Lipid Production from Amino Acid Wastes by the Oleaginous Yeast Rhodosporidium toruloides
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism, Media, and Growth Conditions
2.2. Culture Conditions
2.3. Analytical Methods
2.4. Statistical Analysis
3. Results and Discussion
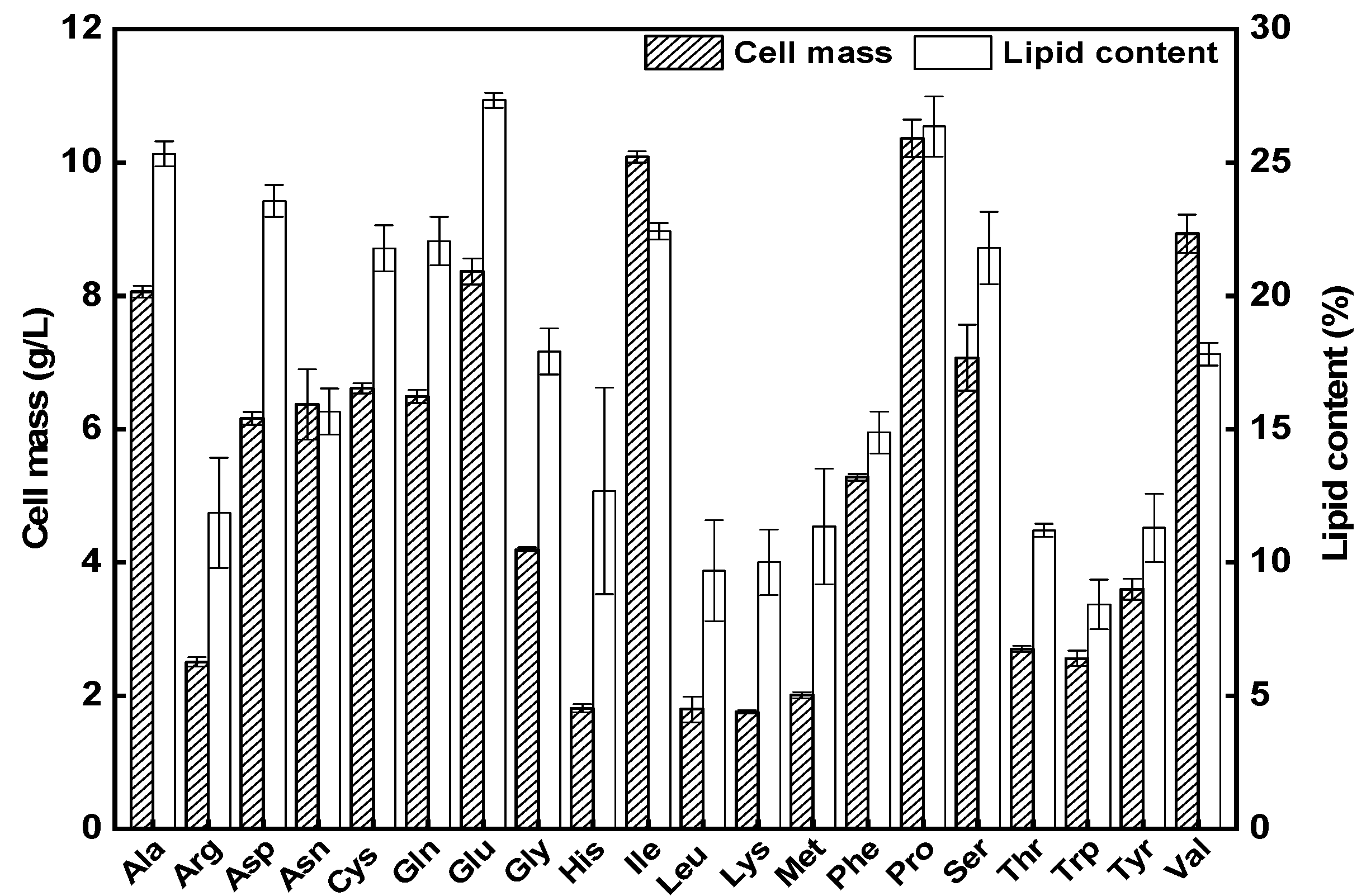
3.1. Evaluating Individual AAs as Carbon Sources for Lipid Production
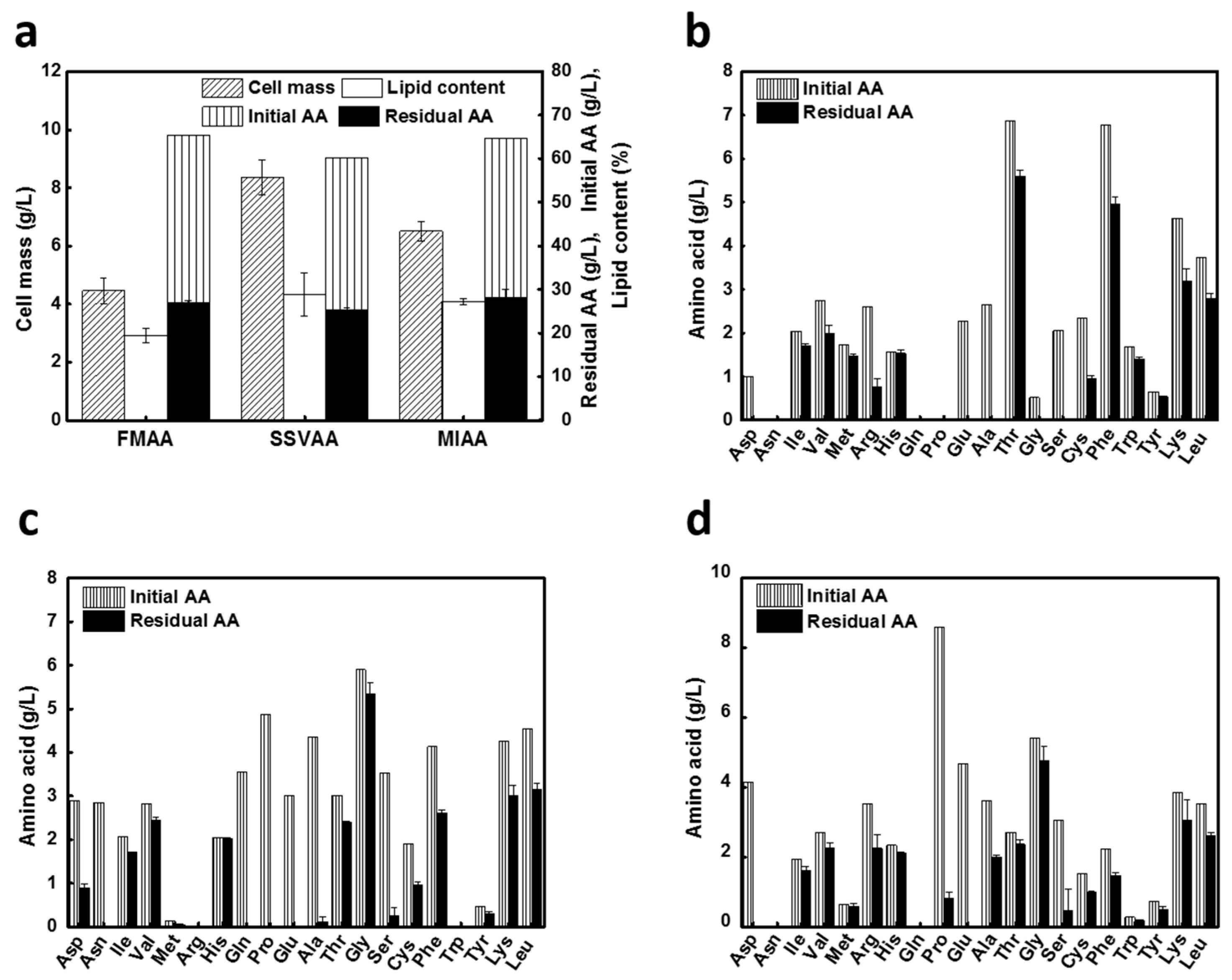
3.2. Lipid Production on AA Blends
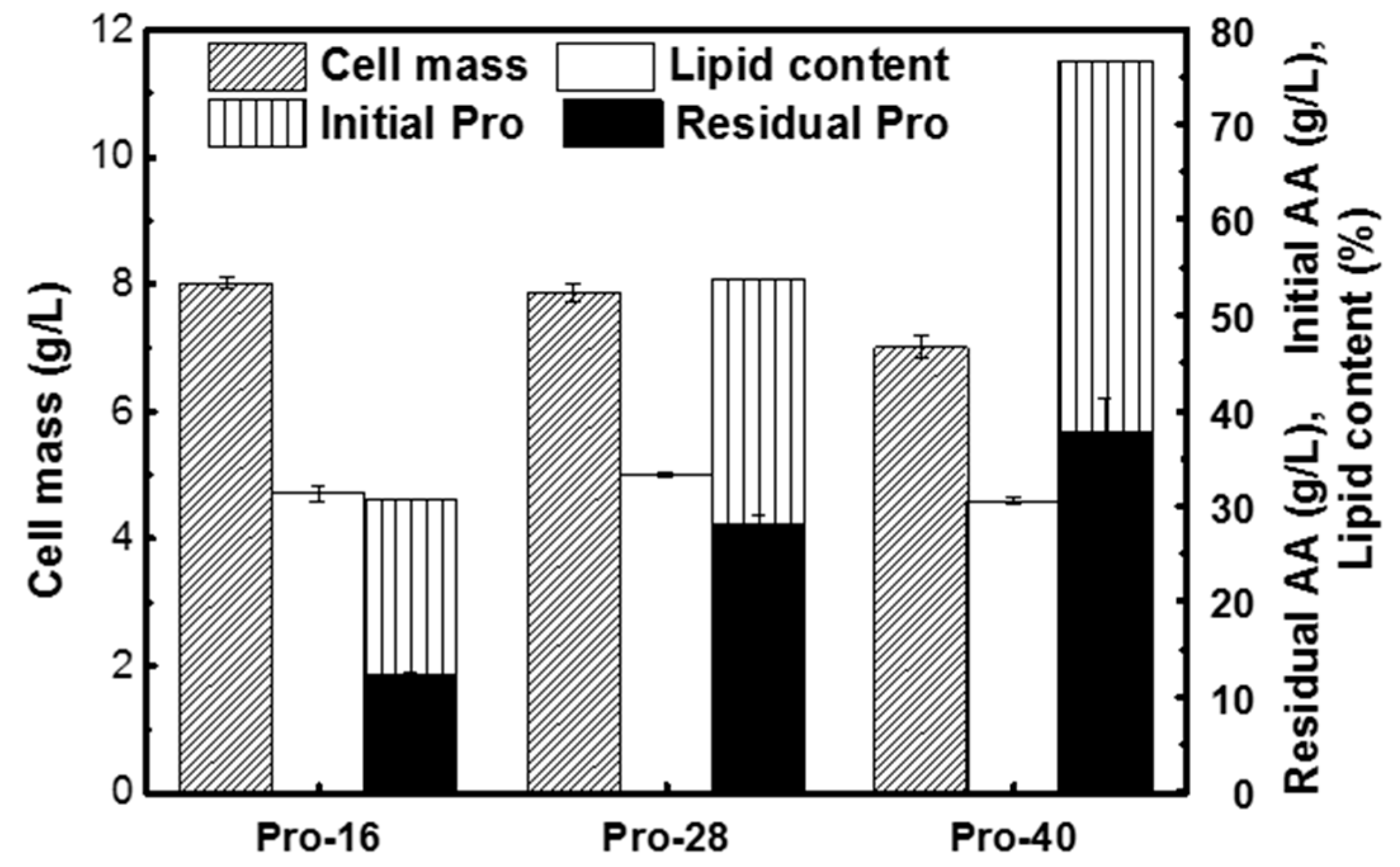
3.3. Lipid Production on L-Proline
3.4. Fatty Acid Compositional Profile of the Lipid Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Luque, R.; Lovett, J.C.; Datta, B.; Clancy, J.; Campeloa, J.M.; Romero, A.A. Biodiesel as feasible petrol fuel replacement: A multidisciplinary overview. Energy Environ. Sci. 2010, 3, 1706–1721. [Google Scholar] [CrossRef]
- Liang, M.H.; Jiang, J.G. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 2013, 52, 395–408. [Google Scholar] [CrossRef]
- Li, Q.; Du, W.; Liu, D. Perspectives of microbial oils for biodiesel production. Appl. Microbiol. Biotechnol. 2016, 80, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar] [PubMed]
- Li, Y.; Zhao, Z.K.; Bai, F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb. Technol. 2007, 41, 312–317. [Google Scholar] [CrossRef]
- Zhao, X.; Hu, C.; Wu, S.; Shen, H.; Zhao, Z.K. Lipid production by Rhodosporidium toruloides Y4 using different substrate feeding strategies. J. Ind. Microbiol. Biotechnol. 2011, 38, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.G.; Rhee, J.S. Kinetic and energetic analyses of lipid accumulation in batch culture of Rhodotorula glutinis. J. Ferment. Technol. 1986, 64, 557–560. [Google Scholar] [CrossRef]
- Hu, C.; Zhao, X.; Zhao, J.; Wu, S.; Zhao, Z.K. Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour. Technol. 2009, 100, 4843–4847. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Papanikolaou, S. Biodiesel production from microbial oil. In Handbook of Biofuels Production; Woodhead Publishing Limited: Cambridge, UK, 2011; pp. 178–198. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Chatzifragkou, A.; Kopsahelis, N.; Papanikolaou, S.; Kookos, I.K. Design and techno-economic evaluation of microbial oil production as a renewable resource for biodiesel and oleochemical froduction. Fuel 2014, 116, 566–577. [Google Scholar] [CrossRef]
- Li, S.Y.; Ng, I.S.; Chen, P.T.; Chiang, C.J.; Chao, Y.P. Biorefining of protein waste for production of sustainable fuels and chemicals. Biotechnol. Biofuels 2018, 11, 256. [Google Scholar] [CrossRef]
- De Schouwer, F.; Claes, L.; Vandekerkhove, A.; Verduyckt, J.; De Vos, D.E. Protein-rich biomass waste as a resource for future biorefineries: State of the art, challenges, and opportunities. ChemSusChem 2019, 12, 1272–1303. [Google Scholar] [CrossRef]
- Pelley, J.W. Elsevier’s Integrated Biochemistry, 1st ed.; Elsevier: Philadelphia, PA, USA, 2007; Chapter 17; pp. 97–104. [Google Scholar]
- Zeng, Y.; Bian, D.; Xie, Y.; Jiang, X.; Li, X.; Li, P.; Zhang, Y.; Xie, T. Utilization of food waste hydrolysate for microbial lipid and protein production by Rhodosporidium toruloides Y2. J. Chem. Technol. Biotechnol. 2017, 92, 666–673. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Wu, S.; Hu, C.; Jin, G.; Zhao, X.; Zhao, Z.K. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour. Technol. 2010, 101, 6124–6129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Zhu, Z.; Shen, H.; Lin, X.; Jin, X.; Jiao, X.; Zhao, Z.K. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol. Biofuels 2018, 11, 148. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hu, C.; Zhao, X.; Zhao, Z.K. Production of lipid from N-acetylglucosamine by Cryptococcus curvatus. Eur. J. Lipid Sci. Technol. 2010, 112, 727–733. [Google Scholar] [CrossRef]
- Toldrá, F.; Mora, L.; Reig, M. New insights into meat by-product utilization. Meat Sci. 2016, 120, 54–59. [Google Scholar] [CrossRef]
- Jayathilakan, K.; Sultana, K.; Radhakrishna, K.; Bawa, A.S. Utilization of byproducts and waste materials from meat, poultry and fish processing industries: A review. J. Food Sci. Technol. 2012, 49, 278–293. [Google Scholar] [CrossRef]
- Bhaskar, N.; Modi, V.K.; Govindaraju, K.; Radha, C.; Lalitha, R.G. Utilization of meat industry by broducts: Protein hydrolysate from sheep visceral mass. Bioresour. Technol. 2007, 98, 388–394. [Google Scholar] [CrossRef]
- Salminen, E.; Rintala, J. Anaerobic digestion of organic solid poultry slaughterhouse waste-a review. Bioresour. Technol. 2002, 83, 13–26. [Google Scholar] [CrossRef]
- Russ, W.; Meyer-Pittroff, R. Utilizing waste products from the food production and processing industries. Crit. Rev. Food Sci. Nutr. 2004, 44, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Guo, R.; Dai, Z.; Zhang, Y. Investigation of enzymatic hydrolysis conditions on the properties of protein hydrolysate from fish muscle (Collichthys niveatus) and evaluation of its functional properties. J. Agric. Food Chem. 2012, 60, 5192–5198. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.D.; Ledward, D.A.; Lawrie, R.A. Protein hydrolysates from meat industry by-products. Meat Sci. 1982, 7, 147–157. [Google Scholar] [CrossRef]
- Yang, X.; Jin, G.; Gong, Z.; Shen, H.; Bai, F.; Zhao, Z.K. Recycling biodiesel-derived glycerol by the oleaginous yeast Rhodosporidium toruloides Y4 through the two-stage lipid production process. Biochem. Eng. J. 2014, 91, 86–91. [Google Scholar] [CrossRef]
- Lin, J.; Shen, H.; Tan, H.; Zhao, X.; Wu, S.; Hu, C.; Zhao, Z.K. Lipid production by Lipomyces starkeyi cells in glucose solution without auxiliary nutrients. J. Biotechnol. 2011, 152, 184–188. [Google Scholar] [CrossRef]
- Pelley, J.W. Amino Acid and Heme Metabolism. In Elsevier’s Integrated Biochemistry; Elsevier: Philadelphia, PA, USA, 2007; pp. 97–105. [Google Scholar] [CrossRef]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef]
- Jiao, X.; Zhang, Y.; Liu, X.; Zhang, Q.; Zhang, S.; Zhao, Z.K. Developing a CRISPR/Cas9 system for genome editing in the basidiomycetous yeast Rhodosporidium toruloides. Biotechnol. J. 2019, 14, 1900036. [Google Scholar] [CrossRef]
- Turcotte, G.; Kosaric, N. The effect of C/N ratio on lipid production by Rhodosporidium toruloides ATCC 10788. Biotechnol. Lett. 1989, 11, 637–642. [Google Scholar] [CrossRef]
- Ruiz, S.J.; Van Klooster, J.S.; Bianchi, F.; Poolman, B. Growth inhibition by amino acids in Saccharomyces cerevisiae. bioRxiv 2017. [Google Scholar] [CrossRef]
- Dai, X.; Shen, H.; Li, Q.; Rasool, K.; Wang, Q.; Yu, X.; Wang, L.; Bao, J.; Yu, D.; Zhao, Z.K. Microbial lipid production from corn stover by the oleaginous yeast Rhodosporidium toruloides using the PreSSLP process. Energies 2019, 12, 1053. [Google Scholar] [CrossRef]
- Soccol, C.R.; Neto, C.J.D.; Soccol, V.T.; Sydney, E.B.; da Costa, E.S.F.; Medeiros, A.B.P.; Vandenbergh, L.P.S. Pilot scale biodiesel production from microbial oil of Rhodosporidium toruloides DEBB 5533 using sugarcane juice: Performance in diesel engine and preliminary economic study. Bioresour. Technol. 2017, 223, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Zambiazi, R.C.; Przybylski, R.; Zambiazi, M.W.; Menonca, C.B. Fatty acid composition of vegetable oils and fats. Bol. Do Cent. Pesqui. Process. Aliment. 2007, 25, 111–120. [Google Scholar] [CrossRef]
- Patel, A.; Arora, N.; Sartaj, K.; Pruthi, V.; Pruthi, P.A. Sustainable biodiesel production from oleaginous yeasts utilizing hydrolysates of various non-edible lignocellulosic biomasses. Renew. Sustain. Energy Rev. 2016, 62, 836–855. [Google Scholar] [CrossRef]
| AA | Initial Concentration (g/L) | ||
|---|---|---|---|
| SV Blends | FM Blends | MI Blends | |
| L-Aspartic acid (Asp) | 3.22 | 1.14 | 5.02 |
| DL-Asparagine (Asn) | 3.21 | - | - |
| L-Isoleucine (Ile) | 2.62 | 2.54 | 2.32 |
| L-Valine (Val) | 3.45 | 3.07 | 2.90 |
| L-Methionine (Met) | 0.82 | 2.51 | 1.36 |
| L-Arginine (Arg) | - | 3.47 | 4.15 |
| L-Histidine (His) | 1.46 | 1.16 | 1.64 |
| L-Glutamine (Gln) | 3.82 | - | - |
| L-Proline (Pro) | 4.56 | 0.43 | 8.77 |
| L-Glutamic acid (Glu) | 7.30 | 6.50 | 8.96 |
| L-Alanine (Ala) | 5.39 | 3.12 | 3.96 |
| L-Threonine (Thr) | 2.69 | 6.54 | 2.51 |
| L-Glycine (Gly) | 7.95 | 1.20 | 6.17 |
| L-Serine (Ser) | 3.18 | 2.01 | 2.90 |
| L-Cysteine (Cys) | 0.82 | 1.49 | 0.66 |
| L-Phenylalanine (Phe) | 3.65 | 6.46 | 2.22 |
| L-Tryptophan (Trp) | - | 2.13 | 0.65 |
| L-Tyrosine (Tyr) | 0.73 | 5.91 | 1.64 |
| L-Lysine (Lys) | 5.06 | 5.85 | 4.73 |
| L-Leucine (Leu) | 5.28 | 4.37 | 3.95 |
| Total | 65.23 | 59.88 | 64.52 |
| Media | Lipid Content (%) | Relative Fatty Acid Content (%, w/w) | |||||
|---|---|---|---|---|---|---|---|
| Myristic (14:0) | Palmitic (16:0) | Palmitoleic (16:1) | Stearic (18:0) | Oleic (18:1) | Linoleic (18:2) | ||
| Pro | 27.7 | 3.0 | 40.9 | 0.7 | 15.1 | 36.9 | 3.5 |
| FMAA | 29.5 | 2.2 | 44.6 | 0.8 | 16.3 | 34.4 | 1.7 |
| MIAA | 27.7 | 3.1 | 43.0 | 0.6 | 16.3 | 35.6 | 1.4 |
| SVAA | 23.7 | 2.1 | 46.1 | 0.8 | 14.7 | 35.0 | 1.4 |
| Glucose [5] | 67.5 | 1.3 | 20.0 | 0.6 | 14.6 | 46.9 | 13.1 |
| Glycerol [26] | 35.0 | 1.4 | 27.8 | 0.6 | 21.8 | 43.8 | 2.9 |
| Corn stover [33] | - | 2.6 | 44.6 | 1.0 | 15.8 | 36.0 | 0.7 |
| Sugarcane juice [34] | 45.0 | 1.0 | 21.5 | 0.7 | 4.6 | 62.1 | 7.6 |
| Palm [35] | - | - | 42.7 | - | 2.1 | 38.4 | 10.6 |
| Canola [35] | - | - | 3.7 | 0.2 | 1.9 | 62.4 | 20.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Kamal, R.; Wang, Q.; Yu, X.; Zhao, Z.K. Lipid Production from Amino Acid Wastes by the Oleaginous Yeast Rhodosporidium toruloides. Energies 2020, 13, 1576. https://doi.org/10.3390/en13071576
Li Q, Kamal R, Wang Q, Yu X, Zhao ZK. Lipid Production from Amino Acid Wastes by the Oleaginous Yeast Rhodosporidium toruloides. Energies. 2020; 13(7):1576. https://doi.org/10.3390/en13071576
Chicago/Turabian StyleLi, Qiang, Rasool Kamal, Qian Wang, Xue Yu, and Zongbao Kent Zhao. 2020. "Lipid Production from Amino Acid Wastes by the Oleaginous Yeast Rhodosporidium toruloides" Energies 13, no. 7: 1576. https://doi.org/10.3390/en13071576
APA StyleLi, Q., Kamal, R., Wang, Q., Yu, X., & Zhao, Z. K. (2020). Lipid Production from Amino Acid Wastes by the Oleaginous Yeast Rhodosporidium toruloides. Energies, 13(7), 1576. https://doi.org/10.3390/en13071576






