Abstract
Skeletal muscle mitochondrial dysfunction, evidenced by incomplete beta oxidation and accumulation of fatty acid intermediates in the form of long and medium chain acylcarnitines, may contribute to ectopic lipid deposition and insulin resistance during high fat diet (HFD)-induced obesity. The present review discusses the roles of anterograde and retrograde communication in nucleo-mitochondrial crosstalk that determines skeletal muscle mitochondrial adaptations, specifically alterations in mitochondrial number and function in relation to obesity and insulin resistance. Special emphasis is placed on the effects of high fat diet (HFD) feeding on expression of nuclear-encoded mitochondrial genes (NEMGs) nuclear receptor factor 1 (NRF-1) and 2 (NRF-2) and peroxisome proliferator receptor gamma coactivator 1 alpha (PGC-1α) in the onset and progression of insulin resistance during obesity and how HFD-induced alterations in NEMG expression affect skeletal muscle mitochondrial adaptations in relation to beta oxidation of fatty acids. Finally, the potential ability of acylcarnitines or fatty acid intermediates resulting from mitochondrial beta oxidation to act as retrograde signals in nucleo-mitochondrial crosstalk is reviewed and discussed.
Keywords:
mitochondria; skeletal muscle; obesity; insulin resistance; high fat diet; PGC1α; NRF-1; NRF-2; TFAM; acylcarnitine; metabolomics 1. Introduction
Obesity and type 2 diabetes (T2D) represent a major healthcare issue. Recent data suggest that about 35% of the adult US population is considered obese [1], whereas 9.3% of the population, i.e., 29.1 million people, have diabetes [2]. Unhealthy lifestyle choices such as low physical activity, excess caloric intake, and consumption of high fat diets (HFD) contribute to metabolic disorders such as obesity, T2D, diabetic dyslipidemia, and non-alcoholic fatty liver disease [3,4,5]. On the other hand, positive changes in lifestyle such as reduction of fat intake and increasing physical activity have been shown to decrease the incidence of obesity and T2D [6,7]. In the absence of positive lifestyle modifications, obese subjects are at a higher risk of development of other metabolic disorders such as hypertension, cardiovascular diseases, and T2D [8,9], whereas T2D patients are at a risk of developing other complications such as nephropathy, cardiovascular diseases, retinopathy, and neuropathy [10].
Mitochondrial dysfunction, systemic inflammation, aberrant adipokine signaling, intramyocellular lipid accumulation, and ectopic lipid deposition are some of the underlying mechanisms that may contribute to the development and progression of metabolic disorders including obesity and T2D, as well as their complications [11,12,13,14,15,16,17,18]. For example, fatty acid oxidation (FAO) and the activity of enzymes involved in mitochondrial fatty acid transport and oxidation, such as carnitine palmitoyltransferase 1 (CPT-1) and citrate synthase (CS), are lower in the skeletal muscle during obesity, and higher intramyocellular lipid (IMCL) content is observed in insulin resistant subjects [18,19]. Mitochondrial dysfunction may be further compromised in T2D obese skeletal muscle, as T2D skeletal muscle exhibits decreased respiration rates and lower oxidative enzyme levels compared to obese, non-diabetic or lean muscle [11,20]. Tissue mitochondrial dysfunction, such as decreased rates in FAO, may be attributed to the dysfunction of the mitochondrion itself or to a decrease in total mitochondrial number within the tissue, as diabetic subjects also exhibit a decrease in mitochondrial content in skeletal muscle [21]. Nuclear encoded mitochondrial genes (NEMGs) regulate both mitochondrial biogenesis and function [22] and may play a role in the development of insulin resistance during obesity and T2D [23,24]. Here, we discuss the role of NEMGs in regulating mitochondrial adaptations in the skeletal muscle in relation to HFD-induced obesity and insulin resistance.
2. Nucleo-Mitochondrial Crosstalk
Mitochondria play a significant role in determining cellular physiology, as they are responsible for production of cellular energy, essential metabolites, and the regulation of apoptosis [25]. Mitochondrial integrity, biogenesis, and function, which contribute to the cellular physiology, are dependent on gene expression from both the mitochondrion as well as the nucleus [26]. Communication between the nuclear to mitochondrial genomes is a two-way process involving both anterograde (from the nucleus to the mitochondria) and retrograde (from the mitochondria to the nucleus) communication [27]. Thus, many fundamental cellular processes are dependent on this crosstalk between the nuclear and mitochondrial genomes [22], which also plays an essential role in determining the cellular response to environmental cues [27].
2.1. Nucleo-Mitochondrial Crosstalk and Evolution
Although mitochondria have their own genomes, they require nuclear-encoded proteins for their biogenesis and function [28]. Evolutionarily, mitochondria are thought to have originated from an α-proteobacterium or a protomitochondrion [29]. The protomitochondria may have either invaded a primitive eukaryotic cell or been engulfed by a primitive eukaryotic cell [29]. Originally, this α-proteobacterium had all the genes necessary for its independent existence; however, over time, functionally essential genes transferred from its genome to the nuclear genome [29]. Currently, ~1500 proteins that are necessary for mitochondrial function are encoded by the nuclear genome [27,30]. These nuclear proteins are imported into the mitochondria, where they perform their functions. The mitochondrial genome encodes for a total of 37 genes, two of these encode rRNAs and 22 encode tRNAs that are used for translation of mitochondrial proteins and 13 genes encode respiratory chain enzyme subunits [22]. Thus, evolutionary processes that lead to the transfer of mitochondrial genes to the nucleus have rendered nuclear-mitochondrial crosstalk essential for mitochondrial function. Additionally, nuclear-mitochondrial communication may have itself played a crucial role in evolution [31,32]. Blockage of this crosstalk leads to reduced mitochondrial activity [33,34]. Transfer of mitochondrial DNA from one species to cells containing nuclear DNA from other species leads to lowered mitochondrial activity [33,34]. Additionally, cybrid experiments in human cells have also shown altered mitochondrial activity [35]. These experiments point to the fact that nucleo-mitochondrial communication may have played a role in evolutionary processes and is essential in maintaining correct cellular functioning.
2.2. Anterograde Signaling
Anterograde signaling coordinates mitochondrial gene expression or mitochondrial DNA replication in response to cellular or environmental cues that are detected by the nucleus [36]. While mitochondria are essential for practically all cell types, various cell types may contain mitochondria in different number and shapes, which may be dependent on the energy demand [37,38]. Besides, a specific cell type may adapt their mitochondria to various environmental cues such as physical activity, nutrition, or temperature [39]. Mitochondrial biogenesis and function are regulated by the nuclear genome via complex mechanisms in response to various physiological and pathological states [40,41,42]. Coordinate regulation of the mitochondrial genome through the nuclear genome is necessary for mitochondrial function, as 13 electron transport chain genes encoded in the mitochondrial genome as well as hundreds of proteins encoded by the nuclear genome are required for the activity of the electron transport chain [43]. Various transcriptional factors and coactivators regulate nuclear and mitochondrial gene expression in response to temperature [44], caloric intake [45], and exercise [46]. These factors activate genes required for mitochondrial DNA replication and the genes responsible for mitochondrial function. For example, the nuclear transcription factor nuclear respiratory factor-1 (NRF-1) activates transcription factor A, mitochondrial (TFAM) [47], which is essential for transcriptional activation of mitochondrial DNA replication and for its organization [48]. The nuclear-encoded transcriptional-coactivator, peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α), may also act as a master regulator of mitochondrial biogenesis through nucleo-mitochondrial communication by activating many nuclear-encoded mitochondrial genes (NEMGs) that regulate mitochondrial adaptations [49,50,51] and inducing NRF-1 and NRF-2 activity to also stimulate Tfam [40]. In addition to the effects of Tfam, PGC-1α itself may also translocate to the mitochondria to regulate mitochondrial DNA replication and transcription needed for biogenesis and function [52]. NRF-1 and NRF-2 transcriptionally activate many other NEMGs as well mitochondrial genes [53,54]. Specifically, NRF-1 binding sites are found in promoters of genes encoding oxidative phosphorylation subunits [53], while NRF-2 binds to promoters of genes encoding cytochrome c oxidase subunits [54]. Mechanistic target of rapamycin (mTOR), a sensor of cellular physiology is also a regulator of mitochondrial function [55,56]. mTOR inhibition reduces PGC-1α and Yin Yang 1 (YY1) interaction resulting in reduced mitochondrial gene expression [55]. These nuclear genes control distinct import, export, and assembly pathways that are followed by nuclear and mitochondrial proteins in order to form multimeric proteins needed for the biogenesis of mitochondria [27]. Thus, NEMGs regulate nucleo-mitochondrial epistasis by enhancing activity of nuclear as well as mitochondrial genes necessary for mitochondrial adaptations.
2.3. Retrograde Communication
Retrograde communication involves signaling from organelles such as the mitochondria, which can lead to altered nuclear gene expression [57]. ROS can influence nuclear gene expression through retrograde signaling [58]. Specifically, ROS has been shown to recruit DNA methyltransferases (DNMTs) and sirtuin-1 (SIRT-1), which leads to DNA hypermethylation of nuclear genes [59]. This would lead to downregulation of these genes. In yeast, low functioning mitochondria can signal the transcription factors, retrograde signaling proteins 1–3 (RTG 1–3), which translocate to the nucleus and activate expression of many genes including citrate synthase, peroxisomal (Cit-1) [60]. These genes are responsible for regulating cellular homeostasis in an energy deficient situation crested due to low functioning mitochondria [60]. Knockout of Complex IV gene in Drosophila leads to transformation of cellular metabolism from oxidative to glycolytic through transcriptional regulation of p53 and hypoxia-inducible factor-α (Hif-α) [61]. Lower mitochondrial membrane potential leads to increased cytosolic Ca2+ which leads to alteration of levels of various transcription factors and an upregulation of cytochrome c oxidase V b gene [62]. Mitochondrial encoded protein, humanin, is known to affect cellular metabolism in various models of cellular stress and disease states leading to cytoprotective effects [63]. Various studies have demonstrated that mitochondrial function can regulate cellular differentiation, where blockage of mitochondrial translation leads to inhibition of cellular differentiation [64,65,66]. Additionally, inhibition of fatty acid uptake by CPT-1 leads to activation of the nuclear receptor, peroxisome proliferator-activated receptors (PPAR) [67]. It is possible that the metabolites accumulated as result of inhibition of CPT-1 may act as signaling molecules to activate nuclear PPAR [67]. Indeed, it has been shown that fatty acid binding proteins (FABPs) transfer fatty acids to the nucleus where these fatty acids act as ligands for peroxisome proliferator-activated receptor α (PPAR-α) [68]. Additionally, low mitochondrial number lead to hypermethylation of nuclear genes in prostate and breast cancer cells and this hypermethylation was reversed when cellular mitochondrial DNA levels were restored to normal [69,70]. Isogenic nuclear cybrid cells containing either T2D susceptible or T2D resistant mitochondrial DNA haplogroups showed that the cybrid cells containing susceptible mitochondrial DNA have lower levels of oxidative phosphorylation gene expression and higher levels of glycolytic gene expression compare to cybrid cells containing resistant mitochondrial DNA [71]. Thus, mitochondrial signaling can be crucial in regulating nuclear gene expression in response to disease states [71]. Recently, mitochondrial genomes have been found to encode small-RNAs [72]. Although their function is yet to be deciphered, they may affect nuclear gene expression [72]. In summary, altered mitochondrial metabolism leading to retrograde signaling can be of significance in various physiological and pathophysiological states.
3. HFD-Induced Obesity, Insulin Resistance and Anterograde Communication
HFD feeding results in differential effects on mitochondrial adaptations, with many studies (summarized in Table 1) reporting increases in NEMG expression and mitochondrial function, and others reporting decreased NEMG expression and mitochondrial maladaptations coinciding with the onset and progression of insulin resistance [73,74,75,76,77]. These seemingly apposing effects of HFD feeding may be explained by compensatory alterations in NEMG expression and beneficial mitochondrial adaptations that act in some conditions to prevent insulin resistance or act in other conditions in which increased lipid is present (i.e., obesity) or during repeated stimulation with HFD to inhibit and/or desensitize FAO and lead to fatty acid accumulation and mitochondrial maladaptations that contribute to insulin resistance. Several studies support this conclusion. First, after a single high fat meal (65% kcal), Pgc-1α expression is upregulated in skeletal muscle of lean subjects, but not in obese subjects [73]; Second, 5 days of HFD (65% kcal) feeding upregulates gene expression of NEMGs such as pyruvate dehydrogenase lipoamide kinase isozyme 4 (Pdk-4), uncoupling protein 3 (Ucp-3), Ppar-α and Pgc-1α in the skeletal muscle of lean subjects; whereas no effect was observed in obese subjects [73]; Lastly, in C57Bl/6J fed HFD (45% kcal) for three days, there was an increase in oxidative phosphorylation gene expression. However, this effect was no longer observed after 28 days of HFD feeding [78]. In addition to changes in NEMG expression, mitochondrial adaptations may also be dependent on the length of HFD feeding. For example, 15 days of HFD (50% kcal) increases fatty acid oxidative capacity in subsarcolemmal and intermyofibrillar mitochondria [74], and four weeks of HFD (60% kcal) feeding increases mtDNA copy number, FAO capacity, mitochondrial oxidative enzymes, and respiratory chain enzymes in Wistar rats [79].

Table 1.
Effects of dietary fat interventions or lipid treatment on skeletal muscle nuclear-encoded mitochondrial gene (NEMG) or protein expression and mitochondrial function and number. ↑indicates an increase and ↓ indicates a decrease in respective measured outcomes.
Other studies support the conclusion that HFD feeding leads to obesity and insulin resistance, with decreased NEMG expression, impaired mitochondrial function and decreased mitochondrial number observed in various tissues including the skeletal muscle [75,76,77]. For example, three weeks of HFD (45% kcal) feeding in C57BL/6J mice downregulates oxidative phosphorylation gene expression as well as expression of Pgc-1α [75]. HFD (65% kcal) feeding for 10 weeks results in downregulation of several NEMG, including peroxisome proliferator-activated receptor γ (Ppar-γ) and Pgc-1α expression in skeletal muscle of C57BL/6J mice [76]. In L6 myotubes, 24 h treatment with palmitate decreased the protein level and activity of PGC-1α and the protein level of TFAM [80]. HFD (60% kcal) feeding for one year significantly lowered mitochondrial function in skeletal muscle [81], and HFD (45% kcal) feeding for 12 weeks in Wistar rats reduced tricarboxylic cyclic acid (TCA) intermediates and increased incomplete beta-oxidation in skeletal muscle, leading to the accumulation of beta oxidation intermediates [82]. Similarly, eight weeks of HFD (45% kcal) feeding in C57BL/6J mice lead to incomplete beta oxidation of fatty acids in skeletal muscle [83], indicating mitochondrial dysfunction [82].
Thus, diet-induced regulation of specific NEMG determines anterograde communication between the nucleus and mitochondria, and may play a significant role in determining mitochondrial adaptations that contribute to obesity and insulin resistance. The roles of the NEMGs, NRF-1, NRF-2, PGC-1α, and others in determining mitochondrial number and function in regards to FAO and their potential roles in obesity and insulin resistance are further discussed below.
3.1. NRF-1 and NRF-2
NRF-1 and NRF-2 are transcription factors that have been shown to regulate many mitochondrial and FAO genes [47,84]. For example, NRF-1 and/or NRF-2 responsive regulatory elements are found in the promoter region of cytochrome c, which is involved in mitochondrial respiration [85], and other mitochondrial respiration genes [86]; and NRF-1 transcriptionally activates genes which encode subunits of the five respiratory complexes [53]. NRF-1 target genes are generally related to oxidative phosphorylation, mitochondrial transcription and replication and heme biosynthesis; whereas NRF-2 target genes are related to mitochondrial complex II, IV, V, and mitochondrial transcription and replication [47]. NRFs are also known to induce the expression of Tfam [87], a key regulator of mitochondrial DNA replication [88], and play a role in PGC-1α-induced mitochondrial biogenesis [40,87]. Interestingly, Nrf-1 and Nrf-2α gene expression is enhanced by ectopic expression of PGC-1α in C2C12 cells [10], and PGC-1α coactivates transcription of several NRF-1 target genes [84], such as cytochrome c [89]. Loss of Nrf-1 specifically leads to reduced mtDNA levels and proves to be lethal during embryo development [90]. Furthermore, treatments that deplete mtDNA increase both Nrf-1 and Tfam expression, likely to induce compensatory mitochondrial biogenesis [91,92]. Thus, NRFs play a major role in determining mitochondrial adaptations.
Indeed, skeletal muscle NRF-1 and NRF-2 seem to play a significant role in determining metabolic dysregulation contributing to T2D [93]. Decreased skeletal muscle Nrf-1 and Nrf-2 expression have been noted during HFD-induced obesity [94], and Nrf-1 is downregulated in T2D subjects compared to non-diabetic subjects [93]. NRF-1 dependent genes involved in oxidative metabolism and mitochondrial function are also downregulated in T2D subjects [93]. Nrf-1 expression has also shown to be inversely related to fasting blood glucose levels [93] and expression of NRF-1 dependent genes has been observed in prediabetes subjects in conjunction with lower Pgc-1α [93].
3.2. PGC-1α
PGC-1α, a NEMG, is a transcriptional co-activator that responds to environmental cues and activates other NEMGs [95,96]. It is upregulated by exercise, caloric restriction, and cold exposure while it is reduced by high-fat diet feeding [76,97,98]. Induction of Pgc-1α is associated with increased expression of peroxisome proliferator-activated receptor α (Ppar-α), which in turn induces the expression of beta-oxidation pathway enzymes [98]. PGC-1α is involved in regulation of lipid metabolism in the skeletal muscle, liver, brown adipose tissue and the heart [51]. In the skeletal muscle, it regulates lipid metabolism by enhancing mitochondrial biogenesis and function [99]. It increases fatty acid oxidation and leads to muscle fiber-type switching to type I fibers, which have higher mitochondrial density and a higher capacity for fatty acid oxidation [100,101].
Pgc-1α activation results in an increase in mitochondrial number in various tissues, and this has been demonstrated in humans as well as animal models [40,102,103]. Puigserver et al. first cloned Pgc-1α from a brown fat cDNA library and provided evidence that Pgc-1α overexpression leads to increased mitochondrial content, suggesting that PGC-1α might play a role in promotion of mitochondrial biogenesis [44]. Similarly, C2C12 cells overexpressing Pgc-1α exhibit higher mtDNA content and mitochondrial proliferation in both myoblast and myotube states [40]. In skeletal muscle, exercise-induced upregulation of nuclear PGC-1α protein levels coincide with increased expression of genes related to mitochondrial biogenesis, suggesting that PGC-1α is responsible for inducing mitochondrial biogenesis in human skeletal muscle [104]. Pgc-1α activation may also lead to muscle fiber type conversion, with PGC-1α leading to conversion of type II to type I [101]. Type I muscle fibers are dark red in color, have higher mitochondrial content, store less glycogen, and have more oxidative enzyme content compared to type II fibers [105]. While PGC-1α may regulate the fiber type switching, it is not necessary for the development of skeletal muscle and fiber type determination, as mice lacking PGC-1α in skeletal muscle exhibit a normal distribution of fiber types [106]. However, PGC-1α may be essential for determining mitochondrial biogenesis, as Pgc-1α deficiency leads to lower mitochondrial number in skeletal muscle [107].
PGC-1α not only increases mitochondrial number but also improves mitochondrial function and increases beta-oxidation of fatty acids, which is particularly beneficial in obese and T2D subjects. For example, overexpression of Pgc-1α in insulin-resistant obese Zucker rats increases mtDNA content, CS activity, and palmitate oxidation, and decreases intramyocellular lipid content [108]. One mechanism through which PGC-1α improves mitochondrial function may be by FAO enzyme activity through PPAR-α binding in order to enhance PPAR-α transcriptional activity [100]. PGC-1α coactivation of PPAR-α leads to increases in CPT-1 activity in skeletal muscle, which in turn increases long-chain fatty acid import into the mitochondria for oxidation [100,109]. The overexpression of both Ppar-α and Pgc-1α results in greater upregulation of mitochondrial FAO genes and increased palmitate oxidation rate compared to overexpression of Ppar-α alone [100]. The effects of PGC-1α on FAO occur through a SIRT-1-dependent mechanism [110], as knockdown of Sirt-1 results in reduction of the effects of PGC-1α on the mitochondrial oxidative gene expression of cytochrome c, medium-chain acyl-CoA dehydrogenase, isocitrate dehydrogenase 3α, Cpt-1b, and Pdk-4 in primary skeletal muscle cells [110]. SIRT-1 activation also prevents the development of HFD-induced insulin resistance and to improve insulin sensitivity in T2D animal models through deacetylation of PGC-1α and by increasing mitochondrial function [111,112]. SIRT-1 acts to post-translationally modify PGC-1α through deacetylation, resulting in its activation and subsequent expression of target genes related to mitochondrial FAO [113]. Similarly, PGC-1α protein phosphorylation by AMP-activated protein kinase (AMPK) or p38 mitogen-activated protein kinases (p38-MAPK) increases PGC-1α stability and enhances its activity [114,115]. In addition to enhancing PGC-1α activity through phosphorylation, AMPK also acts indirectly to enhance PGC-1α activity through SIRT-1 in response to increased NAD+ levels and a low-energy status, such as that created during exercise or fasting [110,113,116]. Phosphorylation of PGC-1α by AMPK may be the first step necessary for deacetylation by SIRT-1, as PGC-1α protein lacking AMPK phosphorylation sites is not deacetylated by SIRT-1 [116]. Thus, phosphorylation and acetylation may work sequentially to activate PGC-1α [116] and increase transcription of target genes responsible for regulation for mitochondrial function and FAO in the skeletal muscle [110,116].
In addition to increasing FAO rate, Pgc-1α upregulation also acts to improve the completeness of beta oxidation of fatty acids. Koves et al. show that Pgc-1α overexpression in rat L6 myocytes promotes complete oxidation of fatty acids [117]. During beta oxidation of fatty acids, each round of oxidation results in a shortening of the fatty acid by two carbons until the final two carbon product, acetyl-coA, is produced and further oxidized into ATP through the TCA cycle and electron transport chain (ETC) [118]. Disruption in FAO at any step leads to incomplete oxidation of fatty acids and possible accumulation of specific chain lengths of these fatty acids intracellularly, which may contribute to ectopic lipid accumulation and insulin resistance [82,119]. In the skeletal muscle during obesity and insulin resistance, an accumulation of medium and long chain fatty acids has been noted and may contribute to the onset and progression of insulin resistance during obesity [120]. Pgc-1α may attenuate insulin resistance by increasing the completeness of fatty acid beta-oxidation [117]. Importantly, T2D skeletal muscle and HFD-fed obese muscle exhibit mitochondrial dysfunction in the form of incomplete beta oxidation of fatty acids in the skeletal muscle [119]. Furthermore, Pgc-1α expression is reduced in patients with T2D and obesity, and this reduction seems to be one of the factors responsible for the development and progression of T2D [93,121,122,123], as downstream PGC-1α target genes involved in oxidative phosphorylation are also downregulated in the skeletal muscle of T2D patients [122]. Pgc-1α gene polymorphisms are associated with altered lipid oxidation and T2D development in many populations [124,125,126,127]. These studies point to the fact that PGC-1α is involved in the development of obesity and T2D. Lower Pgc-1α levels that are commonly seen in obese and T2D patients could lead to progression of these disorders possibly as a result of impaired mitochondrial metabolism.
3.3. Other NEMGs
According to the MitoCarta2.0 database, there are 1158 genes that play a role in determining mitochondrial number and function [128]. Most notable among these with respect to obesity and insulin resistance (in addition to those previously discussed), which are also coactivated by PGC-1α, are Ppar-α, Ppar-γ, and estrogen-related receptor-α (Err-α). The nuclear receptor PPAR-α is activated by prostaglandin derivatives, eicosanoids, and long chain fatty acids, and is responsible for transcriptional regulation of genes related to lipid metabolism and regulates cellular lipid utilization [129,130,131]. PPAR-α target genes such as medium-chain acyl-CoA dehydrogenase (Mcad) and Cpt-1 are responsible for determining cellular FAO [67,130,132]. Studies utilizing Ppar-α null mice have shown that PPAR-α is essential for expression of mitochondrial FAO genes [133]. PPAR-ɣ is known to enhance insulin sensitivity in peripheral tissues [134], and synthetic PPAR-ɣ agonists, such as pioglitazone and rosiglitazone, are approved by the FDA for treatment of T2D [134]. Muscle specific deletion of Ppar-γ in mice leads to insulin resistance [135,136], whereas overexpression of Ppar-γ upregulates skeletal muscle cytochrome c and cytochrome oxidase and prevents diet-induced insulin resistance [137]. ERR-α is involved in PGC-1α-induced mitochondrial biogenesis, and Err-α inhibition prevents PGC-1α-induced expression of genes responsible for stimulating mitochondrial biogenesis, such as Tfam, and decreases mtDNA content [138]. Putative ERR-α binding sites were found in the regulatory region of mitochondrial biogenesis regulating genes that are induced by ERR-α and PGC-1α [138]. Err-α expression in concert with Pgc-1α regulates substrate utilization to promotes FAO rather than glucose oxidation in skeletal muscle [139], which may act to prevent ectopic lipid deposition and attenuate or prevent insulin resistance during obesity.
4. HFD-Induced Obesity, Insulin Resistance, and Retrograde Communication
HFD feeding resulting in obesity and insulin resistance leads to mitochondrial dysfunction that is partially evidenced by incomplete beta oxidation of fatty acids and ectopic lipid deposition [76,82,119]. Specifically in the skeletal muscle, HFD-induced obesity is associated with the accumulation of FAO intermediates in the form of long and medium chain acylcarnitines in the skeletal muscle that occurs as a result of incomplete beta oxidation [76,82,119]. Accumulation of acylcarnitines as a consequence of incomplete mitochondrial beta oxidation of fatty acids is associated with development of insulin resistance in the skeletal muscle [140]. While diets high in dietary fat content induce obesity and insulin resistance in association with incomplete FAO [76,82,119], dietary supplementation with mono- or polyunsaturated fatty acids may prevent insulin resistance through alterations in intraceullular fatty acid metabolites and mitochondrial morphology and function [141,142,143,144]. Thus, dietary fat composition may differentially affect the completeness of beta oxidation and specific levels or ratios of acylcarnitines that accumulate within the skeletal muscle and is an area worthy of further investigation. Because studies investigating the effects of dietary fat composition are lacking, this study focuses on the effects of dietary fat content and discuss the formation, transport, and localization of acylcarnitines, and the role of acylcarnitines in HFD-induced obesity and T2D.
4.1. Mitochondrial Transport of Intracellular Fatty Acids for Oxidation
Mitochondrial beta oxidation of fatty acids is essential for the correct functioning of many cell types [145]. The inability of fatty acids to enter the mitochondria to undergo beta oxidation is associated with several disease states and eventually leads to death [145]. However, fatty acids, which occur intracellularly in the form of acyl-CoAs, cannot initially participate in beta oxidation as they are unable to traverse the impermeable inner membrane of the mitochondrion [118]. CPT-1, a NEMG and enzyme that is present on the surface of the outer mitochondrial membrane, acts to convert fatty acyl-CoAs to acylcarnitines that can easily traverse the membrane [146]. Acylcarnitines that cross the outer mitochondrial membrane subsequently translocate through the inner mitochondrial membrane and into the mitochondrial matrix by carnitine/acylcarnitine translocase in exchange for intramitochondrial free carnitine [147,148]. Inside the matrix, carnitine palmitoyltransferase 2 (CPT-2) converts acylcarnitines into free carnitine and fatty acyl CoAs, which can then be oxidized [149].
4.2. Incomplete Beta-Oxidation and Acylcarnitines
Incomplete beta oxidation has been shown to play a role in development of insulin resistance in the skeletal muscle [82]. During complete beta oxidation of fatty acids, the two carbon fatty acid acetyl-CoA is produced during each step, leading to a reduction of the input acyl-CoA chain length by two carbon atoms and forming acetyl-coA as the end product of mitochondrial beta oxidation [118]. Acetyl-CoA may be further metabolized in the TCA cycle and the NADH and FADH2 formed during the TCA cycle is utilized to produce ATP in the ETC [150]. If the rate of acetyl-CoA production through beta oxidation exceeds the functional capacity of the TCA cycle or ETC, acetyl-coA may feedback to inhibit beta oxidation resulting in incomplete oxidization of acyl-CoAs that accumulate intracellularly [82]. Because translocation of acylcarnitines into the mitochondrial matrix is maintained at an equilibrium, while levels of total CoA and carnitine inside the matrix are equal [151], if acyl-CoAs are incompletely oxidized and accumulate, their respective acylcarnitines are formed [151]. Thus, metabolomics approaches that detect and measure specific species of acylcarnitines can help detect acyl-CoA accumulation of varying chain lengths, represented by acylcarnitines, and thus determine the completeness of beta oxidation of fatty acids [152]. Indeed, accumulation of long and medium chain acylcarnitines, indicating incomplete beta oxidation of fatty acids, is observed during HFD-induced obesity and insulin resistance [82].
4.3. Role of Acylcarnitines in Obesity and T2D
Various studies have shown that incomplete beta oxidation and the accumulation of long and medium chain acylcarnitines is involved in the development of insulin resistance [82,140,153]. In obese Zucker diabetic rats, long chain acylcarnitines are increased in the skeletal muscle [82]. Human subjects with obesity and T2D also show acylcarnitine accumulation and incomplete beta oxidation in the skeletal muscle [119,140]. When primary myocytes obtained from subjects with obesity were cultured with lipolytically active adipocytes, medium- and long-chain acylcarnitines accumulated [154]. This accumulation was not observed in primary myocytes cultured without these adipocytes [154]. In the skeletal muscle, knockdown of the enzyme carnitine acetyltransferase (CrAT), which preferentially converts short- and medium-chain fatty acids to acylcarnitines [155], leads to increased amounts long chain acylcarnitines in association with glucose intolerance [153]. Conversely, when incomplete beta oxidation and accumulation of acylcarnitines in the mitochondrial matrix is inhibited through inhibition of CPT-1, HFD fed mice are protected from the development of insulin resistance [156].
Acylcarnitines may affect the insulin signaling pathway in the skeletal muscle through retrograde communication that contributes to nucleo-mitochondrial crosstalk [22,62,76,157]. Several products of mitochondrial metabolism are known to affect NEMG expression. For example, increased cytosolic Ca2+ concentrations, resulting from compromised ATP synthesis or disruption of the mitochondrial membrane potential, alters NEMG expression [62]. Furthermore, reactive oxygen species (ROS), which may be produced as a byproduct of mitochondrial respiration, and glucose or fatty acid levels also regulate NEMG [60,158,159]. mtDNA mutation or depletion is known to alter NEMG expression while also compromising respiration to effect changes in metabolic products/byproducts which may act as signaling molecules in retrograde communication [27,60,160]. Because acylcarnitines result as products/byproducts/intermediates of mitochondrial beta oxidation of fatty acids that may be transported throughout the cell, they may play a role in retrograde communication to determine NEMG expression, obesity and insulin resistance [22,62,76,157]. Interestingly, in a study by Aguer et al., myocytes treated with acylcarnitines (C4:0, C14:0, or C16:0) exhibited increased oxidative stress, and insulin sensitivity was reduced by 20–30% [161]. Furthermore, inhibition of carnitine acyltransferases CPT-1 and CPT-2, resulting in inhibition of medium and long chain acylcarnitine synthesis and accumulation, prevents fatty acid-induced insulin resistance in muscle [161]. However, upregulation of CrAT, which produces short chain acylcarnitines, allows for increased efflux of short chain fatty acids from the mitochondria and may alleviate inhibition of glucose catabolism to increase insulin sensitivity [153,162]. Previous studies have shown that acetyl-coA availability determines histone acetylation and is linked to DNA methylation and that fatty acids can be targeted to the nucleus, although the mechanisms of action for fatty acid transport into the nucleus through a carnitine shuttle in mammalian cells remain unknown [157,163,164,165]. These data are compelling given that retrograde communication that determines NEMG expression may occur through an epigenetic mechanism involving acyl-coAs, as several NEMG have recently been shown to be epigenetically regulated by HFD, fatty acids, and exercise [76,166,167]. Thus, HFD feeding leading to incomplete beta oxidation and accumulation of long and medium chain acylcarnitines or increased efflux of short chain acylcarnitines from the mitochondria may play a contributory role in the development of obesity and insulin resistance in the skeletal muscle through retrograde communication to regulate NEMG expression and mitochondrial adaptations.
5. Conclusions
Decreased NEMG expression and low mitochondrial number and dysfunction are observed in obesity and T2D [18,19,21,121,122,123]. Based on the literature discussed in this review, it can be hypothesized that medium and long chain fatty acids accumulated as a result of HFD-induced incomplete beta oxidation could negatively regulate NEMG expression via retrograde communication with the nucleus (Figure 1). This in turn would further reduce mitochondrial number and function through anterograde communication, creating a vicious aberrant metabolic cycle (Figure 1). Investigation of this hypothesis can potentially provide insights about the mechanisms responsible for progressive loss of mitochondrial function and development of insulin resistance seen in obesity and T2D.
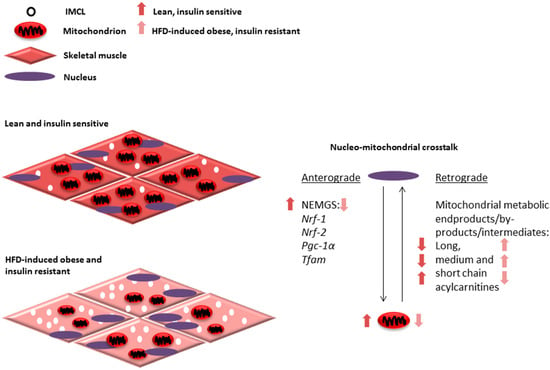
Figure 1.
Mitochondrial adaptations, such as alterations in number and function, are determined through coordinated communication that occurs between the nucleus and mitochondria. This communication or nucleo-mitochondrial crosstalk is a two-way process involving anterograde communication from the nucleus to the mitochondria and retrograde communication from the mitochondria to the nucleus (right). The expression of several nuclear-encoded mitochondrial genes (NEMGs) are known to regulate skeletal muscle mitochondrial number and function, including Nrf-1, Nrf-2, Pgc-1α and Tfam and contribute to anterograde communication. Alternatively, mitochondrial protein or metabolic products are known to play a role in retrograde communication to determine NEMG expression. We propose that mitochondrial beta oxidation intermediates, by-products and products, such as long, medium and short acylcarnitines, may act as part of this retrograde communication. Interestingly, in response to high fat diet (HFD) feeding and during obesity and insulin resistance, a downregulation in NEMG expression is seen. Alterations in acylcarnitine profiles suggesting incomplete beta oxidation with accumulation of long and medium chain acylcarnitines with respect to short chain acylcarnitines (arrows represent ratios of skeletal muscle acylcarnitines in comparison between lean, insulin sensitive, and obese, insulin resistant states) and decreased mitochondrial number and function. These alterations may contribute to increased intramyocellular lipid (IMCL) accumulation and insulin resistance during HFD-induced obesity (left).
Acknowledgments
This work was funded in part by Purdue University Ralph W. and Grace M. Showalter Trust Fund (Tara M. Henagan).
Author Contributions
Prasad P. Devarshi reviewed the literature and wrote the manuscript; Sean M. McNabney helped edit and write the manuscript; Tara M. Henagan reviewed the literature, conceptualized the manuscript, and helped write and edit the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ogden, C.L.; Carroll, M.D.; Kit, B.K.; Flegal, K.M. Prevalence of childhood and adult obesity in the united states, 2011–2012. JAMA 2014, 311, 806–814. [Google Scholar] [CrossRef] [PubMed]
- 2014 Statistics report|Data & Statistics|Diabetes|cdc. Available online: http://www.cdc.gov/diabetes/data/statistics/2014statisticsreport.html (accessed on 13 April 2017).
- Laaksonen, D.E.; Lakka, H.M.; Salonen, J.T.; Niskanen, L.K.; Rauramaa, R.; Lakka, T.A. Low levels of leisure-time physical activity and cardiorespiratory fitness predict development of the metabolic syndrome. Diabetes Care 2002, 25, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Haapanen, N.; Miilunpalo, S.; Vuori, I.; Oja, P.; Pasanen, M. Association of leisure time physical activity with the risk of coronary heart disease, hypertension and diabetes in middle-aged men and women. Int. J. Epidemiol. 1997, 26, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Despres, J.P.; Hu, F.B. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation 2010, 121, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Tuomilehto, J.; Lindstrom, J.; Eriksson, J.G.; Valle, T.T.; Hamalainen, H.; Ilanne-Parikka, P.; Keinanen-Kiukaanniemi, S.; Laakso, M.; Louheranta, A.; Rastas, M.; et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N. Engl. J. Med. 2001, 344, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Wadden, T.A.; Webb, V.L.; Moran, C.H.; Bailer, B.A. Lifestyle modification for obesity: New developments in diet, physical activity, and behavior therapy. Circulation 2012, 125, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Rahmouni, K.; Correia, M.L.; Haynes, W.G.; Mark, A.L. Obesity-associated hypertension: New insights into mechanisms. Hypertension 2004, 45, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, N.; Kern, P.A. Adipocytokines and the metabolic complications of obesity. J. Clin. Endocrinol. Metab. 2008, 93, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.M.; Cooper, M.E. Mechanisms of diabetic complications. Physiol. Rev. 2013, 93, 137–188. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, M.; Sahlin, K.; Fernstrom, M.; Glintborg, D.; Vind, B.F.; Beck-Nielsen, H.; Hojlund, K. Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes 2007, 56, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Hojlund, K.; Mogensen, M.; Sahlin, K.; Beck-Nielsen, H. Mitochondrial dysfunction in type 2 diabetes and obesity. Endocrinol. Metab. Clin. N. Am. 2008, 37, 713–731. [Google Scholar] [CrossRef] [PubMed]
- Dandona, P.; Aljada, A.; Chaudhuri, A.; Mohanty, P.; Garg, R. Metabolic syndrome: A comprehensive perspective based on interactions between obesity, diabetes, and inflammation. Circulation 2005, 111, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Antuna-Puente, B.; Feve, B.; Fellahi, S.; Bastard, J.P. Adipokines: The missing link between insulin resistance and obesity. Diabetes Metab. 2007, 34, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Kersten, S.; Hesselink, M.K.; Schrauwen, P. Re-evaluating lipotoxic triggers in skeletal muscle: Relating intramyocellular lipid metabolism to insulin sensitivity. Prog. Lipid Res. 2011, 51, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Sivitz, W.I.; Yorek, M.A. Mitochondrial dysfunction in diabetes: From molecular mechanisms to functional significance and therapeutic opportunities. Antioxid. Redox Signal. 2010, 12, 537–577. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Wei, Y.; Sowers, J.R. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 2008, 102, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Hickner, R.C.; Cortright, R.L.; Dohm, G.L.; Houmard, J.A. Lipid oxidation is reduced in obese human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E1039–E1044. [Google Scholar] [PubMed]
- Petersen, K.F.; Dufour, S.; Befroy, D.; Garcia, R.; Shulman, G.I. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 2004, 350, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Simoneau, J.A.; Kelley, D.E. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in niddm. J. Appl. Physiol. 1997, 83, 166–171. [Google Scholar] [PubMed]
- Boushel, R.; Gnaiger, E.; Schjerling, P.; Skovbro, M.; Kraunsoe, R.; Dela, F. Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia 2007, 50, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Garesse, R.; Vallejo, C.G. Animal mitochondrial biogenesis and function: A regulatory cross-talk between two genomes. Gene 2001, 263, 1–16. [Google Scholar] [CrossRef]
- Montgomery, M.K.; Turner, N. Mitochondrial dysfunction and insulin resistance: An update. Endocr. Connect. 2014, 4, R1–R15. [Google Scholar] [CrossRef] [PubMed]
- Lowell, B.B.; Shulman, G.I. Mitochondrial dysfunction and type 2 diabetes. Science 2005, 307, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Grivell, L.A. Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 121–164. [Google Scholar] [CrossRef] [PubMed]
- Poyton, R.O.; McEwen, J.E. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 1996, 65, 563–607. [Google Scholar] [CrossRef] [PubMed]
- Angelini, C.; Bello, L.; Spinazzi, M.; Ferrati, C. Mitochondrial disorders of the nuclear genome. Acta Myol. 2009, 28, 16–23. [Google Scholar] [PubMed]
- Lang, B.F.; Gray, M.W.; Burger, G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 2000, 33, 351–397. [Google Scholar] [CrossRef] [PubMed]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Gershoni, M.; Templeton, A.R.; Mishmar, D. Mitochondrial bioenergetics as a major motive force of speciation. BioEssays 2009, 31, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Das, J. The role of mitochondrial respiration in physiological and evolutionary adaptation. BioEssays 2006, 28, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, A.; Moraes, C.T. Simultaneous transfer of mitochondrial DNA and single chromosomes in somatic cells: A novel approach for the study of defects in nuclear-mitochondrial communication. Hum. Mol. Genet. 1998, 7, 1801–1808. [Google Scholar] [CrossRef] [PubMed]
- Mastromonaco, G.F.; Favetta, L.A.; Smith, L.C.; Filion, F.; King, W.A. The influence of nuclear content on developmental competence of gaur × cattle hybrid in vitro fertilized and somatic cell nuclear transfer embryos. Biol. Reprod. 2006, 76, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Kazuno, A.A.; Munakata, K.; Nagai, T.; Shimozono, S.; Tanaka, M.; Yoneda, M.; Kato, N.; Miyawaki, A.; Kato, T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006, 2, e128. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.P.; Zuckerbraun, B.S. Mitochondrial signaling: Forwards, backwards, and in between. Oxid Med. Cell Longev. 2013, 2013, 351613. [Google Scholar] [CrossRef] [PubMed]
- Campello, S.; Scorrano, L. Mitochondrial shape changes: Orchestrating cell pathophysiology. EMBO Rep. 2010, 11, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Veltri, K.L.; Espiritu, M.; Singh, G. Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol. 1990, 143, 160–164. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.A.; Griffin, J.L.; Murray, A.J.; Edwards, L.M. Mitochondrial responses to extreme environments: Insights from metabolomics. Extrem. Physiol. Med. 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Nisoli, E.; Tonello, C.; Cardile, A.; Cozzi, V.; Bracale, R.; Tedesco, L.; Falcone, S.; Valerio, A.; Cantoni, O.; Clementi, E.; et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of enos. Science 2005, 310, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Pulakat, L.; Whaley-Connell, A.; Sowers, J.R. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J. Mol. Med. 2010, 88, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Dominy, J.E.; Puigserver, P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a015008. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Yoon, J.C.; Puigserver, P.; Chen, G.; Donovan, J.; Wu, Z.; Rhee, J.; Adelmant, G.; Stafford, J.; Kahn, C.R.; Granner, D.K.; et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001, 413, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Baar, K.; Wende, A.R.; Jones, T.E.; Marison, M.; Nolte, L.A.; Chen, M.; Kelly, D.P.; Holloszy, J.O. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002, 16, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.P.; Scarpulla, R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.B.; Lovely, G.A.; Phillips, R.; Chan, D.C. Distinct structural features of tfam drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 2014, 5, 3077. [Google Scholar] [CrossRef] [PubMed]
- Scarpulla, R.C. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 2002, 286, 81–89. [Google Scholar] [CrossRef]
- Safdar, A.; Little, J.P.; Stokl, A.J.; Hettinga, B.P.; Akhtar, M.; Tarnopolsky, M.A. Exercise increases mitochondrial PGC-1α content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J. Biol. Chem. 2011, 286, 10605–10617. [Google Scholar] [CrossRef] [PubMed]
- Finck, B.N.; Kelly, D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Investig. 2006, 116, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Aquilano, K.; Vigilanza, P.; Baldelli, S.; Pagliei, B.; Rotilio, G.; Ciriolo, M.R. Peroxisome proliferator-activated receptor gamma co-activator 1α (PGC-1α) and sirtuin 1 (SIRT1) reside in mitochondria: Possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010, 285, 21590–21599. [Google Scholar] [CrossRef] [PubMed]
- Virbasius, C.A.; Virbasius, J.V.; Scarpulla, R.C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993, 7, 2431–2445. [Google Scholar] [CrossRef] [PubMed]
- Virbasius, J.V.; Virbasius, C.A.; Scarpulla, R.C. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993, 7, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Schieke, S.M.; Phillips, D.; McCoy, J.P., Jr.; Aponte, A.M.; Shen, R.F.; Balaban, R.S.; Finkel, T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006, 281, 27643–27652. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Butow, R.A. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999, 19, 6720–6728. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T. Oxygen radicals and signaling. Curr. Opin. Cell Biol. 1998, 10, 248–253. [Google Scholar] [CrossRef]
- O’Hagan, H.M.; Wang, W.; Sen, S.; Destefano Shields, C.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Van Neste, L.; Easwaran, H.; et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter cpg islands. Cancer Cell 2011, 20, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Sekito, T.; Thornton, J.; Butow, R.A. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 2000, 11, 2103–2115. [Google Scholar] [CrossRef] [PubMed]
- Freije, W.A.; Mandal, S.; Banerjee, U. Expression profiling of attenuated mitochondrial function identifies retrograde signals in drosophila. G3 2012, 2, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Biswas, G.; Adebanjo, O.A.; Freedman, B.D.; Anandatheerthavarada, H.K.; Vijayasarathy, C.; Zaidi, M.; Kotlikoff, M.; Avadhani, N.G. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: A novel mode of inter-organelle crosstalk. EMBO J. 1999, 18, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yen, K.; Cohen, P. Humanin: A harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metab. 2013, 24, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, N.H.; Zwart, R.; Wolterman, R.A.; Ruiter, J.P.; Wanders, R.J.; Bolhuis, P.A.; van den Bogert, C. Differentiation and proliferation of respiration-deficient human myoblasts. Biochim. Biophys. Acta 1993, 1181, 63–67. [Google Scholar] [CrossRef]
- Laeng, H.; Schneider, E.; Bolli, R.; Zimmermann, A.; Schaffner, T.; Schindler, R. Participation of mitochondrial proliferation in morphological differentiation of murine mastocytoma cells. Exp. Cell Res. 1988, 179, 222–232. [Google Scholar] [CrossRef]
- Rochard, P.; Rodier, A.; Casas, F.; Cassar-Malek, I.; Marchal-Victorion, S.; Daury, L.; Wrutniak, C.; Cabello, G. Mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression and activity of myogenic factors. J. Biol. Chem. 2000, 275, 2733–2744. [Google Scholar] [CrossRef] [PubMed]
- Gulick, T.; Cresci, S.; Caira, T.; Moore, D.D.; Kelly, D.P. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. USA 1994, 91, 11012–11016. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.; Petrescu, A.D.; Huang, H.; Atshaves, B.P.; McIntosh, A.L.; Martin, G.G.; Hostetler, H.A.; Vespa, A.; Landrock, D.; Landrock, K.K.; et al. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids 2007, 43, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.H.; Naito, A.; Mizumachi, T.; Evans, T.T.; Douglas, M.G.; Cooney, C.A.; Fan, C.Y.; Higuchi, M. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem. Biophys. Res. Commun. 2007, 364, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Smiraglia, D.J.; Kulawiec, M.; Bistulfi, G.L.; Gupta, S.G.; Singh, K.K. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 2008, 7, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kwak, S.H.; Bhak, J.; Kang, H.S.; Lee, Y.R.; Koo, B.K.; Park, K.S.; Lee, H.K.; Cho, Y.M. Gene expression pattern in transmitochondrial cytoplasmic hybrid cells harboring type 2 diabetes-associated mitochondrial DNA haplogroups. PLoS ONE 2011, 6, e22116. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.E.; Canham, J.P.; Consitt, L.A.; Zheng, D.; Koves, T.R.; Gavin, T.P.; Holbert, D.; Neufer, P.D.; Ilkayeva, O.; Muoio, D.M.; et al. A high-fat diet elicits differential responses in genes coordinating oxidative metabolism in skeletal muscle of lean and obese individuals. J. Clin. Endocrinol. Metab. 2010, 96, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Iossa, S.; Mollica, M.P.; Lionetti, L.; Crescenzo, R.; Botta, M.; Liverini, G. Skeletal muscle oxidative capacity in rats fed high-fat diet. Int. J. Obes. Relat. Metab. Disord. 2002, 26, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Sparks, L.M.; Xie, H.; Koza, R.A.; Mynatt, R.; Hulver, M.W.; Bray, G.A.; Smith, S.R. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 2005, 54, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Henagan, T.M.; Cefalu, W.T.; Ribnicky, D.M.; Noland, R.C.; Dunville, K.; Campbell, W.W.; Stewart, L.K.; Forney, L.A.; Gettys, T.W.; Chang, J.S.; et al. In vivo effects of dietary quercetin and quercetin-rich red onion extract on skeletal muscle mitochondria, metabolism, and insulin sensitivity. Genes Nutr. 2015, 10, 451. [Google Scholar] [CrossRef] [PubMed]
- Winzell, M.S.; Ahren, B. The high-fat diet-fed mouse: A model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes 2004, 53, 215–219. [Google Scholar] [CrossRef]
- De Wilde, J.; Mohren, R.; van den Berg, S.; Boekschoten, M.; Dijk, K.W.; de Groot, P.; Muller, M.; Mariman, E.; Smit, E. Short-term high fat-feeding results in morphological and metabolic adaptations in the skeletal muscle of C57BL/6J mice. Physiol. Genom. 2007, 32, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Roves, P.; Huss, J.M.; Han, D.H.; Hancock, C.R.; Iglesias-Gutierrez, E.; Chen, M.; Holloszy, J.O. Raising plasma fatty acid concentration induces increased biogenesis of mitochondria in skeletal muscle. Proc. Natl. Acad. Sci. USA 2007, 104, 10709–10713. [Google Scholar] [CrossRef] [PubMed]
- Yuzefovych, L.; Wilson, G.; Rachek, L. Different effects of oleate vs. palmitate on mitochondrial function, apoptosis, and insulin signaling in l6 skeletal muscle cells: Role of oxidative stress. Am. J. Physiol. Endocrinol. Metab. 2010, 299, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, T.; Grunnet, N.; Quistorff, B. One-year high fat diet affects muscle-but not brain mitochondria. J. Cereb. Blood Flow Metab. 2015, 35, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Koves, T.R.; Ussher, J.R.; Noland, R.C.; Slentz, D.; Mosedale, M.; Ilkayeva, O.; Bain, J.; Stevens, R.; Dyck, J.R.; Newgard, C.B.; et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008, 7, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.K.; Wang, Z.; Ribnicky, D.; Soileau, J.L.; Cefalu, W.T.; Gettys, T.W. Failure of dietary quercetin to alter the temporal progression of insulin resistance among tissues of C57BL/6J mice during the development of diet-induced obesity. Diabetologia 2009, 52, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Lehman, J.J.; Barger, P.M.; Kovacs, A.; Saffitz, J.E.; Medeiros, D.M.; Kelly, D.P. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 2000, 106, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Scarpulla, R.C. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990, 4, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Scarpulla, R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta 2002, 1576, 1–14. [Google Scholar] [CrossRef]
- Virbasius, J.V.; Scarpulla, R.C. Activation of the human mitochondrial transcription factor a gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 1309–1313. [Google Scholar] [CrossRef] [PubMed]
- Ekstrand, M.I.; Falkenberg, M.; Rantanen, A.; Park, C.B.; Gaspari, M.; Hultenby, K.; Rustin, P.; Gustafsson, C.M.; Larsson, N.G. Mitochondrial transcription factor a regulates mtdna copy number in mammals. Hum. Mol. Genet. 2004, 13, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Scarpulla, R.C. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001, 21, 3738–3749. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Scarpulla, R.C. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol. Cell. Biol. 2001, 21, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Miranda, S.; Foncea, R.; Guerrero, J.; Leighton, F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted hela cells. Biochem. Biophys. Res. Commun. 1999, 258, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Suliman, H.B.; Carraway, M.S.; Welty-Wolf, K.E.; Whorton, A.R.; Piantadosi, C.A. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J. Biol. Chem. 2003, 278, 41510–41518. [Google Scholar] [CrossRef] [PubMed]
- Patti, M.E.; Butte, A.J.; Crunkhorn, S.; Cusi, K.; Berria, R.; Kashyap, S.; Miyazaki, Y.; Kohane, I.; Costello, M.; Saccone, R.; et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 2003, 100, 8466–8471. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.S.; Hajira, A.; Pardo, P.S.; Boriek, A.M. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT1/PGC-1α network in skeletal muscle. Diabetes 2014, 63, 1546–1559. [Google Scholar] [CrossRef] [PubMed]
- Mathai, A.S.; Bonen, A.; Benton, C.R.; Robinson, D.L.; Graham, T.E. Rapid exercise-induced changes in PGC-1α mRNA and protein in human skeletal muscle. J. Appl. Physiol. 2008, 105, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Civitarese, A.E.; Carling, S.; Heilbronn, L.K.; Hulver, M.H.; Ukropcova, B.; Deutsch, W.A.; Smith, S.R.; Ravussin, E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007, 4, e76. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Feilchenfeldt, J.; Schreiber, S.; Praz, M.; Crettenand, A.; Gobelet, C.; Meier, C.A.; Bell, D.R.; Kralli, A.; Giacobino, J.P.; et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-γ coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes 2003, 52, 2874–2881. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.A. New insights into PGC-1 coactivators: Redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2014, 282, 647–672. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Huss, J.M.; Kelly, D.P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor-alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000, 20, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, H.; Tarr, P.T.; Zhang, C.Y.; Wu, Z.; Boss, O.; Michael, L.F.; Puigserver, P.; Isotani, E.; Olson, E.N.; et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 2002, 418, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Uldry, M.; Yang, W.; St-Pierre, J.; Lin, J.; Seale, P.; Spiegelman, B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Scarpulla, R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta 2010, 1813, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Little, J.P.; Safdar, A.; Bishop, D.; Tarnopolsky, M.A.; Gibala, M.J. An acute bout of high-intensity interval training increases the nuclear abundance of PGC-1α and activates mitochondrial biogenesis in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.; Stevens, J.; Binder-Macleod, S.A. Human skeletal muscle fiber type classifications. Phys. Ther. 2001, 81, 1810–1816. [Google Scholar] [PubMed]
- Zechner, C.; Lai, L.; Zechner, J.F.; Geng, T.; Yan, Z.; Rumsey, J.W.; Collia, D.; Chen, Z.; Wozniak, D.F.; Leone, T.C.; et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010, 12, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Leone, T.C.; Lehman, J.J.; Finck, B.N.; Schaeffer, P.J.; Wende, A.R.; Boudina, S.; Courtois, M.; Wozniak, D.F.; Sambandam, N.; Bernal-Mizrachi, C.; et al. PGC-1α deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005, 3, 101. [Google Scholar] [CrossRef] [PubMed]
- Benton, C.R.; Holloway, G.P.; Han, X.X.; Yoshida, Y.; Snook, L.A.; Lally, J.; Glatz, J.F.; Luiken, J.J.; Chabowski, A.; Bonen, A. Increased levels of peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1α) improve lipid utilisation, insulin signalling and glucose transport in skeletal muscle of lean and insulin-resistant obese zucker rats. Diabetologia 2010, 53, 2008–2019. [Google Scholar] [CrossRef] [PubMed]
- Jogl, G.; Hsiao, Y.S.; Tong, L. Structure and function of carnitine acyltransferases. Ann. N. Y. Acad. Sci. 2004, 1033, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Gerhart-Hines, Z.; Rodgers, J.T.; Bare, O.; Lerin, C.; Kim, S.H.; Mostoslavsky, R.; Alt, F.W.; Wu, Z.; Puigserver, P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 2007, 26, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Banks, A.S.; Kon, N.; Knight, C.; Matsumoto, M.; Gutierrez-Juarez, R.; Rossetti, L.; Gu, W.; Accili, D. SIRT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.C.; Lambert, P.D.; Schenk, S.; Carney, D.P.; Smith, J.J.; Gagne, D.J.; Jin, L.; Boss, O.; Perni, R.B.; Vu, C.B.; et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007, 450, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Handschin, C.; St-Pierre, J.; Spiegelman, B.M. Amp-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. USA 2007, 104, 12017–12022. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Rhee, J.; Lin, J.; Wu, Z.; Yoon, J.C.; Zhang, C.Y.; Krauss, S.; Mootha, V.K.; Lowell, B.B.; Spiegelman, B.M. Cytokine stimulation of energy expenditure through p38 map kinase activation of PPARγ coactivator-1. Mol. Cell 2001, 8, 971–982. [Google Scholar] [CrossRef]
- Canto, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. Ampk regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Koves, T.R.; Li, P.; An, J.; Akimoto, T.; Slentz, D.; Ilkayeva, O.; Dohm, G.L.; Yan, Z.; Newgard, C.B.; Muoio, D.M. Peroxisome proliferator-activated receptor-gamma co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J. Biol. Chem. 2005, 280, 33588–33598. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H. Beta oxidation of fatty acids. Biochim. Biophys. Acta 1991, 1081, 109–120. [Google Scholar] [CrossRef]
- Adams, S.H.; Hoppel, C.L.; Lok, K.H.; Zhao, L.; Wong, S.W.; Minkler, P.E.; Hwang, D.H.; Newman, J.W.; Garvey, W.T. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic african-american women. J. Nutr. 2009, 139, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Jacob, S.; Machann, J.; Rett, K.; Brechtel, K.; Volk, A.; Renn, W.; Maerker, E.; Matthaei, S.; Schick, F.; Claussen, C.D.; et al. Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes 1999, 48, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Ward, W.F. PGC-1α: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Semple, R.K.; Crowley, V.C.; Sewter, C.P.; Laudes, M.; Christodoulides, C.; Considine, R.V.; Vidal-Puig, A.; O’Rahilly, S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1α is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obes. Relat. Metab. Disord. 2003, 28, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Ek, J.; Andersen, G.; Urhammer, S.A.; Gaede, P.H.; Drivsholm, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia 2002, 44, 2220–2226. [Google Scholar] [CrossRef] [PubMed]
- Andrulionyte, L.; Zacharova, J.; Chiasson, J.L.; Laakso, M. Common polymorphisms of the PPAR-γ2 (Pro12Ala) and PGC-1α (Gly482Ser) genes are associated with the conversion from impaired glucose tolerance to type 2 diabetes in the stop-niddm trial. Diabetologia 2004, 47, 2176–2184. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Tobe, K.; Okada, T.; Kadowaki, H.; Akanuma, Y.; Ito, C.; Kimura, S.; Kadowaki, T. A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to type II diabetes. Diabetologia 2002, 45, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Muller, Y.L.; Bogardus, C.; Pedersen, O.; Baier, L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in pima indians. Diabetes 2003, 52, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. Mitocarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015, 44, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.V.; Torres, N.; Tovar, A.R. PPAR-α as a key nutritional and environmental sensor for metabolic adaptation. Adv. Nutr. 2013, 4, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Lemberger, T.; Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: A nuclear receptor signaling pathway in lipid physiology. Annu. Rev. Cell Dev. Biol. 1996, 12, 335–363. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Mascaro, C.; Acosta, E.; Ortiz, J.A.; Marrero, P.F.; Hegardt, F.G.; Haro, D. Control of human muscle-type carnitine palmitoyltransferase I gene transcription by peroxisome proliferator-activated receptor. J. Biol. Chem. 1998, 273, 8560–8563. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Peters, J.M.; Iritani, N.; Nakajima, T.; Furihata, K.; Hashimoto, T.; Gonzalez, F.J. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARα). J. Biol. Chem. 1998, 273, 5678–5684. [Google Scholar] [CrossRef] [PubMed]
- Yki-Jarvinen, H. Thiazolidinediones. N. Engl. J. Med. 2004, 351, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Hevener, A.L.; He, W.; Barak, Y.; Le, J.; Bandyopadhyay, G.; Olson, P.; Wilkes, J.; Evans, R.M.; Olefsky, J. Muscle-specific pparg deletion causes insulin resistance. Nat. Med. 2003, 9, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.W.; Chen, L.; Fisher, S.J.; Szanto, I.; Ristow, M.; Jozsi, A.C.; Hirshman, M.F.; Rosen, E.D.; Goodyear, L.J.; Gonzalez, F.J.; et al. Muscle-specific PPARγ-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J. Clin. Investig. 2003, 112, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.H.; Mathews, S.T.; Camp, H.S.; Ding, L.; Leff, T. Selective activation of PPARγ in skeletal muscle induces endogenous production of adiponectin and protects mice from diet-induced insulin resistance. Am. J. Physiol. Endocrinol. Metabol. 2009, 298, E28–E37. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, S.N.; Emter, R.; Hock, M.B.; Knutti, D.; Cardenas, J.; Podvinec, M.; Oakeley, E.J.; Kralli, A. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 6472–6477. [Google Scholar] [CrossRef] [PubMed]
- Wende, A.R.; Huss, J.M.; Schaeffer, P.J.; Giguere, V.; Kelly, D.P. PGC-1α coactivates PDK4 gene expression via the orphan nuclear receptor ERRα: A mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell. Biol. 2005, 25, 10684–10694. [Google Scholar] [CrossRef] [PubMed]
- Mihalik, S.J.; Goodpaster, B.H.; Kelley, D.E.; Chace, D.H.; Vockley, J.; Toledo, F.G.; DeLany, J.P. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity 2010, 18, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Tierney, A.C.; Roche, H.M. The potential role of olive oil-derived mufa in insulin sensitivity. Mol. Nutr. Food Res. 2007, 51, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Albert, B.B.; Derraik, J.G.; Brennan, C.M.; Biggs, J.B.; Smith, G.C.; Garg, M.L.; Cameron-Smith, D.; Hofman, P.L.; Cutfield, W.S. Higher ω-3 index is associated with increased insulin sensitivity and more favourable metabolic profile in middle-aged overweight men. Sci. Rep. 2014, 4, 6697. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Pinnamaneni, S.K.; Eo, S.J.; Cho, I.H.; Pyo, J.H.; Kim, C.K.; Sinclair, A.J.; Febbraio, M.A.; Watt, M.J. Saturated, but not n-6 polyunsaturated, fatty acids induce insulin resistance: Role of intramuscular accumulation of lipid metabolites. J. Appl. Physiol. 2005, 100, 1467–1474. [Google Scholar] [CrossRef] [PubMed]
- Putti, R.; Migliaccio, V.; Sica, R.; Lionetti, L. Skeletal muscle mitochondrial bioenergetics and morphology in high fat diet induced obesity and insulin resistance: Focus on dietary fat source. Front. Physiol. 2015, 6, 426. [Google Scholar] [CrossRef] [PubMed]
- Moczulski, D.; Majak, I.; Mamczur, D. An overview of beta-oxidation disorders. Postepy Hig. Med. Dosw. 2009, 63, 266–277. [Google Scholar]
- Lee, K.; Kerner, J.; Hoppel, C.L. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J. Biol. Chem. 2011, 286, 25655–25662. [Google Scholar] [CrossRef] [PubMed]
- Zeth, K.; Thein, M. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem. J. 2010, 431, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, C.; Iacobazzi, V.; Tonazzi, A.; Giangregorio, N.; Infantino, V.; Convertini, P.; Console, L.; Palmieri, F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Asp. Med. 2011, 32, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R.; Gandour, R.D.; van der Leij, F.R. Molecular enzymology of carnitine transfer and transport. Biochim. Biophys. Acta 2001, 1546, 21–43. [Google Scholar] [CrossRef]
- Balaban, R.S. Regulation of oxidative phosphorylation in the mammalian cell. Am. J. Physiol. 1990, 258, C377–C389. [Google Scholar] [PubMed]
- Idell-Wenger, J.A.; Grotyohann, L.W.; Neely, J.R. Coenzyme a and carnitine distribution in normal and ischemic hearts. J. Biol. Chem. 1978, 253, 4310–4318. [Google Scholar] [PubMed]
- Bain, J.R.; Stevens, R.D.; Wenner, B.R.; Ilkayeva, O.; Muoio, D.M.; Newgard, C.B. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes 2009, 58, 2429–2443. [Google Scholar] [CrossRef] [PubMed]
- Muoio, D.M.; Noland, R.C.; Kovalik, J.-P.; Seiler, S.E.; Davies, M.N.; DeBalsi, K.L.; Ilkayeva, O.R.; Stevens, R.D.; Kheterpal, I.; Zhang, J.; et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012, 15, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Kovalik, J.P.; Slentz, D.; Stevens, R.D.; Kraus, W.E.; Houmard, J.A.; Nicoll, J.B.; Lea-Currie, Y.R.; Everingham, K.; Kien, C.L.; Buehrer, B.M.; et al. Metabolic remodeling of human skeletal myocytes by cocultured adipocytes depends on the lipolytic state of the system. Diabetes 2011, 60, 1882–1893. [Google Scholar] [CrossRef] [PubMed]
- Violante, S.; Ijlst, L.; Ruiter, J.; Koster, J.; van Lenthe, H.; Duran, M.; de Almeida, I.T.; Wanders, R.J.; Houten, S.M.; Ventura, F.V. Substrate specificity of human carnitine acetyltransferase: Implications for fatty acid and branched-chain amino acid metabolism. Biochim. Biophys. Acta 2013, 1832, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Keung, W.; Ussher, J.R.; Jaswal, J.S.; Raubenheimer, M.; Lam, V.H.; Wagg, C.S.; Lopaschuk, G.D. Inhibition of carnitine palmitoyltransferase-1 activity alleviates insulin resistance in diet-induced obese mice. Diabetes 2012, 62, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.M.; Jones, A.D.; Henagan, T.M. A review of mitochondrial-derived fatty acids in epigenetic regulation of obesity and type 2 diabetes. J. Nut. Health Food Sci. 2014, 2, 1–4. [Google Scholar]
- Schmidt, K.N.; Amstad, P.; Cerutti, P.; Baeuerle, P.A. The roles of hydrogen peroxide and superoxide as messengers in the activation of transcription factor NF-κB. Chem. Biol. 1995, 2, 13–22. [Google Scholar] [CrossRef]
- Richardson, D.K.; Kashyap, S.; Bajaj, M.; Cusi, K.; Mandarino, S.J.; Finlayson, J.; DeFronzo, R.A.; Jenkinson, C.P.; Mandarino, L.J. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J. Biol. Chem. 2005, 280, 10290–10297. [Google Scholar] [CrossRef] [PubMed]
- Parikh, V.; Morgan, M.; Scott, R.; Clements, L.; Butow, R. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 1987, 235, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Aguer, C.; McCoin, C.S.; Knotts, T.A.; Thrush, A.B.; Ono-Moore, K.; McPherson, R.; Dent, R.; Hwang, D.H.; Adams, S.H.; Harper, M.E. Acylcarnitines: Potential implications for skeletal muscle insulin resistance. FASEB J. 2014, 29, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Noland, R.C.; Koves, T.R.; Seiler, S.E.; Lum, H.; Lust, R.M.; Ilkayeva, O.; Stevens, R.D.; Hegardt, F.G.; Muoio, D.M. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J. Biol. Chem. 2009, 284, 22840–22852. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; McCaffery, J.M.; Irizarry, R.A.; Boeke, J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 2006, 23, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Morrish, F.; Noonan, J.; Perez-Olsen, C.; Gafken, P.R.; Fitzgibbon, M.; Kelleher, J.; van Gilst, M.; Hockenbery, D. Myc-dependent mitochondrial generation of acetyl-coa contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J. Biol. Chem. 2010, 285, 36267–36274. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Starodub, O.; McIntosh, A.; Kier, A.B.; Schroeder, F. Liver fatty acid-binding protein targets fatty acids to the nucleus: Real time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 2002, 277, 29139–29151. [Google Scholar] [CrossRef] [PubMed]
- Barrès, R.; Osler, M.E.; Yan, J.; Rune, A.; Fritz, T.; Caidahl, K.; Krook, A.; Zierath, J.R. Non-cpg methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009, 10, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Barrès, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).