R | It is downregulated by a high-fat diet in mice pancreatic islets | [114,121,122,123,124] |
| miR-130b | Decreased in aging | PI3K/AKT (through direct inhibition of PTEN) | Circulating levels increase after an intervention with polyunsaturated fatty acids | [114,118,121] |
| miR-181a | Decreased in aging | PI3K/AKT (through direct inhibition of PTEN) SIRT1 | Its levels increase in skeletal muscle of monkeys under a CR regime | [114,119,148] |
| miR-223 | Decreased in CD4+ T cells of old donors and reduced in postmenopausal women | IGF1R/FOXO3A PI3K/AKT mTOR | A high-protein regime decreases High-Density Lipoproteins-associated miR-223 levels | [132,133,159,168] |
| miR-17-92 cluster | Decreased with age | mTOR | The expression of this cluster is downregulated by CR in a mouse model of breast cancer | [134,135,165] |
| miR-145 | Decreased in peripheral blood mononuclear cells with aging | IRS1/IRS2 | Upregulated by a high-fat diet, although not by CR | [137,138,166] |
| miR-199 | Decreased in aging rhesus monkeys | SIRT1 | Unknown | [142,143] |
| miR-19b | Lower levels in octogenarians as compared with centenarians and young individuals | PGC1α | Circulating levels increase after an intervention with polyunsaturated fatty acids | [111,121] |
| miR-455 | Decreased in old mice | AMPK | Upregulated in the liver of mice fed a high-fat diet | [145,146,147] |
| miR-451 | Increased in aging primates | AMPK, mTORC1 | Increased in primates under a CR regime | [148,149,164] |
| miR-142-3p | Decreased in postmenopausal women | IGF1R/FOXO3A PI3K/AKT mTOR | Upregulated by a high-fat diet | [130,159] |
| miR-182 | Decreased in postmenopausal women | IGF1R/FOXO3A PI3K/AKT mTOR | Increased in the liver of mice fed a high-fat diet | [159,160] |
| miR-144 | Increased in the skeletal muscle of old rhesus monkeys | PI3K/AKT (through direct inhibition of PTEN and IRS1) mTOR AMPK | Decreased in rats and monkeys under a CR regime | [149,155,156,157,158,163,164] |
| miR-221 | Decreased by CR | PGC1α | It is downregulated by CR and polyunsaturated fatty acids | [121,149,151] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Micó, V.; Berninches, L.; Tapia, J.; Daimiel, L. NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging. Int. J. Mol. Sci. 2017, 18, 915. https://doi.org/10.3390/ijms18050915
Micó V, Berninches L, Tapia J, Daimiel L. NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging. International Journal of Molecular Sciences. 2017; 18(5):915. https://doi.org/10.3390/ijms18050915
Chicago/Turabian StyleMicó, Víctor, Laura Berninches, Javier Tapia, and Lidia Daimiel. 2017. "NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging" International Journal of Molecular Sciences 18, no. 5: 915. https://doi.org/10.3390/ijms18050915
APA StyleMicó, V., Berninches, L., Tapia, J., & Daimiel, L. (2017). NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging. International Journal of Molecular Sciences, 18(5), 915. https://doi.org/10.3390/ijms18050915