Abstract
With growing demand for clean and cheap energy resources, biogas production is emerging as an ideal solution, as it provides relatively cheap and clean energy, while also tackling the problematic production of excessive organic waste from crops and animal agriculture. Behind this process stands a variety of anaerobic microorganisms, which turn organic substrates into valuable biogas. The biogas itself is a mixture of gases, produced mostly as metabolic byproducts of the microorganisms, such as methane, hydrogen, or carbon dioxide. Hydrogen itself figures as a potent bio-fuel, however in many bioreactors it serves as the main substrate of methanogenesis, thus potentially limiting biogas yield. With help of modern sequencing techniques, we tried to evaluate the composition in eight bioreactors using different input materials, showing shifts in the microbial consortia depending on the substrate itself. In this paper, we provide insight on the occurrence of potentially harmful microorganisms such as Clostridium novyi and Clostridium septicum, as well as key genera in hydrogen production, such as Clostridium stercorarium, Mobilitalea sp., Herbinix sp., Herbivorax sp., and Acetivibrio sp.
1. Introduction
Biogas production appears to be a brilliant solution to tackle common energetics, waste management, and environmental pollution problems. Production of biogas provides us with clean energy in a form of gas mixture, utilizing common organic wastes as an input substrate and thus helping to maintain the biomass cycle without the formation of unnecessary byproducts [1,2]. The elegance of biogas production lies in the complex microbial consortia capable of anaerobic fermentation, which ultimately leads to the final step, methanogenesis. Methanogenesis itself is a dominant metabolic pathway typical for the group of microorganisms referred to as methanogenic archaea. These microorganisms rely on the end-products of bacterial fermentation, such as short-chain fatty acids (SCFA), carbon dioxide, hydrogen, and even methyl–amines and methanol [3,4,5,6]. However, relatively little is known about the microbial consortia responsible for the production of methanogenesis precursors. The composition of the bioreactor ecosystem has a direct impact on the overall fitness of biogas production [7,8]. Unbalance in the microbial consortia may lead to the collapse of fermentative processes or contamination of biogas by undesired products, such as H2S [9].
In order to keep biogas production sustainable for a long period of time, with high yields of gas, one must be able to identify key groups of microorganisms that help to maintain steady conditions and precursors for biogas production [3,10]. Many anaerobic microorganisms were identified as potential hydrogen producers, figuring in many natural and anthropogenic processes. This knowledge then helps not only to ensure methanogenesis occurs in bioreactors but is also exploited in terms of biohydrogen production [11,12,13]. Hydrogen itself is one of the many final products of bacterial fermentation and also serves as a proton that is used in various microbial enzymatic pathways. In connection with SCFAs, it is also associated with interspecies hydrogen-electron transfer, which further serves as a connection among fermenting bacterial taxa [14,15,16]. In anaerobic digesters and bioreactors, the microbial community is greatly dependent on the inoculum and the origin of used substrate. These factors have a substantial effect on the behaviour of the microbial community and yields of the biogas plant [7,8]. With crop waste and animal manure being used as an input substrate, a great portion of the microbial community is connected to soil and gastrointestinal tract consortia. Understanding the diversity of primary hydrogen producers can foretell if a correlation exists between production, environment, and the microbial community, and if this has some effect on biogas yield. Bearing this knowledge in mind, the Clostridiales order was chosen as the desired microbial group. Many species belonging to this clade are considered potent hydrogen producers. These microorganisms create a substantial portion of the Gram-positive microbial community in mesophilic bioreactors [17,18]. Results of this study illustrate the variability in the composition of the Clostridiales order depending on the different input substrates.
2. Materials and Methods
Eight biogas plants throughout the Czech Republic were chosen based on known input material with information about the substrate provided as the ratio of substrate components (w/w %). Names of the plants, as well as information about substrate components, are summarized in Table 1. Fermenters used in these biogas plants had the operational volume of 2500–3500 m3. Sampling was carried out anoxically and material was directly transferred from the fermenter into sterile sampling vessels. Afterwards, the samples were stored in a thermo-isolating box and were transported for immediate analysis to our laboratory.

Table 1.
Characteristics of evaluated anaerobic digesters (Reproduced with permission of Kushkevych [6]).
Prior to DNA analysis, chemical and physical parameters were determined. The pH, redox potential, and temperature were measured, as well as total solids content, volatile solids content, and biogas composition. Measurements were conducted for each anaerobic digester of a biogas plant with data summarized in Table 1. For pH and redox, potential measurement was used pH/Cond meter 3320 (WTW GmbH, Dinslaken, Germany) in accordance with standard procedures [19]. The sample temperature was assessed by high accuracy PT100 RTD thermometer HH804U (OMEGA Engineering, Stamford, CT 06907-0047, USA). Total solids (TS) content was determined by drying at 105 ± 5 °C, using EcoCELL 111 (BMT Medical Technology Ltd., Brno, Czech Republic) according to Czech Standard Method [20]. After drying, samples were cooled in a desiccator, until they reached constant weight and the value could be determined. Volatile solids content (VS) was assessed by combustion of the samples in a muffle furnace LMH 11/12 (LAC, Ltd., Rajhrad, Czech Republic) at 550 ± 5 °C according to Czech Standard Method [21].
According to our previous studies, we used the QIAamp Fast DNA Stool Mini Kit (QIAGEN GmbH, Hilden, Germany). This was appropriate for a fast and easy purification process of total DNA from fresh or frozen samples and was used for DNA extraction from samples of anaerobic fermenters in previous studies [9]. DNA extractions were conducted according to manufacturer protocol, only with minor adjustments, which we describe below. We extracted 100 mg of each sample, which we mixed with 1.4 mL of ASL buffer (QIAGEN GmbH, Hilden, Germany) and incubated at 95 °C for 10 min. After centrifugation, we added InhibitEX tablets to the supernatant, according to protocol, to remove impurities and possible PCR inhibitors. After another centrifugation, 200 μL of supernatant was mixed with 15 μL of proteinase K solution and 200 μL of buffer AL (QIAGEN GmbH, Hilden, Germany). Then, the solution was incubated at 70 °C for 10 min, cooled down and 200 μL of ethanol (96–100%) was added to the mixture. Following the procedure, the supernatant was centrifuged through the QIAamp kit column and was washed twice by buffers AW1 and AW2 (QIAGEN GmbH, Hilden, Germany). Finally, the DNA elution was conducted by 200 μL of elution buffer.
For amplification of the desired product, V3 and V4 variable regions of the 16S rRNA gene were targeted by universal primers [22]. Primers were marked for sample identification by molecular barcoding. According to our previous research, the Maxima™ Probe qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) was used for PCR reaction. Initial denaturation was conducted at 95 °C for 10 min, followed by 30 cycles of incubation at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 120 s, with a final extension step at 72 °C with a 2 min duration. Visualization of PCR products was performed using 1.5% agarose gel. After the reaction, DNA was extracted from the gel using the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). DNA was quantified using the Quant-iTPicoGreen dsDNA Assay (Thermo Fisher Scientific, Waltham, MA, USA) and equimolar amounts of PCR products were pooled together. Paired-end amplicons were sequenced via Illumina Mi-Seq platform. Data analysis of 16S rRNA sequences was carried out using QIIME data analysis package [23].
According to base quality score distributions, average base content per reading and the guanosine-cytosine pairs (GC) distribution in the reads, quality filtering on raw sequences was conducted. Chimeras and reads that did not cluster with other sequences were removed. The obtained sequences with quality scores higher than 20 were shortened to the same length of 350 bp and classified with RDP Seqmatch with an operational taxonomic unit (OTU) discrimination level set to 97%. The relative abundance of taxonomic groups was calculated for microorganisms detected in this study. Sequences were compared using the BLAST feature of the National Center for Biotechnology Information (NCBI) [24]. The sequences were uploaded to Geneious (Geneious 7.1.9) for comparative genomic analyses [25]. Alignments of sequences were performed in Geneious 7.1.9 using Muscle (Clustal W) with the BLOSUM cost matrix and clustering was performed by the neighbor-joining method [26].
Sequences of selected microorganisms were then deposited in GenBank under accession numbers: MH045949, MH045950, MH045951, MH045952, MH045953, MH045954, MH045955, MH045956, MH045957, MH045958, MH045959, MH045960, MH045961, MH045962, MH045963, MH045964, MH045965, MH045966, MH045967, MH045968, MH045969, MH045970, MH045971, MH045972, MH045973, MH045974, MH045975, MH045976, MH045977. Origin7.0 software (www.origin-lab.com) was used for further processing data and analysis of the obtained results.
3. Results
3.1. Commenting on the Control
After the analysis of raw data, the most abundant genera were investigated thoroughly. First, the overall abundance of the Clostridiales order was compared against the total microbial background (Figure 1A). The mean ratio was 5.84% of the whole microbial background, with a minimum of 0.64% in the case of the Modřice bioreactor, which serves as a control, and a maximum of 9.51% in the case of the Čejč bioreactor. This discrepancy is even more visible on the graph visualizing the ratios of detected OTUs in the manner of the whole set of raw data (Figure 1B). Variation in the structure of the microbial community of the Modřice digester is probably a consequence of a significant difference in input substrate. The input material, in this case, consists of biological and waste sludge, which has a slightly less alkaline pH and a higher redox potential compared to other reactors, with crop silage or manure being used as the main input substrate. Thus, the explanation probably lies in the different substrate, its properties, and microbial load [27,28]. Biological sludge is a complex microbial ecosystem, comprised of many bacterial, archaeal, and eukaryotic taxa such as bacteria Acinetobacter sp., Flavobacterium sp., Cloacibacterium sp., Desulfovibrio sp., Desulfomicrobium sp. or archaea such as Methanobrevibacter sp. and Methanosarcina sp. [8,29,30,31,32]. Microorganisms coming from this environment may favour these conditions, leading to a significant shift in the Clostridiales or total bacterial ratio due to different conditions (Figure 1A).
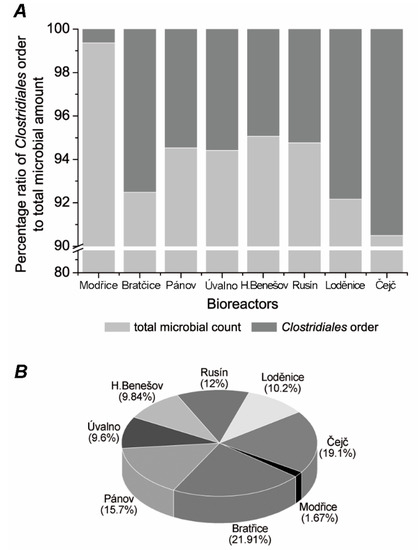
Figure 1.
Ratio of Clostridiales representative composition: ratio between Clostridiales order and total microbial amount (A); pie-graph indicating portions of each biogas plant to Clostridiales order ratio (B).
3.2. Comparison of the Clostridiales Order Communities
Comparing the rest of the bioreactors, it seems for the Figure 1A that there is a shift from the Clostridiales order microorganisms towards other bacterial species. The lower ratios detected in Pánov, Horní Benešov, Úvalno, and Rusín (Figure 1B) may be connected to the optimal growth temperature and temperature threshold for many mesophilic microorganisms. Many Clostridiales order organisms manifest poor growth above 44 °C [33]. Judging by this, it is probable that higher temperatures may lead to changes in the taxa, moving from Clostridiales candidates to more thermophilic organisms from different taxonomic groups. Analysis of the most abundant genera revealed domination of the Clostridiales group by the genus Clostridium. The mean value for the Clostridium genus is 58.98%. In addition to Figure 1, Figure 2 shows that in the case of extreme results, the microbial consortium in the Čejč bioreactor was dominated by this genus with the ratio of 89.44%. In the case of the Čejč bioreactor, the Clostridium genus was dominated by the Clostridium novyi species, belonging to cluster I, which comprised 65.63% of all OTUs retrieved from this bioreactor sample, deposited in GenBank under accession number MH045969.
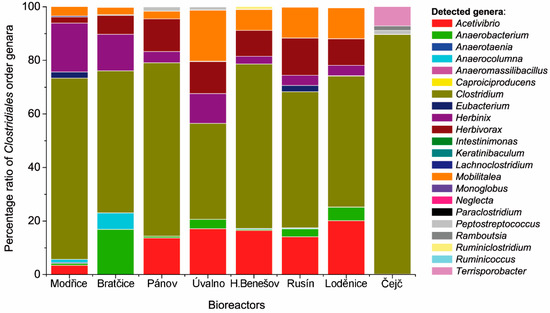
Figure 2.
Visualization of the genera composition in each biogas plant and mean of its abundance to total microbial count.
Analysis by BLAST gives 99% similarity to the sequence with GenBank accession number LC193834.1 (Figure 3). C. novyi is characterized as 0.6–1.4 × 1.6–17 μm long gram-positive rods, which usually occur singly or in pairs with good gas production. Among the common substrates belong carbohydrates such as raffinose, glucose, melibiose, and cellobiose, which in turn produce products such as acetate, propionate, isobutyrate, butyrate, isovalerate, and more importantly hydrogen, and CO2. C. novyi is a sporulating rod with sub-terminally to terminally localized spores, which may cause swelling of the cell. Optimal growth temperatures range from 30–45 °C, with an alkaline pH threshold of 8.5. However, C. novyi is regarded as a soil-dwelling microorganism and a known pathogen of animals and even humans. It is the first time that C. novyi was detected in such a high ratio compared to other data. C. novyi was formerly detected in bioreactors only in low abundances as described by Fröschle et al., 2015. Its potential to pose a threat to biogas plant operators remains unclear [33,34,35,36]. Another notable species is Clostridium septicum from cluster I and Clostridium stercorarium belonging to cluster II. As in the case of C. novyi, C. septicum may also be considered a human pathogen. The optimal temperature is 44 °C. However, its growth is inhibited by temperatures above 46 °C. It forms straight to curved cells that are motile. This species creates oval spores, located sub-terminally within the cell, which may swell if present. It is considered a strong gas producer [33].

Figure 3.
Phylogenetic relationship of detected species in terms of their source.
C. stercorarium is a representative of thermophilic clostridia. Its temperature optimum lies around 65 °C and under given conditions forms relatively small rods 0.3–0.4 × 2–4 μm and produces oval terminal spores. This species is able to ferment cellulose, cellobiose, melibiose, raffinose, fructose, and sucrose in turn for the production of acetate, lactate, ethanol, hydrogen, and CO2. A possible explanation for the occurrence of pathogenic clostridia may be their connection with the gastrointestinal tract of farm animals and soil. In the case of C. stercorarium, it naturally occurs in compost, thus probably promoting anaerobic digestion of silage in the bioreactor [18]. Other notable species detected in this study are C. clariflavum, C. propionicum, C. neopropionicum, and C. sulfidigenes (Figure 3). The importance of these species lies mainly in the formation of hydrogen and SCFAs, which are connected with methanogenesis and linked to interspecies hydrogen transfer. Their metabolism ensures the thermodynamic stability of anaerobic fermentation, providing a favorable environment for deeper and fine digestion of the given substrate [33].
Apart from the genus Clostridium, minor clades such as Acetivibrio sp. (10.64%), Herbinix sp. (8.26%), Herbivorax sp. (8.38%), and Mobilitalea sp. (8.40%) were detected in the bioreactors, comprising 10–20% of all retrieved OTUs (Figure 2 and Figure 3). Acetivibrio cellulolyticus was the sole member of this genus detected in this study, which was similar from 98% to GenBank sequence NR_025917.1 and was deposited under accession number MH045953 (Figure 3). Its morphology consists of slightly curved gram-negative rods with a single flagellum having dimensions of 0.5–0.8 × 4–10 µm. Major substrates consisted of cellulose, cellobiose, and salicin, producing acetate, hydrogen, and CO2 in return [36]. Similar to Acetivibrio, the genus Herbinix is represented by the sole species H. luporum (NR_152095.1), with a sequence similarity of 99% (MH045949). This thermophilic cellulose-degrading bacterium can be found in bioreactors, where it is connected with plant biomass degradation. H. luporum represents a novel genus in the family Lachnospiraceae. It is a non-motile rod, growing to 2.0–6.0 × 0.5 µm, with an optimal growth temperature of 55 °C and main fermentation products such as ethanol, acetate, butyrate, and hydrogen [37]. Herbivorax saccinicola is a key player in plant biomass remineralization during anaerobic digestion processes and a member of the family Ruminococcaceae. These non-motile long and thin rods with dimensions of 5–10 × 0.2 µm grow between 45–65 °C. However, the growth of this microorganism was observed even in lower temperatures (40–44 °C), one may deduce that temperature specificity is not so strict. The main fermentation products are acetate, ethanol, and hydrogen [38]. This organism (NR_152684.1) was similar to the sequence deposited in GenBank under the accession number MH045957 (Figure 3). Mobilitalea sibirica is the last of the moderately abundant members of curved rod-shaped anaerobic bacteria with the main products of fermentation being ethanol, acetate, hydrogen, and CO2 [39]. Our sequence (MH045951) possessed 99% similarity with type strain P3M-3 sequence under accession number NR_134091.1 (Figure 3). Although many genera were detected in this study, the remaining clades created only a negligible minority which abundance ranged mostly in order of tenths of percentages compared to other taxa (Figure 2). Thus, we didn´t evaluated their occurrence in a deeper context.
4. Discussion
As for methanogenesis and methanogenic archaea, understanding fine arrangements in microbial community relationships are key to maintaining steady and profitable biogas production. As it may seem from some studies, the amount of hydrogen may be misjudged as a smaller portion of gas production. However, it is essential for CO2 to be reduced into methane by methanogenic archaea and naturally turned into biogas. The smaller portion of produced hydrogen also serves as a mediator in interspecies hydrogen transfer, resulting in a high level of well-being of the anaerobic microbial community [3,4,6]. Our data suggests that the composition of anaerobic fermenters depends on the input substrate and may vary greatly because of the given condition. The impact of these findings remains to be evaluated, also whether this knowledge can be exploited to the artificially potent microenvironment, as well as to design inoculum, and knowing possible impacts of using different input substrates. The stable and diverse microbiota of the bioreactor may prevent colonization and succession of undesired microorganisms such as sulfate reducing bacteria. Their metabolic products may damage not only the microbial consortia by strong competition for hydrogen, originally utilized by methanogens, but may also lead to corrosion of the bioreactor itself [40,41,42]. Additionally, this study confirmed the presence of potentially harmful microorganism such as C. septicum and C. novyi, which are connected to myonecrosis and other dangerous illnesses, this raises questions about aseptic and sanitation protocols when operating bioreactor processes. Their presence agrees with previous findings, however, we detected C. novyi in a much higher ratio, comprising 65.63% of the whole Clostridiales microbiota in the bioreactor for the first time [35]. Apart from the genus Clostridium, families Lachnospiraceae and Ruminococcaceae, also have their place in the anaerobic consortia. These microorganisms have similar physiological and biochemical properties as the Clostridium genus, thus also significantly contributing to the methane production process. On the contrary, the input substrate has a direct impact on the microbial community, as well as the well-being of the reactor. It is not only modulating accessibility to essential nutrients, but also bears an additional microbial load with which native microbes must compete for resources.
Author Contributions
M.Č. and I.K. conceived and contributed to the writing of the paper. Together with, M.V., T.V., and M.B. who further designed and performed the experiments and measurements described in this paper. M.Č. together with T.V. analyzed the data and optimized process parameters. T.V. performed analysis of the data and validation of the results.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hobson, P.N. Biogas production from agricultural wastes. Experientia 1982, 38, 206–209. [Google Scholar] [CrossRef]
- Papurello, D.; Silvestri, S.; Tomasi, L.; Belcari, I.; Biasioli, F.; Santarelli, M. Biowaste for SOFCs. Energy Procedia 2016, 101, 424–431. [Google Scholar] [CrossRef]
- Oppermann, R.A.; Nelson, W.O.; Brown, R.E. In vivo studies of methanogenesis in the bovine rumen: Dissimilation of acetate. J. Gen. Microbiol. 1961, 25, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.L.; Wolin, M.J. Methanosphaera stadtmaniae gen. Nov., sp. nov.: A species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 1985, 141, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Papurello, D.; Soukoulis, C.; Schuhfried, E.; Cappellin, L.; Gasperi, F.; Silvestri, S.; Santarelli, M.; Biasioli, F. Monitoring of volatile compound emissions during dry anaerobic digestion of the Organic Fraction of Municipal Solid Waste by Proton Transfer Reaction Time-of-Flight Mass Spectrometry. Bioresour. Technol. 2012, 126, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Kováč, J.; Kaucká, P.; Jesionek, W.; Bartoš, M.; Barton, L. A new combination of substrates: Biogas production and diversity of the methanogenic microorganisms. Open Life Sci. 2018, 13, 119–128. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017, 12, 82–91. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kováč, J.; Vítězová, M.; Vítěz, T.; Bartoš, M. The diversity of sulfate-reducing bacteria in the seven bioreactors. Arch. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Nath, K.; Das, D. Improvement of fermentative hydrogen production: Various approaches. Appl. Microbiol. Biotechnol. 2004, 65, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, F.R.; Dinsdale, R.; Hawkes, D.L.; Hussy, I. Sustainable fermentative hydrogen production: Challenges for process optimisation. Int. J. Hydrogen Energy. 2002, 27, 1339–1347. [Google Scholar] [CrossRef]
- Levin, D.B.; Pitt, L.; Love, M. Biohydrogen production: Prospects and limitations to practical application. Int. J. Hydrogen Energy. 2004, 29, 173–185. [Google Scholar] [CrossRef]
- Nanqi, R.; Wanqian, G.; Bingfeng, L.; Guangli, C.; Jie, D. Biological hydrogen production by dark fermentation: Challenges and prospects towards scaled-up production. Curr. Opin. Biotechnol. 2011, 22, 365–370. [Google Scholar] [CrossRef]
- Baek, G.; Kim, J.; Kim, J.; Lee, C. Role and potential of direct interspecies electron transfer in anaerobic digestion. Energies 2018, 11, 107. [Google Scholar] [CrossRef]
- Ozturk, S.S.; Palsson, B.O.; Thiele, J.H. Control of interspecies electron transfer flow during anaerobic digestion: Dynamic diffusion reaction models for hydrogen gas transfer in microbial flocs. Biotechnol. Bioeng. 1989, 33, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Cord-Ruwisch, R.; Seitz, H.-J.; Conrad, R. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 1988, 149, 350–357. [Google Scholar] [CrossRef]
- Kong, X.; Yu, S.; Fang, W.; Liu, J.; Li, H. Enhancing syntrophic associations among Clostridium butyricum, Syntrophomonas and two types of methanogen by zero valent iron in an anaerobic assay with high organic loading. Bioresour. Technol. 2018, 257, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.H.P.; Zhang, T.; Liu, H. Microbial diversity of a mesophilic hydrogen-producing sludge. Appl. Microbiol. Biotechnol. 2002, 58, 112–118. [Google Scholar] [CrossRef] [PubMed]
- CSN EN 12176, Characterization of Sludge—Determination of pH-Value; Czech Standards Institute: Prague, Czech Republic, 1999.
- CSN EN 14346, Characterization of Waste—Calculation of Dry Matter by Determination of Dry Residue or Water Content; Czech Standards Institute: Prague, Czech Republic, 2007.
- CSN EN 15169, Characterization of Waste—Determination of Loss on Ignition in Waste, Sludge and Sediments; Czech Standards Institute: Prague, Czech Republic, 2007.
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; DeSantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World. J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Mille, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Venkiteshwaran, K.; Bocher, B.; Maki, J.; Zitomer, D. Relating anaerobic digestion microbial community and process function. Microbiol. Insights 2015, 8, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Moestedt, J.; Nilsson Påledal, S.; Schnürer, A. The effect of substrate and operational parameters on the abundance of sulphate-reducing bacteria in industrial anaerobic biogas digesters. Bioresour. Technol. 2013, 132, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Shchegolkova, N.M.; Krasnov, G.S.; Belova, A.A.; Dmitriev, A.A.; Kharitonov, S.L.; Klimina, K.M.; Melnikova, N.V.; Kudryavtseva, A.V. Microbial community structure of activated sludge in treatment plants with different wastewater compositions. Front. Microbiol. 2016, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.V. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochim. Polonica 2015, 62, 103–108. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kollar, P.; Suchy, P.; Parak, K.; Pauk, K.; Imramovsky, A. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuroendocrinol. Lett. 2015, 36, 106–113. [Google Scholar] [PubMed]
- Kushkevych, I.; Vítězová, M.; Kos, J.; Kollár, P.; Jampílek, J. Effect of selected 8-hydroxyquinoline-2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J. Appl. Biomed. 2018, 16, 241–246. [Google Scholar] [CrossRef]
- Rainey, F.A. Clostridiales. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–5. [Google Scholar] [CrossRef]
- Kim, M.D.; Song, M.; Jo, M.; Shin, S.G.; Khim, J.H.; Hwang, S. Growth condition and bacterial community for maximum hydrolysis of suspended organic materials in anaerobic digestion of food waste-recycling wastewater. Appl. Microbiol. Biotechnol. 2010, 85, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Fröschle, B.; Messelhäusser, U.; Höller, C.; Lebuhn, M. Fate of Clostridium botulinum and incidence of pathogenic clostridia in biogas processes. J. Appl. Microbiol. 2015, 119, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.B.; Khan, A.W.; Agnew, B.J.; Colvin, J.R. Isolation and characterization of an anaerobic, cellulolytic microorganism, Acetivibrio cellulolyticus gen. nov., sp. nov. Int. J. Syst. Bacteriol. 1980, 30, 179–185. [Google Scholar] [CrossRef]
- Koeck, D.E.; Hahnke, S.; Zverlov, V.V. Herbinix luporum sp. nov., a thermophilic cellulose-degrading bacterium isolated from thermophilic biogas reactor. Int. J. Syst. Evol. Microbiol. 2016, 66, 4132–4137. [Google Scholar] [CrossRef] [PubMed]
- Koeck, D.E.; Mechelke, M.; Zverlov, V.V.; Liebl, W.; Schwarz, W.H. Herbivorax saccincola gen. nov., sp. nov., a cellulolytic, anaerobic, thermophilic bacterium isolated via in sacco enrichments from a lab-scale biogas reactor. Int. J. Syst. Evol. Microbiol. 2016, 66, 4458–4463. [Google Scholar] [CrossRef] [PubMed]
- Podosokorskaya, O.A.; Bonch-Osmolovskaya, E.A.; Beskorovaynyy, A.V.; Toshchakov, S.V.; Kolganova, T.V.; Kublanov, I.V. Mobilitalea sibirica gen. nov., sp. nov., a halotolerant polysaccharide-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 2657–2661. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Polish J. Microbiol. 2015, 64, 107–114. [Google Scholar]
- Kushkevych, I.; Fafula, R.; Parak, T.; Bartos, M. Activity of Na+/K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Acta Vet Brno 2015, 84, 3–12. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Fedrová, M.; Vochyanová, Z.; Paráková, L.; Hošek, J. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet Brno 2017, 86, 405–411. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).