Microbial Degradation of Epoxy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Soil Samples
2.3. Enrichment and Growth of Bacteria
2.4. Turbidity (OD600nm) Measurements
2.5. Identification of Bacteria
2.6. Demonstration of the Biodegradation of Epoxy Resin by Bacteria
3. Results and Discussion
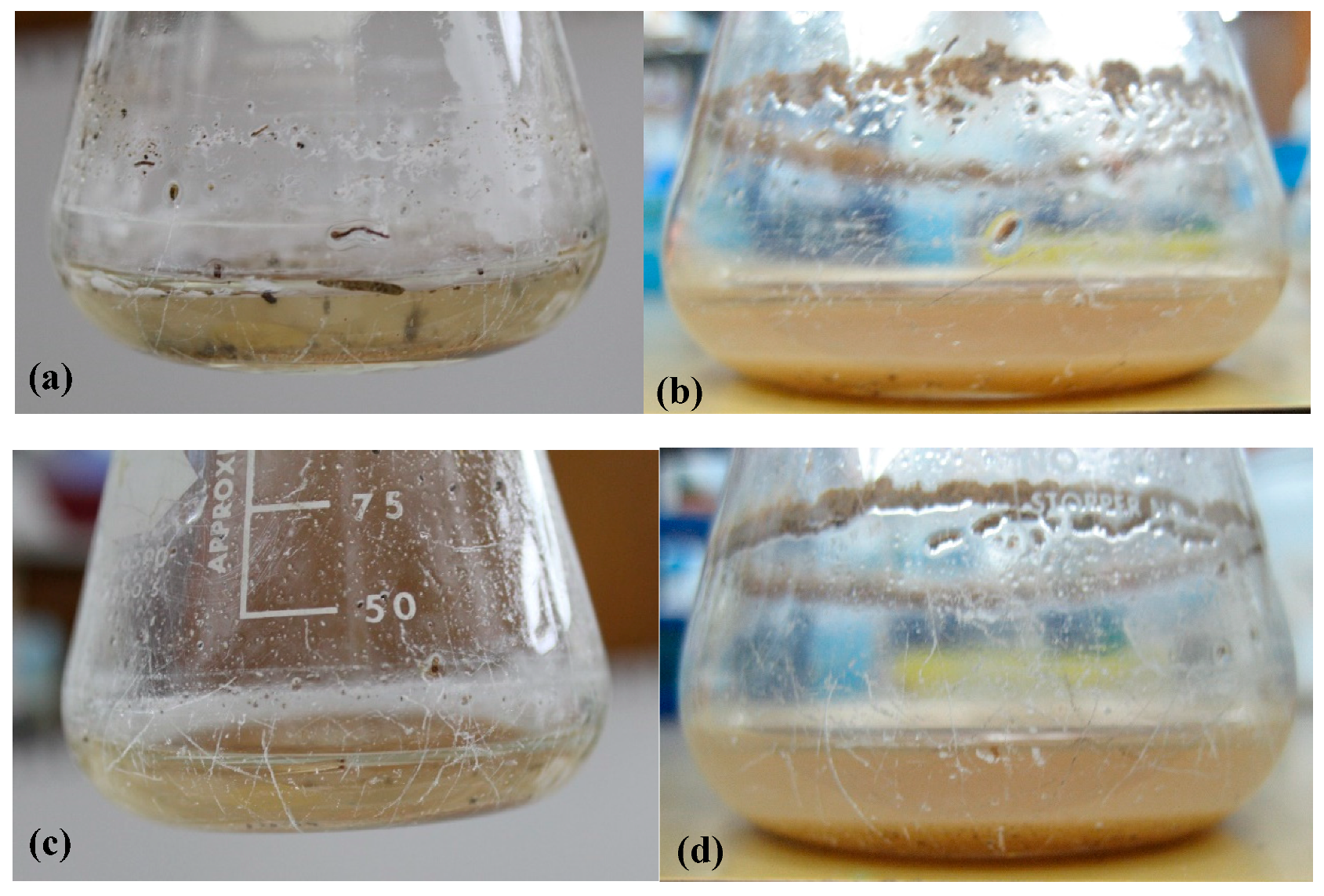
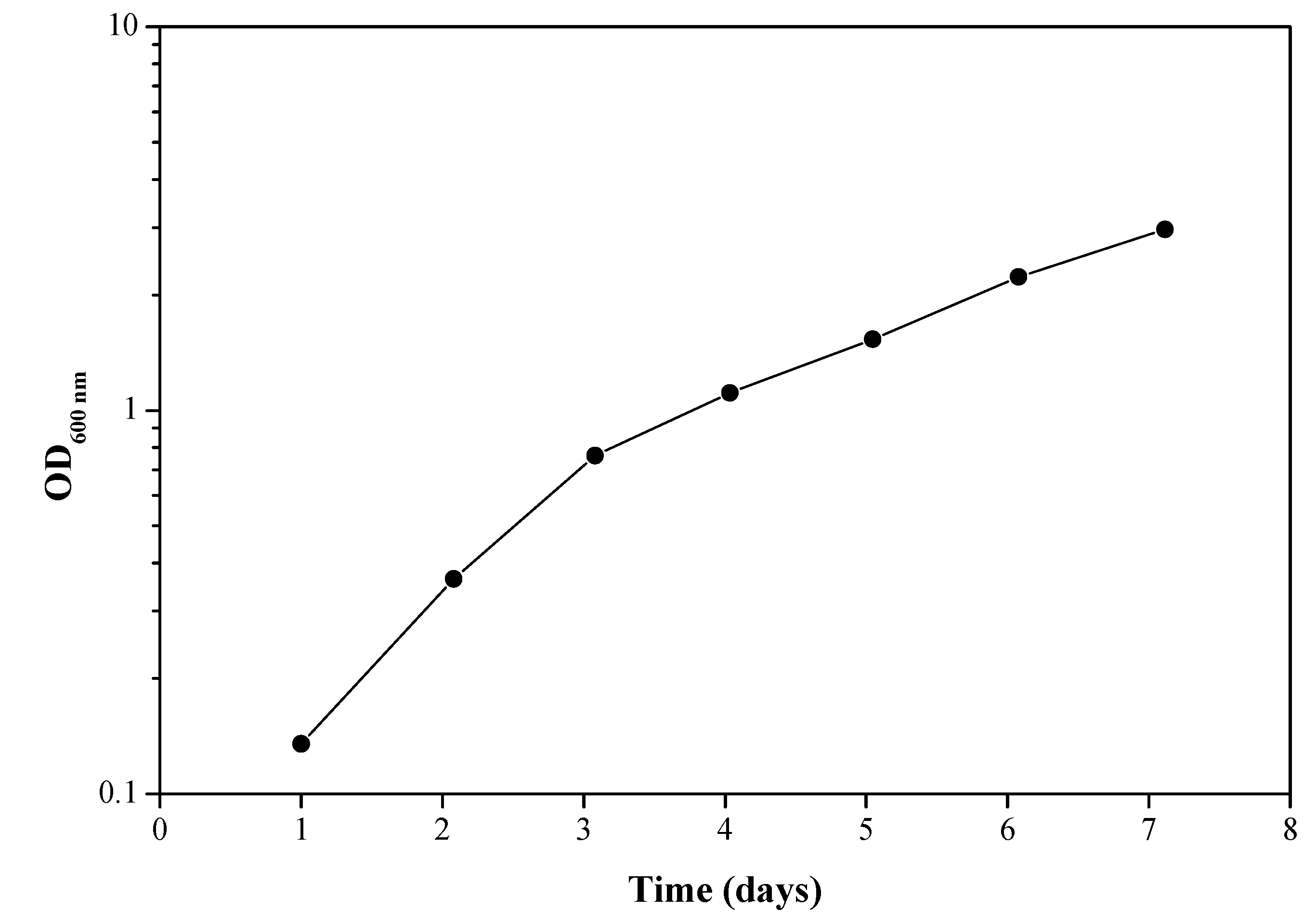
3.1. Enrichment for Bacteria Capable of Degrading Epoxy Resin
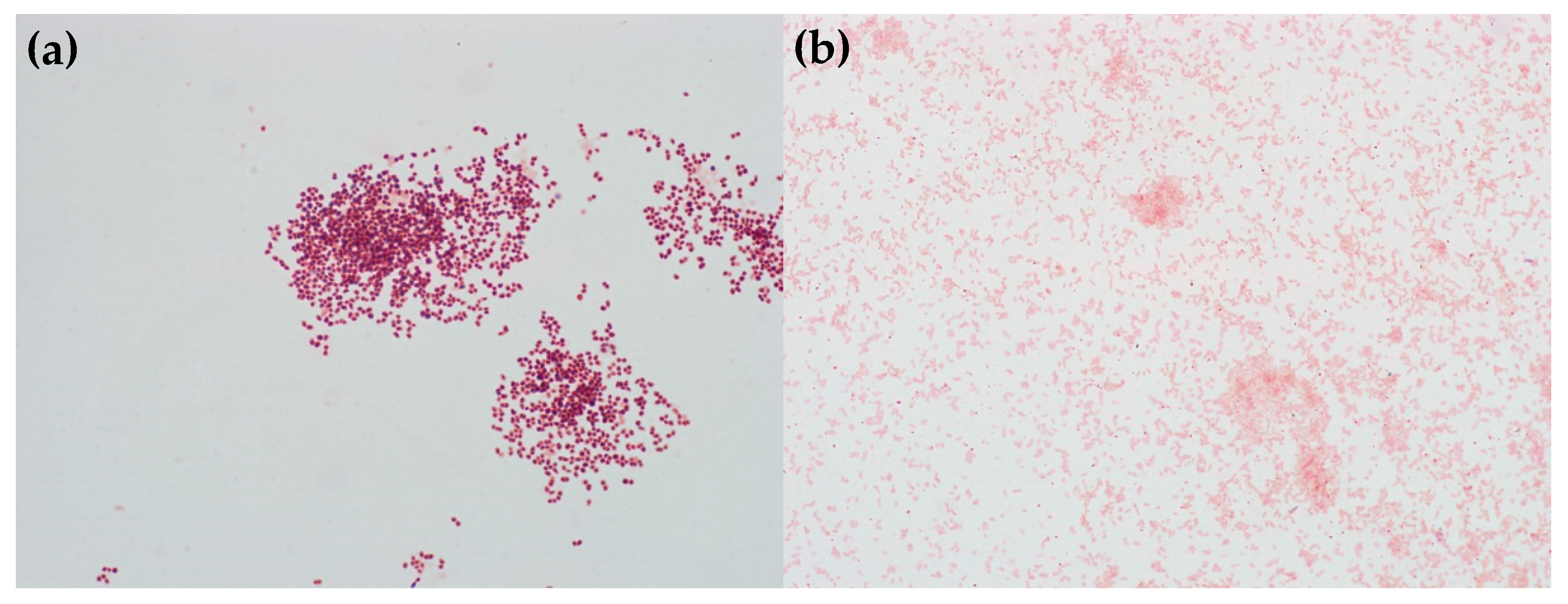
3.2. Characterization of the Bacteria
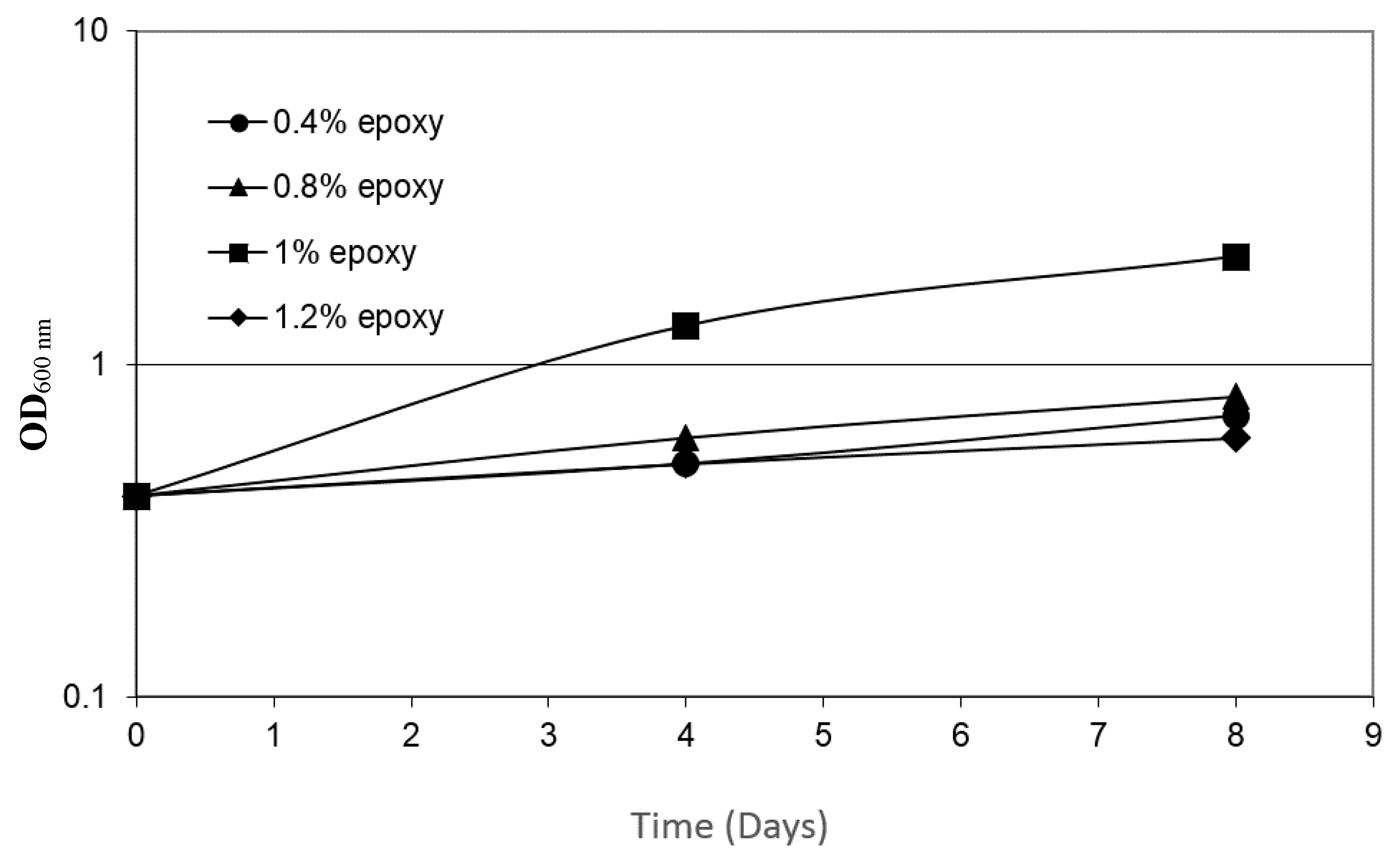
3.3. Growth on Epoxy Requires the Presence of the Two Bacteria
3.4. The Bacteria Change the Surface Properties of the Epoxy Resin
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Acmite Market Intelligence. Global Epoxy Resin Market, 3rd ed.; Acmite Market Intelligence: Ratingen, Germany, 2017. [Google Scholar]
- Shah, A.A.; Hasan, F.; Hameed, A.; Ahmed, S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008, 26, 246–265. [Google Scholar] [CrossRef] [PubMed]
- Pangallo, D.; Buckova, M.; Krakova, L.; Puskarova, A.; Sakova, N.; Grivalsky, T.; Chovanova, K.; Zemankova, M. Biodeterioration of epoxy resin: A microbial survey through culture-independent and culture-dependent approaches. Environ. Microbiol. 2015, 17, 462–479. [Google Scholar] [CrossRef] [PubMed]
- Little, B.J.; Lee, J.S. Microbiologically Influenced Corrosion; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Wang, G.; Chai, K.; Wu, J.; Liu, F. Effect of Pseudomonas putida on the degradation of epoxy resin varnish coating in seawater. Int. Biodeterior. Biodegrad. 2016, 115, 156–163. [Google Scholar] [CrossRef]
- Davis, B.D.; Mingioli, E.S. Mutants of Escherichia coli requiring metliionine or vitamin B12. J. Bacteriol. 1950, 60, 17–28. [Google Scholar] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tse, H.; Yuen, K.-Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef] [PubMed]
- Teyssier, C.; Marchandin, H.; Jean-Pierre, H.; Diego, I.; Darbas, H.; Jeannot, J.L.; Gouby, A.; Jumas-Bilak, E. Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J. Med. Microbiol. 2005, 54, 945–953. [Google Scholar] [CrossRef] [PubMed]
- James, G. Universal bacterial identification by PCR and DNA sequencing of 16S rRNA gene. In PCR for Clinical Microbiology; Schuller, M., Sloots, T., James, G., Halliday, C., Carter, I., Eds.; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Jeong, S.J.; Kwon, G. Cloning of fibrinolytic enzyme gene from Bacillus subtilis isolated from Cheonggukjung and its expression in protease-deficient Bacillus subtilis. J. Microbiol. Biotechnol. 2007, 17, 1018–1023. [Google Scholar] [PubMed]
- Ligozzi, M.; Bernini, C.; Grazia Bonora, M.; de Fatima, M.; Zuliani, J.; Fontana, R. Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant Gram-positive cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Deak, E.; Charlton, C.L.; Bobenchik, A.M.; Miller, S.A.; Pollett, S.; McHardy, I.H.; Wu, M.T.; Garner, O.B. Comparison of the Vitek MS and Bruker Microflex LT MALDI-TOF MS platforms for routine identification of commonly isolated bacteria and yeast in the clinical microbiology laboratory. Diagn. Microbiol. Infect. Dis. 2015, 81, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Carbonnelle, E.; Mesquita, C.; Bille, E.; Day, N.; Dauphin, B.; Beretti, J.L.; Ferroni, A.; Gutmann, L.; Nassif, X. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 2011, 44, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Martiny, D.; Busson, L.; Wybo, I.; El Haj, R.A.; Dediste, A.; Vandenberg, O. Comparison of the MICROFLEX LT and VITEK® MS systems for the routine identification of bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Shapira, Y.; Multanen, V.; Whyman, G.; Bormashenko, Y.; Chaniel, G.; Barkay, Z.; Bormashenko, E. Plasma treatment switches the regime of wetting and floating of pepper seeds. Colloids Surf. B Biointerfaces 2017, 157, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 1990, 54, 305–315. [Google Scholar] [PubMed]
- Cheremisinoff, N.P. Biotechnology for Waste and Wastewater Treatment; Noyes Publications: Westwood, NJ, USA, 1996. [Google Scholar]
- Cerny, G. Method for the distinction of Gram-negative from Gram-positive bacteria. Eur. J. Appl. Microbiol. 1976, 3, 223–225. [Google Scholar] [CrossRef]
- Chester, B.; Cooper, L.H. Achromobacter species (CDC group Vd): Morphological and biochemical characterization. J. Clin. Microbiol. 1979, 9, 425–436. [Google Scholar] [PubMed]
- Laffineur, K.; Janssens, M.; Charlier, J.; Avesani, V.; Wauters, G.; Delmée, M. Biochemical and susceptibility tests useful for identification of nonfermenting gram-negative rods. J. Clin. Microbiol. 2002, 40, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.; Popoff, M.; Kiredjian, M.; Kersters, K. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int. J. Syst. Evol. Microbiol. 1988, 38, 406–416. [Google Scholar] [CrossRef]
- Hagiya, H.; Ohnishi, K.; Maki, M.; Watanabe, N.; Murase, T. Clinical characteristics of Ochrobactrum anthropi bacteremia. J. Clin. Microbiol. 2013, 51, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Chain, P.S.G.; Lang, D.M.; Comerci, D.J.; Malfatti, S.A.; Vergez, L.M.; Shin, M.; Ugalde, R.A.; Garcia, E.; Tolmasky, M.E. Genome of Ochrobactrum anthropi ATCC 49188T, a versatile opportunistic pathogen and symbiont of several eukaryotic hosts. J. Bateriol. 2011, 193, 4274–4275. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.H.; Gómez, J.R.; Vázquez, E.G.; Gómez, J.G. Ochrobactrum anthropi bacteraemia: Report of six cases and review of the literature. Intern. Med. 2014, 4, 134. [Google Scholar]
- Mastroianni, A.; Cancellieri, C.; Montiní, G. Ochrobactrurn anthropi bacteremia: Case report and review of the literature. Clin. Microbiol. Infect. 1999, 5, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Alnor, D.; Frimodt-Meller, N.; Espersen, F.; Frederiksen, W. Infections with the unusual human pathogens Agrobacterium species and Ochrobactrum anthropi. Clin. Infect. Dis. 1994, 18, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.V.; Chiang, B.W.; Yuan, S.Y. Biodegradation of nonylphenol in soil. Chemosphere 2007, 66, 1857–1862. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M. Genus Rhodococcus. In Bergey’s Manual of Systematic Bacteriology; Williams, S.T., Sharpe, M.E., Holt, J.G., Eds.; Williams and Wilkins: Baltimore, MD, USA, 1989; Volume 4, pp. 2362–2371. [Google Scholar]
- Bell, K.S.; Philp, J.C.; Aw, D.W.J.; Christofi, N. The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.S.; Otten, L.G.; Resch, V.; Muyzer, G.; Hanefeld, U. Draft genome sequence of Rhodococcus rhodochrous strain ATCC 17895. Stand. Genom. Sci. 2013, 9, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.J.; Kulakov, L.A.; Allen, C.C. Biodegradation and Rhodococcus–masters of catabolic versatility. Curr. Opin. Biotechnol. 2005, 16, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.J.; Kulakov, L.A.; Allen, C.C.R. Genomes and plasmids in Rhodococcus. In Biology of Rhodococcus. Microbiology Monographs; Alvarez, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 16, pp. 73–90. [Google Scholar]
- Larkin, M.J.; De Mot, R.; Kulakov, L.A.; Nagy, I. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 1998, 74, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, K.; Fernandes, M.M.; Francesko, A.; Mendoza, E.; Guezguez, J.; Burnet, M.; Tzanov, T. Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces 2015, 7, 27066–27077. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B. Quorum-sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Pan, D.; Zheng, J.; Cheng, Y.; Ma, X.; Huang, F.; Lin, Z. Microscopic investigations of the Cr(VI) uptake mechanism of living Ochrobactrum anthropi. Langmuir 2008, 24, 9630–9635. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.; Cheng, X.; Feng, L.; Xi, C.; Zhang, Y. Immobilization of Rhodococcus rhodochrous BX2 (an acetonitrile-degrading bacterium) with biofilm-forming bacteria for wastewater treatment. Bioresour. Technol. 2013, 131, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Kanga, J.H.; Katayama, Y.; Kondo, F. Biodegradation or metabolism of bisphenol A: From microorganisms to mammals. Toxicology 2006, 217, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Danz, E.; Sei, K.; Soda, S.; Ike, M.; Fujita, M. Biodegradation of bisphenol A, bisphenol F and bisphenol S in seawater. Int. J. Environ. Res. Public Health 2009, 6, 1472–1484. [Google Scholar] [CrossRef] [PubMed]
- Eio, E.J.; Kawai, M.; Tsuchiya, K.; Yamamoto, S.; Toda, T. Biodegradation of bisphenol A by bacterial consortia. Int. Biodeterior. Biodegrad. 2014, 96, 166–173. [Google Scholar] [CrossRef]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eliaz, N.; Ron, E.Z.; Gozin, M.; Younger, S.; Biran, D.; Tal, N. Microbial Degradation of Epoxy. Materials 2018, 11, 2123. https://doi.org/10.3390/ma11112123
Eliaz N, Ron EZ, Gozin M, Younger S, Biran D, Tal N. Microbial Degradation of Epoxy. Materials. 2018; 11(11):2123. https://doi.org/10.3390/ma11112123
Chicago/Turabian StyleEliaz, Noam, Eliora Z. Ron, Michael Gozin, Sara Younger, Dvora Biran, and Noam Tal. 2018. "Microbial Degradation of Epoxy" Materials 11, no. 11: 2123. https://doi.org/10.3390/ma11112123
APA StyleEliaz, N., Ron, E. Z., Gozin, M., Younger, S., Biran, D., & Tal, N. (2018). Microbial Degradation of Epoxy. Materials, 11(11), 2123. https://doi.org/10.3390/ma11112123





