Effect of Copper Ion Concentration on Bacteria and Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inductively Coupled Plasma (ICP) Spectrometry Investigation
2.2. Experimental Procedure
2.3. Bacteria Luminescence
2.4. Bacteria CFU Study
2.5. Cell Study
2.6. Statistical Analysis
3. Results
3.1. Inductively Coupled Plasma (ICP) Spectrometry Study for Cu
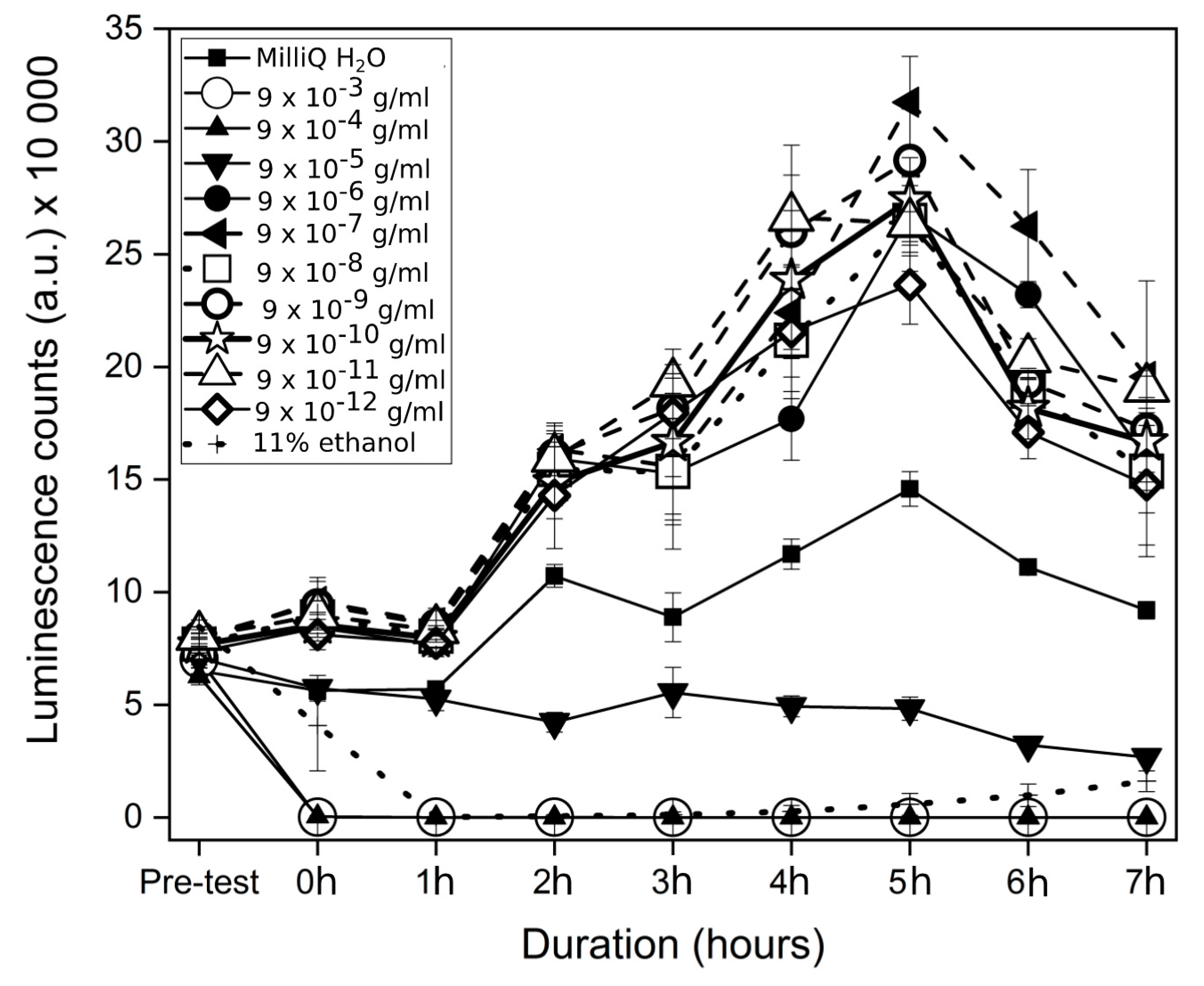
3.2. Bacteria Luminescence
3.3. CFU Counts for XEN43
3.4. Cell Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakka, S.; Coulthard, P. Implant failure: Etiology and complications. Med. Oral. 2011, e42–e44. [Google Scholar] [CrossRef]
- Moy, P.K.; Medina, D.; Shetty, V.; Aghaloo, T.L. Dental implant failure rates and associated risk factors. Int. J. Oral Maxillofac. Implant. 2005, 20, 569–577. [Google Scholar]
- Kärrholm, J.; Lindahl, H.; Malchau, H.; Mohaddes, M.; Nemes, S.; Rogmark, C.; Rolfson, O. Swedish Hip Arthroplasty Register Annual Report; Gothenburg, Sweden, 2016. Available online: https://registercentrum.blob.core.windows.net/shpr/r/Annual-Report-2016-B1eWEH-mHM.pdf (accessed on 26 July 2018).
- Lee, C.-T.; Huang, Y.-W.; Zhu, L.; Weltman, R. Prevalences of peri-implantitis and peri-implant mucositis: Systematic review and meta-analysis. J. Dent. 2017, 62, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rakic, M.; Galindo-Moreno, P.; Monje, A.; Radovanovic, S.; Wang, H.-L.; Cochran, D.; Sculean, A.; Canullo, L. How frequent does peri-implantitis occur? A systematic review and meta-analysis. Clin. Oral Investig. 2018, 22, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Persson, G.R.; Renvert, S. Cluster of Bacteria Associated with Peri-Implantitis. Clin. Implant. Dent. Relat. Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- John, V.; Shin, D.; Marlow, A.; Hamada, Y. Peri-Implant Bone Loss and Peri-Implantitis: A Report of Three Cases and Review of the Literature. Case Rep. Dent. 2016. [Google Scholar] [CrossRef]
- Mohajerani, H.; Roozbayani, R.; Taherian, S.; Tabrizi, R. The Risk Factors in Early Failure of Dental Implants: A Retrospective Study. J. Dent. 2017, 18, 298–303. [Google Scholar]
- Li, M.; Wang, Y.; Gao, L.; Sun, Y.; Wang, J.; Qu, S.; Duan, K.; Weng, J.; Feng, B. Porous titanium scaffold surfaces modified with silver loaded gelatin microspheres and their antibacterial behavior. Surf. Coat. Technol. 2016, 286, 140–147. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, J.Y.; Kim, J.; Lee, J.H.; Hahn, J.S.; Gu, M.B.; Yoon, J. Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity. Water Res. 2009, 43, 1027–1032. [Google Scholar] [CrossRef]
- Sotiriou, G.A.; Meyer, A.; Knijnenburg, J.T.N.; Panke, S.; Pratsinis, S.E. Quantifying the Origin of Released Ag + Ions from Nanosilver. Langmuir 2012, 28, 15929–15936. [Google Scholar] [CrossRef]
- Cao, Y.; Su, B.; Chinnaraj, S.; Jana, S.; Bowen, L.; Charlton, S.; Duan, P.; Jakubovics, N.S.; Chen, J. Nanostructured titanium surfaces exhibit recalcitrance towards Staphylococcus epidermidis biofilm formation. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.B.; Wang, Z.M.; Yang, S.Q.; Luo, Z.J.; Fu, R.K.Y.; Chu, P.K. Antibacterial copper-containing titanium nitride films produced by dual magnetron sputtering. Surf. Coat. Technol. 2007, 201, 8606–8609. [Google Scholar] [CrossRef]
- Russel, A.D.; Hugo, W.B. 7 Antimicrobial Activity and Action of Silver. Prog. Med. Chem. 1994, 31, 351–370. [Google Scholar] [CrossRef]
- Unosson, E.; Tsekoura, E.K.; Engqvist, H.; Welch, K. Synergetic inactivation of Staphylococcus epidermidis and Streptococcus mutans in a TiO2/H2O2/UV system. Biomatter 2013, 3, e26727. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, E. Biocorrosion properties of antibacterial Ti–10Cu sintered alloy in several simulated biological solutions. J. Mater. Sci. Mater. Med. 2015, 26. [Google Scholar] [CrossRef]
- Ma, Z.; Li, M.; Liu, R.; Ren, L.; Zhang, Y.; Pan, H.; Zhao, Y.; Yang, K. In vitro study on an antibacterial Ti-5Cu alloy for medical application. J. Mater. Sci.Mater. Med. 2016, 27, 91. [Google Scholar] [CrossRef]
- Zhang, E.; Ren, J.; Li, S.; Yang, L.; Qin, G. Optimization of mechanical properties, biocorrosion properties and antibacterial properties of as-cast Ti–Cu alloys. Biomed. Mater. 2016, 11, 065001. [Google Scholar] [CrossRef]
- Mathews, S.; Hans, M.; Mücklich, F.; Solioz, M. Contact Killing of Bacteria on Copper Is Suppressed if Bacterial-Metal Contact Is Prevented and Is Induced on Iron by Copper Ions. Appl. Environ. Microbiol. 2013, 79, 2605–2611. [Google Scholar] [CrossRef]
- Li, M.; Ma, Z.; Zhu, Y.; Xia, H.; Yao, M.; Chu, X.; Wang, X.; Yang, K.; Yang, M.; Zhang, Y.; et al. Toward a Molecular Understanding of the Antibacterial Mechanism of Copper-Bearing Titanium Alloys against Staphylococcus aureus. Adv. Healthc. Mater. 2016, 5, 557–566. [Google Scholar] [CrossRef]
- Baker, J.; Sitthisak, S.; Sengupta, M.; Johnson, M.; Jayaswal, R.K.; Morrissey, J.A. Copper Stress Induces a Global Stress Response in Staphylococcus aureus and Represses sae and agr Expression and Biofilm Formation. Appl. Environ. Microbiol. 2010, 76, 150–160. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Nogueira-Pedro, A.; Santos, E.W.; Hastreiter, A.; Silva, G.B.; Borelli, P.; Fock, R.A. A review of select minerals influencing the haematopoietic process. Nutr. Res. Rev. 2018, 31, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Memarzadeh, K.; Chang, B.; Zhang, Y.; Ma, Z.; Allaker, R.P.; Ren, L.; Yang, K. Antibacterial effect of copper-bearing titanium alloy (Ti-Cu) against Streptococcus mutans and Porphyromonas gingivalis. Sci. Rep. 2016, 6, 29985. [Google Scholar] [CrossRef] [PubMed]
- Fowler, L.; Janson, O.; Engqvist, H.; Norgren, S.; Öhman-Mägi, C. Antibacterial investigation of titanium-copper alloys using luminescent Staphylococcus epidermidis in a direct contact test. Mater. Sci. Eng. C 2019, 97, 707–714. [Google Scholar] [CrossRef] [PubMed]
- ISO 10993-12:2002-Biological Evaluation of Medical Devices, Part 12: Sample Preparation and Reference materials, n.d. Available online: https://www.iso.org/standard/35331.html (accessed on 30 July 2018).
- ISO 10993-15:2000(en), Biological Evaluation of Medical Devices, Part 15: Identification and Quantification of Degradation Products From Metals and Alloys, n.d. Available online: https://www.iso.org/obp/ui/#iso:std:iso:10993:-15:ed-1:v2:en (accessed on 30 July 2018).
- Vuong, C.; Kocianova, S.; Yu, J.; Kadurugamuwa, J.L.; Otto, M. Development of Real-Time In Vivo Imaging of Device-Related Staphylococcus epidermidis Infection in Mice and Influence of Animal Immune Status on Susceptibility to Infection. J. Infect. Dis. 2008, 198, 258–261. [Google Scholar] [CrossRef]
- Wiedmer, D.; Petersen, F.C.; Lönn-Stensrud, J.; Tiainen, H. Antibacterial effect of hydrogen peroxide-titanium dioxide suspensions in the decontamination of rough titanium surfaces. Biofouling 2017, 33, 451–459. [Google Scholar] [CrossRef]
- EUCAST: MIC Determination of Non-Fastidious and Fastidious Organisms, European Committee on Antibacterial Susceptibility Testing (EUCAST). 2019. Available online: http://www.eucast.org/ast_of_bacteria/mic_determination/?no_cache=1 (accessed on 16 April 2019).
- Hebling, J.; Bianchi, L.; Basso, F.; Scheffel, D.; Soares, D.; Carrilho, M.; Pashley, D.; Tjäderhane, L.; de Souza Costa, C. Cytotoxicity of dimethyl sulfoxide (DMSO) in direct contact with odontoblast-like cells. Dent. Mater. 2015, 31, 399–405. [Google Scholar] [CrossRef]
- Yuan, C.; Gao, J.; Guo, J.; Bai, L.; Marshall, C.; Cai, Z.; Wang, L.; Xiao, M. Dimethyl Sulfoxide Damages Mitochondrial Integrity and Membrane Potential in Cultured Astrocytes. PLoS ONE 2014, 9, e107447. [Google Scholar] [CrossRef]
- Galvao, J.; Davis, B.; Tilley, M.; Normando, E.; Duchen, M.R.; Cordeiro, M.F. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J. 2013, 28, 1317–1330. [Google Scholar] [CrossRef]
- Hanslick, J.L.; Lau, K.; Noguchi, K.K.; Olney, J.W.; Zorumski, C.F.; Mennerick, S.; Farber, N.B. Dimethyl sulfoxide (DMSO) produces widespread apoptosis in the developing central nervous system. Neurobiol. Dis. 2009, 34, 1–10. [Google Scholar] [CrossRef]
- National Center for Health Statistics (U.S.); National Health Examination Survey (U.S.); National Health and Nutrition Examination Survey (U.S.); Hispanic Health and Nutrition Examination Survey (U.S.) (Eds.) Anthropometric Reference Data for Children and Adults: United States, 2007–2010: Data from the Nathional Health and Nutrition Survey; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Hyattsville, MD, USA, 2012.
- Watson, P.E.; Watson, I.D.; Batt, R.D. Total body water volumes for adult males and females estimated from simple anthropometric measurements. Am. J. Clin. Nutr. 1980, 33, 27–39. [Google Scholar] [CrossRef]
- Fatokun, A.A.; Stone, T.W.; Smith, R.A. Hydrogen peroxide-induced oxidative stress in MC3T3-E1 cells: The effects of glutamate and protection by purines. Bone 2006, 39, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Fatokun, A.A.; Stone, T.W.; Smith, R.A. Responses of differentiated MC3T3-E1 osteoblast-like cells to reactive oxygen species. Eur. J. Pharmacol. 2008, 587, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zheng, Y.; Xi, T.; Zhang, C.; Song, W.; Burugapalli, K.; Yang, H.; Ma, Y. Concentration-dependent cytotoxicity of copper ions on mouse fibroblasts in vitro: Effects of copper ion release from TCu380A vs TCu220C intra-uterine devices. Biomed. Microdevices 2012, 14, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. At the Nexus of Antibiotics and Metals: The Impact of Cu and Zn on Antibiotic Activity and Resistance. Trends Microbiol. 2017, 25, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Wang, X.; Chen, M.; Hou, B. Effect of the existing form of Cu element on the mechanical properties, bio-corrosion and antibacterial properties of Ti-Cu alloys for biomedical application. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 69, 1210–1221. [Google Scholar] [CrossRef] [PubMed]





| Cu ion Solutions | Sample Solutions | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CuCl2·2H20 (g/mL) | 0.25 × 10−1 | 0.25 × 10−2 | 0.25 × 10−3 | 0.25 × 10−4 | 0.25 × 10−5 | 0.25 × 10−6 | 0.25 × 10−7 | 0.25 × 10−8 | 0.25 × 10−9 | 0.25 × 10−10 |
| Cu ions (g/mL) | 9 × 10−3 | 9 × 10−4 | 9 × 10−5 | 9 × 10−6 | 9 × 10−7 | 9 × 10−8 | 9 × 10−9 | 9 × 10−10 | 9 × 10−11 | 9 × 10−12 |
| Name | Cu Concentration (mg/L) | Standard Deviation (%) | RSD (%) |
|---|---|---|---|
| 99.5% Cu-200um * | 2.632 | 0.0922 | 3.5 |
| 99.5% Cu-200um * | 2.902 | 0.0204 | 0.7 |
| 99.5% Cu-220um * | 2.72 | 0.0006 | 0.02 |
| TiCu-10-630um * | 0.07 | 0.0012 | 1.69 |
| TiCu-10-590um * | 0.072 | 0.001 | 1.41 |
| TiCu-10-525um * | 0.063 | 0.0012 | 1.94 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fowler, L.; Engqvist, H.; Öhman-Mägi, C. Effect of Copper Ion Concentration on Bacteria and Cells. Materials 2019, 12, 3798. https://doi.org/10.3390/ma12223798
Fowler L, Engqvist H, Öhman-Mägi C. Effect of Copper Ion Concentration on Bacteria and Cells. Materials. 2019; 12(22):3798. https://doi.org/10.3390/ma12223798
Chicago/Turabian StyleFowler, Lee, Håkan Engqvist, and Caroline Öhman-Mägi. 2019. "Effect of Copper Ion Concentration on Bacteria and Cells" Materials 12, no. 22: 3798. https://doi.org/10.3390/ma12223798
APA StyleFowler, L., Engqvist, H., & Öhman-Mägi, C. (2019). Effect of Copper Ion Concentration on Bacteria and Cells. Materials, 12(22), 3798. https://doi.org/10.3390/ma12223798





