Interferon α2–Thymosin α1 Fusion Protein (IFNα2–Tα1): A Genetically Engineered Fusion Protein with Enhanced Anticancer and Antiviral Effect
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
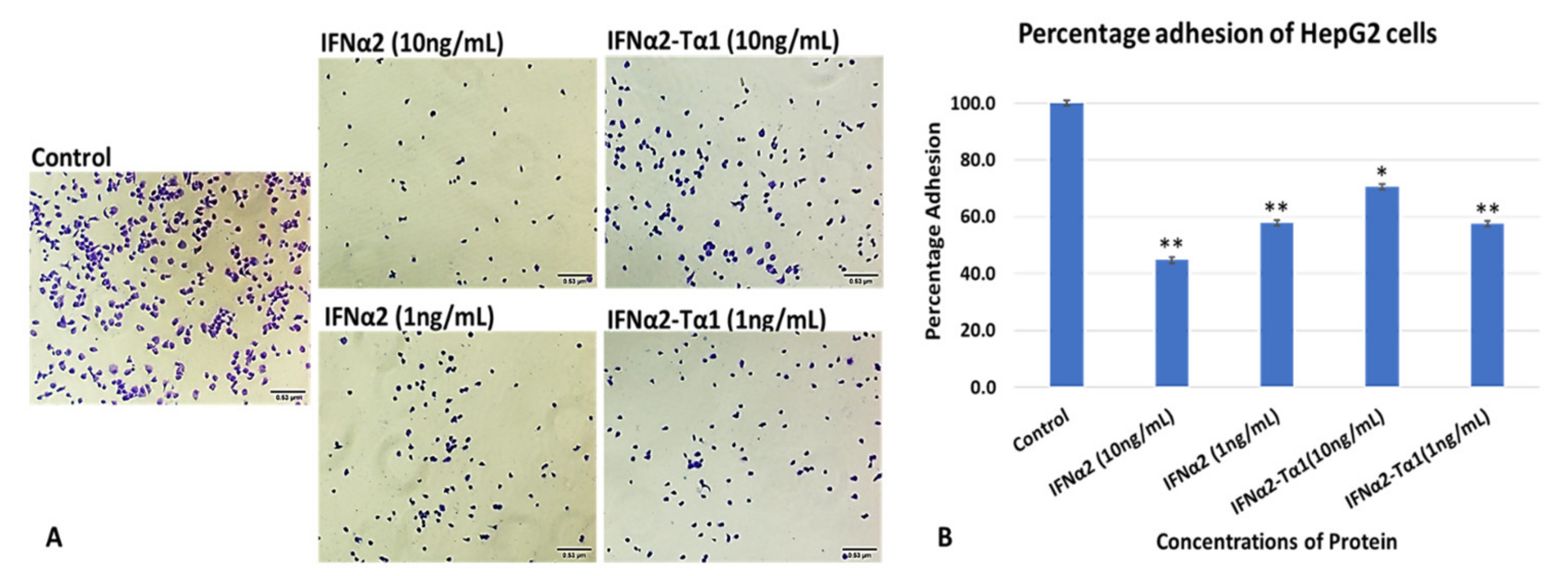
2.2.1. Cell Attachment Assay
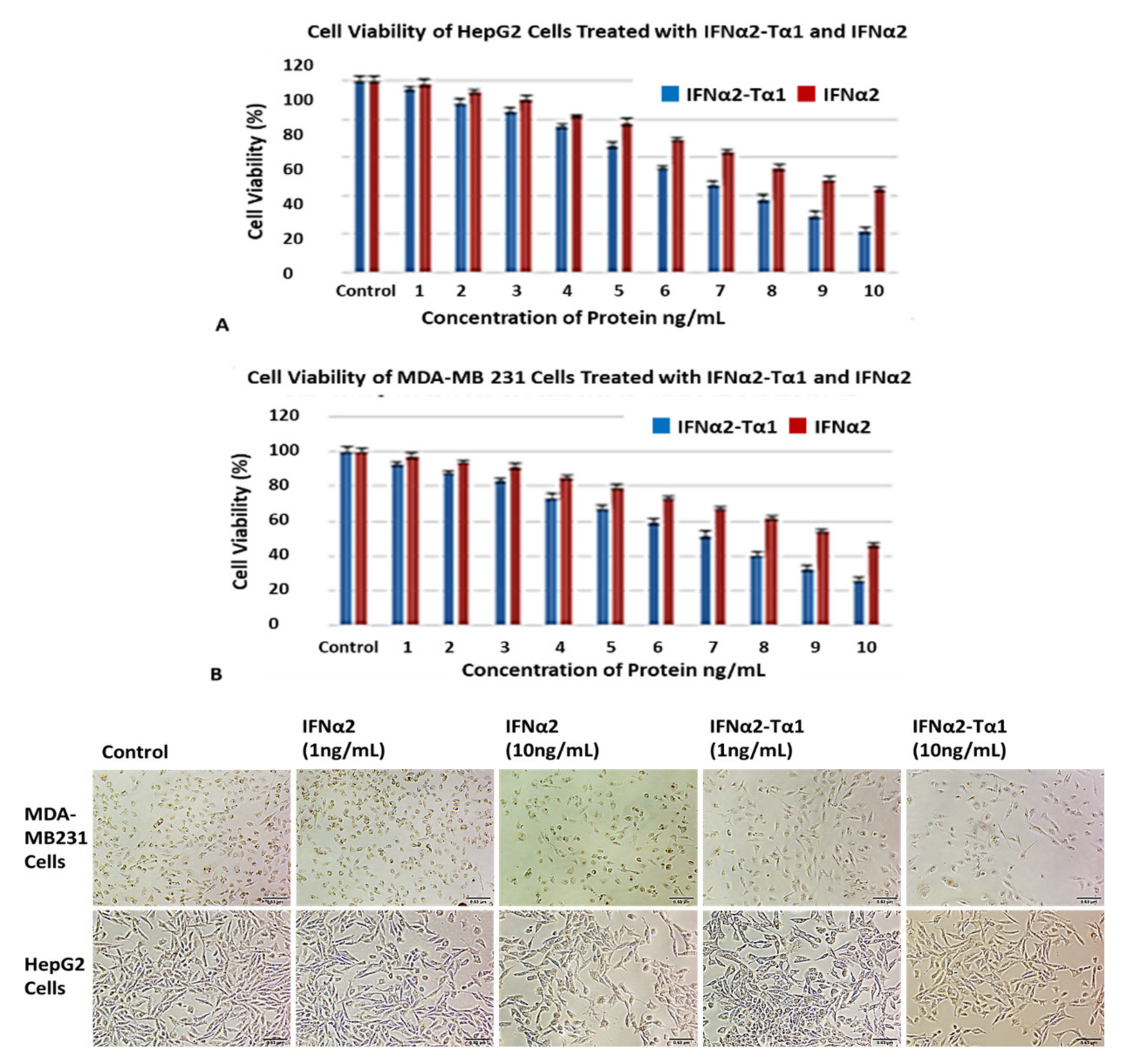
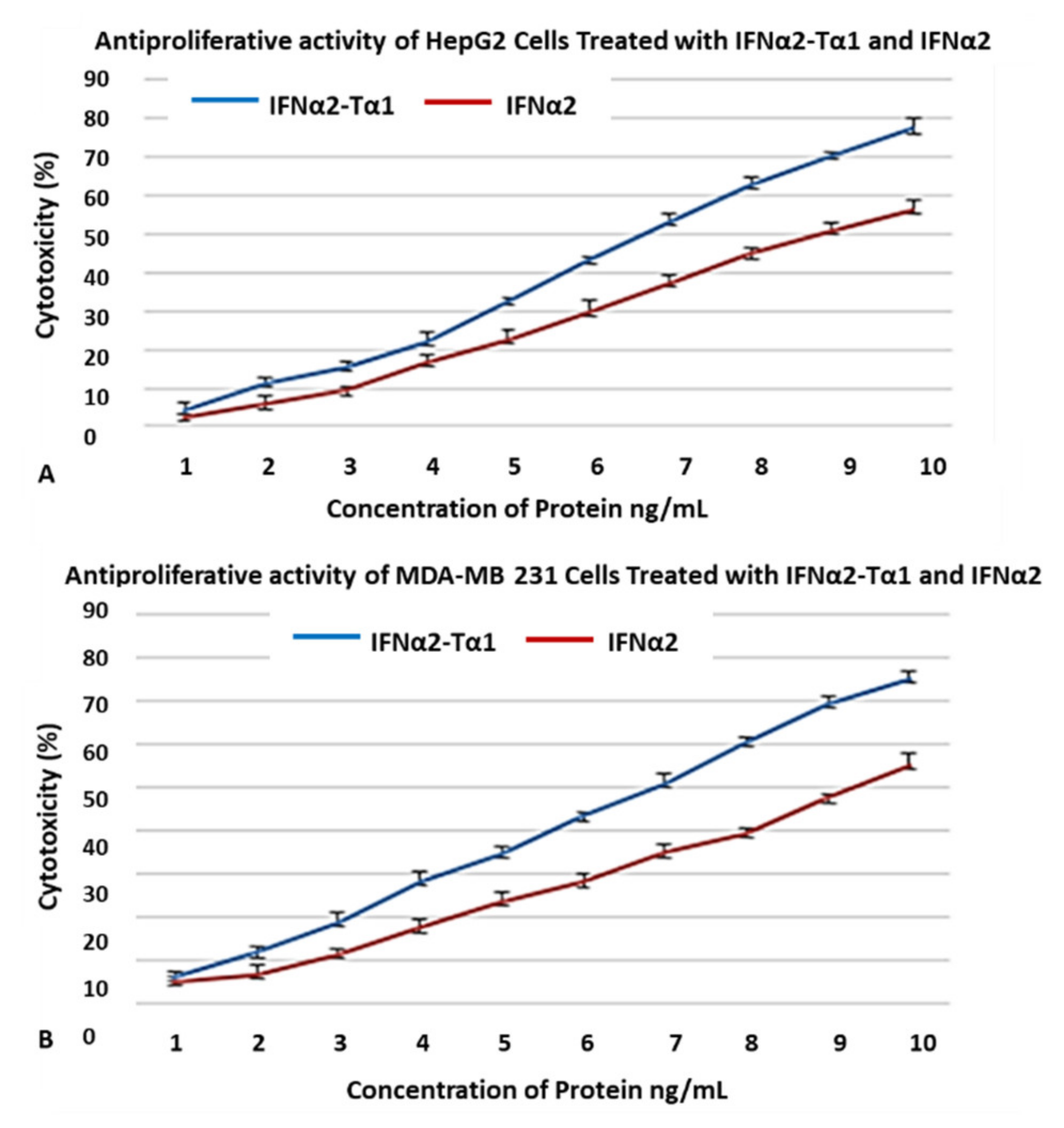
2.2.2. Anti-Proliferative Effect
Neutral Red Assay
MTT Assay
2.2.3. Genotoxic Effect Analysis by Comet Assay
2.2.4. Antiviral Activity
In Vitro Toxicity Analysis
In Vitro Anti-HCV Assay
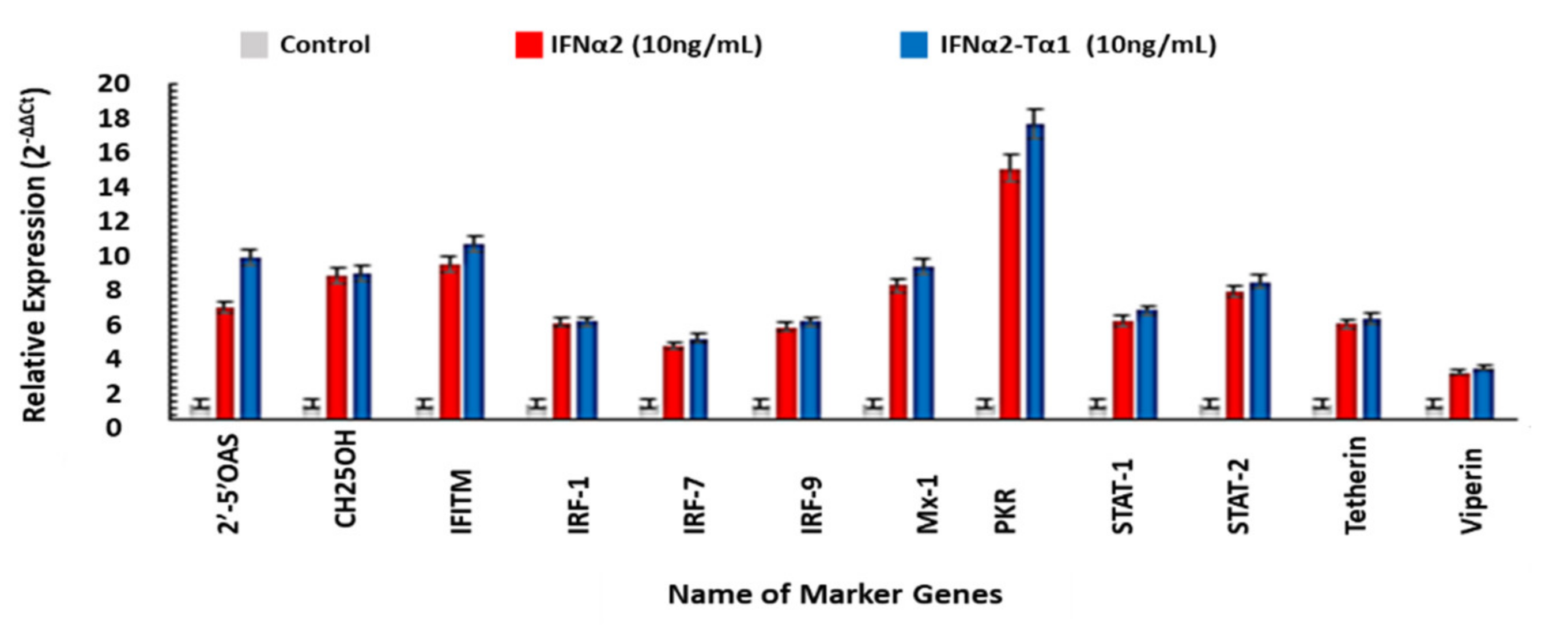
2.2.5. Expression Analysis
2.2.6. Statistical Analysis
3. Results
3.1. In Vitro HepG2 Cells Attachment Assay
3.2. Enhanced Inhibition of HepG2 and MDA-MB-231 Cell Proliferation by IFNα2–Tα1
3.3. Genotoxic Effect of IFNα2–Tα1 in HepG2 Cells and MDA-MB-231 Cells
3.4. Higher Antiviral Effect of IFNα2–Tα1 in HCV-Infected Huh7 Cells
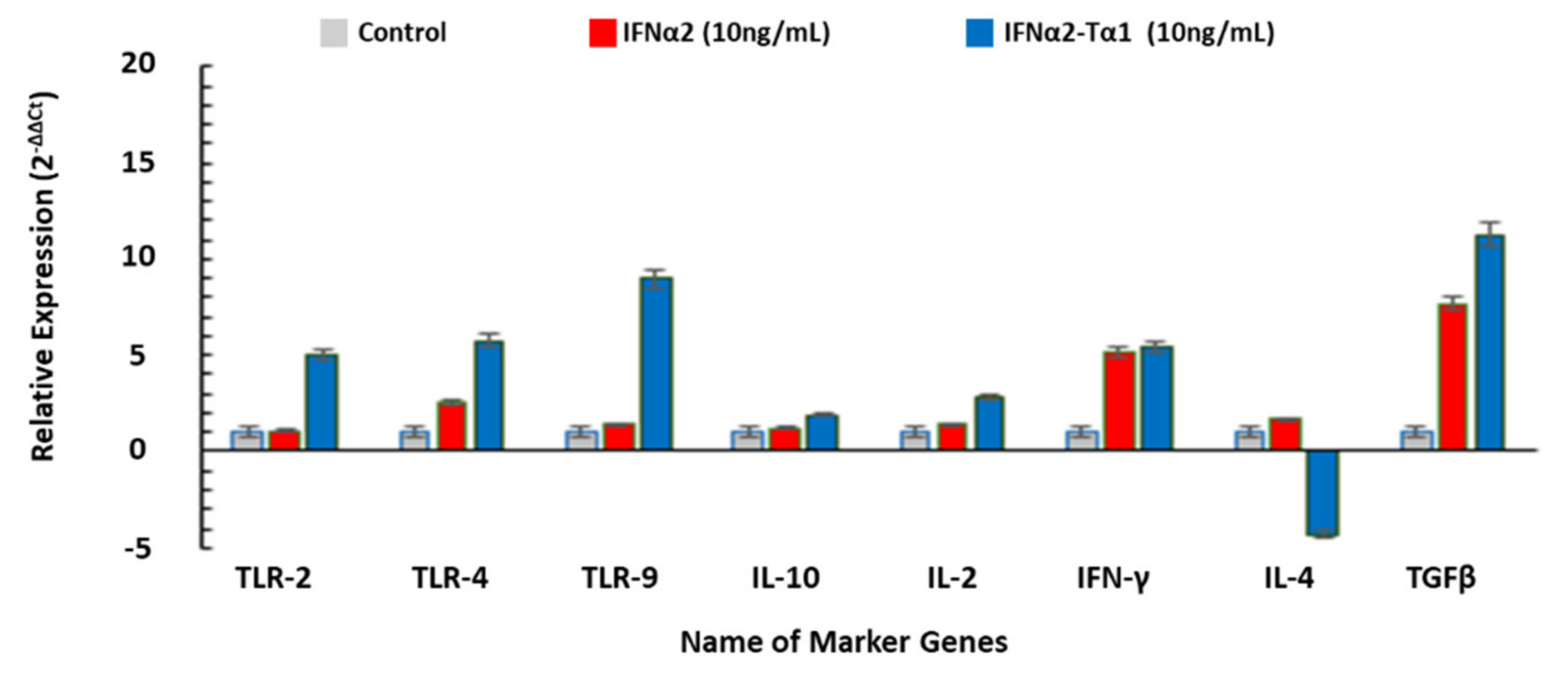
3.5. The Relative Expression of Genes Associated with Anticancer, Antiviral and Immune-modulatory Activities in IFNα2–Tα1 and IFN-α-2 Treated Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| NK | Natural killer cells |
| DCs | Dendritic cells |
| TLR | Toll-like receptor |
| 2′,5′-OAS | 2′,5′-oligoadenylate |
| IRF | Interferon regulatory factor |
| STAT | Signal Transducer and Activator of Transcription |
| PKR | Protein Kinase R |
| JNK-2 | c-Jun N-terminal kinase-2 |
| IFITM | Interferon-inducible transmembrane protein |
| CH25OH | Cholesterol-25-hydroxylase |
| BCL-2 | B-cell lymphoma 2 |
| BAX | BCL2 associated X |
| CDK | Cyclin-dependent kinases |
| Mx-1 | Myxovirus resistance-1 |
| TGF-β | Transforming growth factor-beta |
| TRAIL | TNF-related apoptosis-inducing ligand |
| CASP | Caspase |
References
- Bekisz, J.; Baron, S.; Balinsky, C.; Morrow, A.; Zoon, K.C. Anti-proliferative properties of type I and type II interferon. Pharmaceuticals 2010, 3, 994–1015. [Google Scholar] [CrossRef]
- Belinda, S.P.; Rautela, J.; Hertzog, P.J. Anti-tumor actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 6, 131–144. [Google Scholar]
- Cheon, H.; Borden, E.C.; Stark, G.R. Interferons and Their Stimulated Genes in the Tumor Microenvironment. Semin. Oncol. 2014, 41, 156–173. [Google Scholar] [CrossRef]
- Paola, R.; Franca, M.; Filippo, B. Recent advances on the immunomodulatory effects of IFN-α: Implications for cancer immunotherapy and autoimmunity. Autoimmun 2010, 43, 204–209. [Google Scholar]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Malmgard, L. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 2004, 24, 439–454. [Google Scholar] [CrossRef]
- William, M.; Schneider, M.; Dittmann, C.; Charles, M.R. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar]
- Micco, L.; Dimitra, P.; Elisabetta, L.; Anna, S.; Lucy, J.; Carmela, C.; Arianna, P.; Bernardi, M.; Brander, M.; Bihl, C.; et al. Differential boosting of innate and adaptive antiviral responses during pegylated-interferon alpha therapy of chronic hepatitis B. J. Hepatol. 2013, 58, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Kessler, D.S.; Levy, D.E.; Darnell, J.E. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 1988, 85, 8521–8525. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. The p38 mitogen-activated protein kinase pathway and its role in interferon signaling. Pharmacol. Ther. 2003, 98, 129–142. [Google Scholar] [CrossRef]
- Kaur, S.; Katsoulidis, E.; Platanias, L.C. Akt and mRNA translation by interferons. Cell Cycle 2008, 7, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Sassano, A.; Dolniak, B.; Joshi, S.; Majchrzak-Kita, B.; Baker, D.P.; Hay, N.; Fish, E.N.; Platanias, L.C. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 2008, 105, 4808–4813. [Google Scholar] [CrossRef] [PubMed]
- Martins, E.B.; Tuthill, C. Thymosin alpha 1, a new biologic response modifier for the treatment of viral hepatitis and hepatocellular carcinoma. Rev. Gastroenterol. Peru 2000, 20, 58–63. [Google Scholar]
- Grottesi, A.; Sette, M.; Palamara, A.T.; Rotilio, G.; Garaci, E.; Paci, M. The conformation of peptide thymosin α1 in solution and in a membrane-like environment by circular dichroism and NMR spectroscopy. a possible model for its interaction with the lymphocyte membrane. Peptides 1998, 19, 1731–1738. [Google Scholar] [CrossRef]
- Elizondo-Riojas, M.-A.; Chamow, S.M.; Tuthill, C.W.; Gorenstein, D.G.; Volk, D.E. NMR structure of human thymosin alpha-1. Biochem. Biophys. Res. Commun. 2011, 416, 356–361. [Google Scholar] [CrossRef]
- Billich, A. Thymosin alpha1. SciClone Pharmaceuticals. Expert Opin. Investig. Drugs 2002, 3, 698–707. [Google Scholar]
- Li, J.; Liu, C.H.; Wang, F.S. Thymosin alpha 1: Biological activities, applications and genetic engineering production. Peptides 2010, 31, 2151–2158. [Google Scholar] [CrossRef]
- Xiaoning, W.; Jidong, J.; Hong, Y. Thymosin alpha-1 treatment in chronic hepatitis B. Expert Opin. Biol. Ther. 2015, 15, 129–132. [Google Scholar]
- King, R.; Tuthill, C. Immune Modulation with Thymosin Alpha 1 Treatment. Vitam. Horm. 2016, 102, 151–178. [Google Scholar] [CrossRef]
- Romani, L.; Bistoni, F.; Perruccio, K.; Montagnoli, C.; Gaziano, R.; Bozza, S.; Bonifazi, P.; Bistoni, G.; Rasi, G.; Velardi, A.; et al. Thymosin α1 activates dendritic cell tryptophan catabolism and establishes a regulatory environment for balance of inflammation and tolerance. Blood 2006, 108, 2265–2274. [Google Scholar] [CrossRef]
- Pierluigi, B.; D’Angelo, C.; Fallarino, F.; Moretti, S.; Zelante, T. Thymosin alpha1: The regulator of regulators? Ann. N. Y. Acad. Sci. 2010, 1194, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Singh, S.M.; Singh, N. Effect of thymosin alpha 1 on the anti-tumor activity of tumor-associated macrophage-derived dendritic cells. J. Biomed. Sci. 2004, 11, 623–630. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, F.-D.; Zhou, L.; Gong, X.-G.; Han, Q.-F. Proliferative and anti-proliferative effects of thymosin α1 on cells are associated with manipulation of cellular ROS levels. Chem. Interact. 2009, 180, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shi, Y.; Zhou, J.; Sun, Y.; Piao, H.; Jiang, W.; Ma, A.; Chen, Y.; Xu, M.; Xie, W.; et al. Combination of entecavir with thymosin alpha-1 in HBV-related compensated cirrhosis: A prospective multicenter randomized open-label study. Expert Opin. Biol. Ther. 2018, 18, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Costantini, C.; Bellet, M.M.; Pariano, M.; Renga, G.; Stincardini, C.; Goldstein, A.L.; Garaci, E.; Romani, L. A Reappraisal of Thymosin Alpha1 in Cancer Therapy. Front. Oncol. 2019, 9, 873. [Google Scholar] [CrossRef]
- Garaci, E.; Pica, F.; Serafino, A.; Balestrieri, E.; Matteucci, C.; Moroni, G.; Sorrentino, R.; Zonfrillo, M.; Pierimarchi, P.; Sinibaldi-Vallebona, P. Thymosin α1 and cancer: Action on immune effector and tumor target cells. Ann. N. Y. Acad. Sci. 2012, 1269, 26–33. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Averhoff, F.M.; Glass, N.; Holtzman, D. Global Burden of Hepatitis C: Considerations for Healthcare Providers in the United States. Clin. Infect. Dis. 2012, 55, S10–S15. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Cynthia, W.T.; Robert, S.K. Thymosin Alpha 1-A Peptide Immune Modulator with a Broad Range of Clinical Applications. Clin. Exp. Pharmacol. Physiol. 2013, 3, 1–17. [Google Scholar]
- Xuemei, L.; Jie, W.; Xiaobao, J.; Jiayong, Z. High-level expression of a novel liver-targeting fusion interferon with preferred Escherichia coli codon preference and its anti-hepatitis B virus activity in vivo. BMC Biotechnol. 2015, 15, 1–12. [Google Scholar]
- Young, P.A.; Dang, N.H.; Nastoupil, L.J.; Minning, D.; Gresser, M.J.; Timmerman, J. Antibody-interferon-alpha fusion protein (IGN002) for the treatment of β-cell non-Hodgkin lymphomas: A phase 1, first-in-human, dose-escalation trial. J. Clin. Oncol 2016, 345, 15. [Google Scholar] [CrossRef]
- Wen, Z.; Jia, Q.; Kang, X.; Lou, Y.; Zou, L.; Yang, J.; Gao, J.; Han, L.; Li, X. Anti-tumor activity of recombinant RGD-IFN-α2a-core fusion protein in vitro. Anti-Cancer Drug 2017, 28, 31–39. [Google Scholar] [CrossRef]
- Xingzhen, L.; Meng, L.; Jiao, C.; Heng, Z. A Tumor-Penetrating Peptide Modification Enhances the Anti-tumor Activity of Thymosin Alpha 1. PLoS ONE 2013, 8, e72242. [Google Scholar]
- Fotakis, G.; Timbrell, J.A. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006, 160, 171–177. [Google Scholar] [CrossRef]
- Carmichael, J.; De Graff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [Google Scholar]
- Beedanagari, S.; Vulimiri, S.V.; Bhatia, S.; Mahadevan, B. Genotoxicity Biomarkers: Molecular Basis of Genetic Variability and Susceptibility, Biomarkers in Toxicology; Elsevier Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Dominari, A.; Hathaway, D., III; Pandav, K.; Matos, W.; Biswas, S.; Reddy, G.; Thevuthasan, S.; Khan, M.A.; Mathew, A.; Makkar, S.S.; et al. Thymosin alpha 1: A comprehensive review of the literature. World J. Virol. 2020, 9, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.S.; Gull, I.; Mahmood, M.S.; Iqbal, M.M.; Abbas, Z.; Tipu, I.; Ahmed, A.; Athar, M.A. High yield expression, characterization, and biological activity of IFNα2-Tα1. Prep. Biochem. Biotech. 2020, 50, 281–291. [Google Scholar] [CrossRef]
- Nan-Nan, L.; Ping, L.; Su-Juan, C.; Qing-Ping, L.; Liang-Fan, Z.; Shuang-Quan, Z. Construction and expression of a novel bioactive IFN-a2b/CM4 fusion protein in Escherichia coli. Microbiol. Res. 2010, 165, 116–121. [Google Scholar]
- Hu, J.; Wang, G.; Liu, X.; Gao, W. Enhancing pharmacokinetics, tumor accumulation, and anti-tumor efficacy by elastin-like polypeptide fusion of interferon alpha. Adv. Mater. 2015, 27, 7320–7324. [Google Scholar] [CrossRef]
- Wang, F.; Li, B.; Fu, P.; Li, Q.; Zheng, H.; Lao, X. Immunomodulatory and enhanced anti-tumor activity of a modified thymosin alpha1 in melanoma and lung cancer. Int. J. Pharm. 2018, 547, 611–620. [Google Scholar] [CrossRef]
- Choucroun, P.; Gillet, D.; Dorange, G.; Sawicki, B.; Dewitte, J. Comet assay and early apoptosis. Mutat. Res. Mol. Mech. Mutagen. 2001, 478, 89–96. [Google Scholar] [CrossRef]
- Balachandran, S.; Roberts, P.C.; Brown, L.E.; Truong, H.; Pattnaik, A.K.; Archer, D.R.; Barber, G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 2000, 13, 129–141. [Google Scholar] [CrossRef]
- Chen, Q.; Gong, B.; Mahmoud-Ahmed, A.S.; Zhou, A.; Hsi, E.D.; Hussein, M.; Almasan, A. Apo2L/TRAIL and Bcl-2–related proteins regulate type I interferon–induced apoptosis in multiple myeloma. Blood 2001, 98, 2183–2192. [Google Scholar] [CrossRef] [PubMed]
- Von Marschall, Z.; Scholz, A.; Cramer, T.; Schäfer, G.; Schirner, M.; Öberg, K.; Wiedenmann, B.; Höcker, M.; Rosewicz, S. Effects of Interferon Alpha on Vascular Endothelial Growth Factor Gene Transcription and Tumor Angiogenesis. J. Natl. Cancer Inst. 2003, 95, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Steelman, L.S.; Pohnert, S.C.; Shelton, J.G.; Franklin, R.A.; Bertrand, F.E.; McCubrey, J.A. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 2004, 18, 189–218. [Google Scholar] [CrossRef]
- Fuertes, M.B.; Kacha, A.K.; Kline, J.; Woo, S.R.; Kranz, D.M.; Murphy, K.M.; Gajewski, T.F. Host type I IFN signals are required for anti-tumor CD8+ T cell responses through CD8+ dendritic cells. J. Exp. Med. 2011, 208, 2005–2016. [Google Scholar] [CrossRef]
- Hardwick, J.M.; Soane, L. Multiple Functions of BCL-2 Family Proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008722. [Google Scholar] [CrossRef]
- Ningrum, R.A. Human Interferon Alpha-2b: A Therapeutic Protein for Cancer Treatment. Scientifica 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Lao, X.; Li, B.; Liu, M.; Shen, C.; Yu, T.; Gao, X.; Zheng, H. A modified thymosin alpha 1 inhibits the growth of breast cancer both in vitro and in vivo: Suppressment of cell proliferation, inducible cell apoptosis and enhancement of targeted anticancer effects. Apoptosis 2015, 20, 1307–1320. [Google Scholar] [CrossRef]
- Subramanian, G.M.; Fiscella, M.; Lamousé-Smith, A.; Zeuzem, S.; McHutchison, J.G. Albinterferon α-2b: A genetic fusion protein for the treatment of chronic hepatitis C. Nat. Biotechnol. 2007, 25, 1411–1419. [Google Scholar] [CrossRef]
- Schmeisser, H.; Mejido, J.; Balinsky, C.A.; Morrow, A.N.; Clark, C.R.; Zhao, T.; Zoon, K.C. Identification of Alpha Interferon-Induced Genes Associated with Antiviral Activity in Daudi Cells and Characterization of IFIT3 as a Novel Antiviral Gene. J. Virol. 2010, 84, 10671–10680. [Google Scholar] [CrossRef] [PubMed]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef] [PubMed]
- Isogawa, M.; Robek, M.D.; Furuichi, Y.; Chisari, F.V. Toll-Like Receptor Signaling Inhibits Hepatitis B Virus Replication In Vivo. J. Virol. 2005, 79, 7269–7272. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Seki, E.; Brenner, D.A. Toll-Like Receptor Signaling in the Liver. Gastroenterology 2006, 130, 1886–1900. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, M.; Meng, Z.; Trippler, M.; Broering, R.; Szczeponek, A.; Krux, F.; Dittmer, U.; Roggendorf, M.; Gerken, G.; et al. Toll-like receptor-mediated control of HBV replication by nonparenchymal liver cells in mice. Hepatology 2007, 46, 1769–1778. [Google Scholar] [CrossRef]
- Zhiyong, M.; Qian, C.; Yong, X.; Ejuan, Z.; Mengji, L. Interaction between Hepatitis B Virus and Toll-Like Receptors: Current Status and Potential Therapeutic Use for Chronic Hepatitis B. Vaccines 2018, 6, 6–20. [Google Scholar]
- Andreone, P.; Gramenzi, A.; Cursaro, C.; Felline, F.; Loggi, E.; D’Errico, A.; Spinosa, M.; Lorenzini, S.; Biselli, M.; Bernardi, M. Thymosin-alpha 1 plus interferon-alpha for naive patients with chronic hepatitis C: Results of a randomized controlled pilot trial. J. Viral Hepat. 2004, 11, 69–73. [Google Scholar] [CrossRef]
- Blobe, G.C.; Schiemann, W.P.; Lodish, H.F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000, 342, 1350–1358. [Google Scholar] [CrossRef]
- Iyer, S.S.; Genhong, C. Role of Interleukin 10 Transcriptional Regulation in Inflammation and Autoimmune Disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, J.; Wang, Z.; Zhang, J.; Xiao, M.; Wang, C.; Lu, Y.; Qin, Z. Endogenous Interleukin-4 Promotes Tumor Development by Increasing Tumor Cell Resistance to Apoptosis. Cancer Res. 2008, 68, 8687–8694. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aslam, M.S.; Zaidi, S.Z.J.; Toor, R.H.; Gull, I.; Iqbal, M.M.; Abbas, Z.; Tipu, I.; Ahmed, A.; Athar, M.A.; Harito, C.; et al. Interferon α2–Thymosin α1 Fusion Protein (IFNα2–Tα1): A Genetically Engineered Fusion Protein with Enhanced Anticancer and Antiviral Effect. Materials 2021, 14, 3318. https://doi.org/10.3390/ma14123318
Aslam MS, Zaidi SZJ, Toor RH, Gull I, Iqbal MM, Abbas Z, Tipu I, Ahmed A, Athar MA, Harito C, et al. Interferon α2–Thymosin α1 Fusion Protein (IFNα2–Tα1): A Genetically Engineered Fusion Protein with Enhanced Anticancer and Antiviral Effect. Materials. 2021; 14(12):3318. https://doi.org/10.3390/ma14123318
Chicago/Turabian StyleAslam, Muhammad Shahbaz, Syed Zohaib Javaid Zaidi, Rabail Hassan Toor, Iram Gull, Muhammad Mudassir Iqbal, Zaigham Abbas, Imran Tipu, Aftab Ahmed, Muhammad Amin Athar, Christian Harito, and et al. 2021. "Interferon α2–Thymosin α1 Fusion Protein (IFNα2–Tα1): A Genetically Engineered Fusion Protein with Enhanced Anticancer and Antiviral Effect" Materials 14, no. 12: 3318. https://doi.org/10.3390/ma14123318
APA StyleAslam, M. S., Zaidi, S. Z. J., Toor, R. H., Gull, I., Iqbal, M. M., Abbas, Z., Tipu, I., Ahmed, A., Athar, M. A., Harito, C., & Hassan, S.-u. (2021). Interferon α2–Thymosin α1 Fusion Protein (IFNα2–Tα1): A Genetically Engineered Fusion Protein with Enhanced Anticancer and Antiviral Effect. Materials, 14(12), 3318. https://doi.org/10.3390/ma14123318






