Photocatalytic Generation of Singlet Oxygen by Graphitic Carbon Nitride for Antibacterial Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Preparation
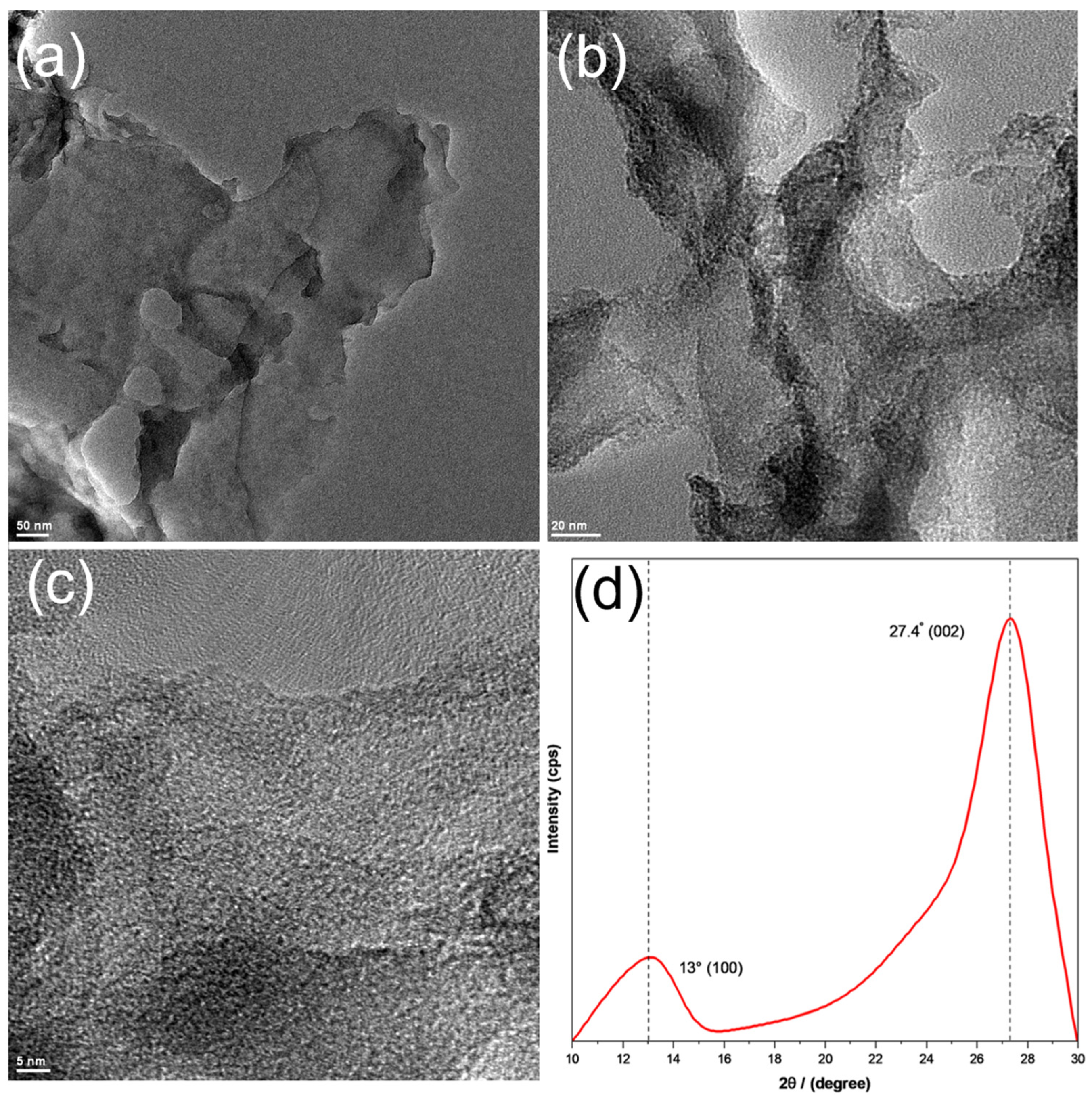
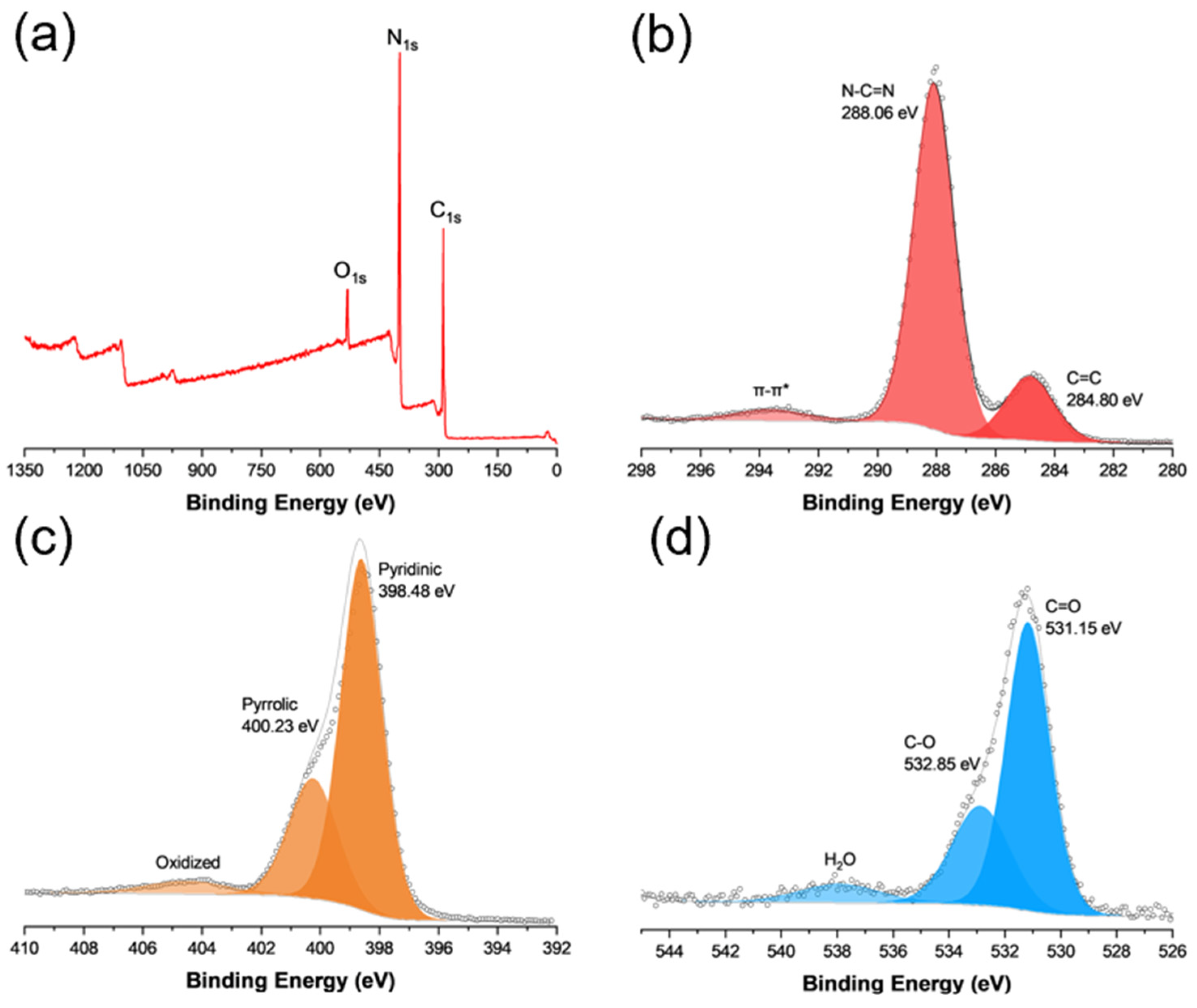
2.3. Characterizations
2.4. Ellman’s Assay
2.5. Photocatalytic Degradation of Methylene Blue
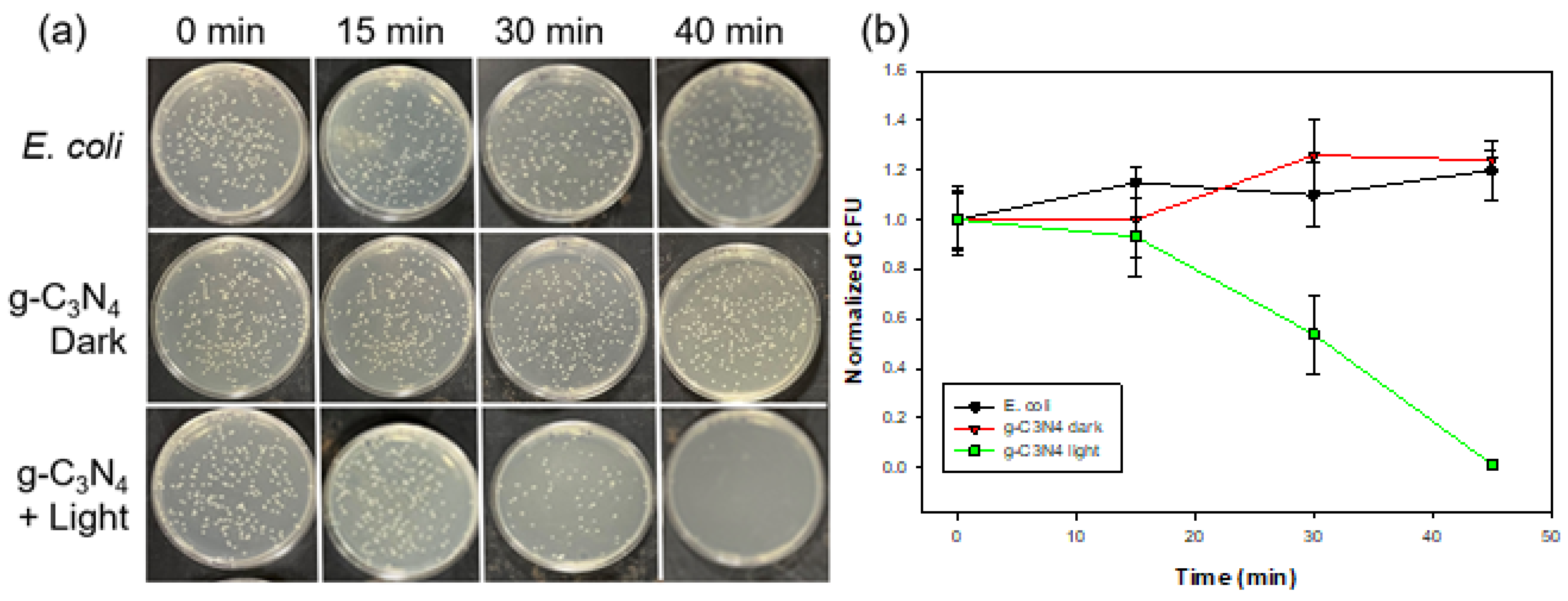
2.6. Photodynamic Studies
2.7. Live/Dead Assay
2.8. SEM Imaging of Bacterial Cells
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, K.Y.; Setyawati, M.I.; Leong, D.T.; Xie, J.P. Antimicrobial silver nanomaterials. Coord. Chem. Rev. 2018, 357, 1–17. [Google Scholar] [CrossRef]
- Piddock, L.J. The crisis of no new antibiotics—What is the way forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Collins, J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 2009, 12, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Andrade, M.D.; Chata, G.; Rouholiman, D.; Liu, J.; Saltikov, C.; Chen, S. Antimicrobial mechanisms of graphene-based composite materials. Nanoscale 2017, 9, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Ristic, B.Z.; Milenkovic, M.M.; Dakic, I.R.; Todorovic-Markovic, B.M.; Milosavljevic, M.S.; Budimir, M.D.; Paunovic, V.G.; Dramicanin, M.D.; Markovic, Z.M.; Trajkovic, V.S. Photodynamic antibacterial effect of graphene quantum dots. Biomaterials 2014, 35, 4428–4435. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Lan, M.; Zhou, B.; Liu, W.; Guo, L.; Wang, H.; Jia, Q.; Niu, G.; Huang, X.; Zhou, H.; et al. A graphene quantum dot photodynamic therapy agent with high singlet oxygen generation. Nat. Commun. 2014, 5, 4596. [Google Scholar] [CrossRef] [PubMed]
- Nonell, S.; Flors, C. Singlet Oxygen: Applications in Biosciences and Nanosciences; Royal Society of Chemistry: London, UK, 2016. [Google Scholar]
- Maisch, T.; Baier, J.; Franz, B.; Maier, M.; Landthaler, M.; Szeimies, R.M.; Bäumler, W. The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria. Proc. Natl. Acad. Sci. USA 2007, 104, 7223–7228. [Google Scholar] [CrossRef]
- Malik, Z.; Hanania, J.; Nitzan, Y. New trends in photobiology bactericidal effects of photoactivated porphyrins—An alternative approach to antimicrobial drugs. J. Photochem. Photobiol. B Biol. 1990, 5, 281–293. [Google Scholar] [CrossRef]
- Hill, E.H.; Pappas, H.C.; Whitten, D.G. Activating the Antimicrobial Activity of an Anionic Singlet-Oxygen Sensitizer through Surfactant Complexation. Langmuir 2014, 30, 5052–5056. [Google Scholar] [CrossRef]
- Yuan, H.; Theresa, M.; Bain, D.; Fakhouri, H.; Sreekanth, K.; Ravi, A.; Thomas, S.; Kalarikkal, N.; Antoine, R.; Radhakrishnan, E.K. Singlet oxygen generation efficiency and antimicrobial ability in glutathione protected Ag31 nanoclusters. Inorg. Chem. Commun. 2024, 159, 111799. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, S.L.; Chen, S.C.; Li, D.D.; Zhang, X.D.; Shao, W.; Sun, X.S.; Xie, J.F.; Zhao, Z.; Zhang, Q.; et al. Enhanced Singlet Oxygen Generation in Oxidized Graphitic Carbon Nitride for Organic Synthesis. Adv. Mater. 2016, 28, 6940–6945. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Che, H.N.; Liu, B.; Ao, Y.H. Unveiling the mechanism on photocatalytic singlet oxygen generation over rationally designed carbonylated carbon nitride. J. Mater. Chem. A 2024, 12, 13427–13434. [Google Scholar] [CrossRef]
- Xu, L.P.; Li, L.J.; Yu, L.; Yu, J.C. Efficient generation of singlet oxygen on modified g-C3N4 photocatalyst for preferential oxidation of targeted organic pollutants. Chem. Eng. J. 2022, 431, 34241. [Google Scholar] [CrossRef]
- Oh, W.D.; Chang, V.W.C.; Hu, Z.T.; Goei, R.; Lim, T.T. Enhancing the catalytic activity of g-CN through Me doping (Me = Cu, Co and Fe) for selective sulfathiazole degradation redox-based advanced oxidation process. Chem. Eng. J. 2017, 323, 260–269. [Google Scholar] [CrossRef]
- Liu, J.L.; Cheng, W.X.; Wang, Y.H.; Fan, X.Y.; Shen, J.H.; Liu, H.; Wang, A.Q.; Hui, A.P.; Nichols, F.; Chen, S.W. Cobalt-Doped Zinc Oxide Nanoparticle-MoS Nanosheet Composites as Broad-Spectrum Bactericidal Agents. Acs Appl. Nano Mater. 2021, 4, 4361–4370. [Google Scholar] [CrossRef]
- Yang, P.; Zuo, S.; Zhang, F.; Yu, B.; Guo, S.; Yu, X.; Zhao, Y.; Zhang, J.; Liu, Z. Carbon Nitride-Based Single-Atom Cu Catalysts for Highly Efficient Carboxylation of Alkynes with Atmospheric CO2. Ind. Eng. Chem. Res. 2020, 59, 7327–7335. [Google Scholar] [CrossRef]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef]
- Khabashesku, V.N.; Zimmerman, J.L.; Margrave, J.L. Powder synthesis and characterization of amorphous carbon nitride. Chem. Mater. 2000, 12, 3264–3270. [Google Scholar] [CrossRef]
- Xiang, Q.J.; Yu, J.G.; Jaroniec, M. Preparation and Enhanced Visible-Light Photocatalytic H-Production Activity of Graphene/CN Composites. J. Phys. Chem. C 2011, 115, 7355–7363. [Google Scholar] [CrossRef]
- Yan, S.C.; Li, Z.S.; Zou, Z.G. Photodegradation of Rhodamine B and Methyl Orange over Boron-Doped g-CN under Visible Light Irradiation. Langmuir 2010, 26, 3894–3901. [Google Scholar] [CrossRef] [PubMed]
- Nichols, F.; Lu, J.E.; Mercado, R.; Rojas-Andrade, M.D.; Ning, S.L.; Azhar, Z.; Sandhu, J.; Cazares, R.; Saltikov, C.; Chen, S.W. Antibacterial Activity of Nitrogen-Doped Carbon Dots Enhanced by Atomic Dispersion of Copper. Langmuir 2020, 36, 11629–11636. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Andrade, M.D.; Nguyen, T.A.; Mistler, W.P.; Armas, J.; Lu, J.E.; Roseman, G.; Hollingsworth, W.R.; Nichols, F.; Millhauser, G.L.; Ayzner, A.; et al. Antimicrobial activity of graphene oxide quantum dots: Impacts of chemical reduction. Nanoscale Adv. 2020, 2, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, J.L.; Chen, X.; Zhao, Y.; Zhao, W.G.; Qian, G.Y.; Li, S.N.; Xiao, Y.G.; Chen, H.; Ye, Y.S.; et al. Highly Dispersed Cobalt Clusters in Nitrogen-Doped Porous Carbon Enable Multiple Effects for High-Performance Li-S Battery. Adv. Energy Mater. 2020, 10, 1903550. [Google Scholar] [CrossRef]
- Lu, Q.; Yu, J.; Zou, X.H.; Liao, K.M.; Tan, P.; Zhou, W.; Ni, M.; Shao, Z.P. Self-Catalyzed Growth of Co, N-Codoped CNTs on Carbon-Encased CoS Surface: A Noble-Metal-Free Bifunctional Oxygen Electrocatalyst for Flexible Solid Zn-Air Batteries. Adv. Funct. Mater. 2019, 29, 1904481. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, P.; Joshi, C.; Manchanda, M.; Boukherroub, R.; Jain, S.L. Nickel Decorated on Phosphorous-Doped Carbon Nitride as an Efficient Photocatalyst for Reduction of Nitrobenzenes. Nanomaterials 2016, 6, 59. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, S.-P. Band gap of C3N4 in the GW approximation. Int. J. Hydrogen Energy 2012, 37, 11072–11080. [Google Scholar] [CrossRef]
- Ong, W.J.; Tan, L.L.; Ng, Y.H.; Yong, S.T.; Chai, S.P. Graphitic Carbon Nitride (g-C3N4)-Based Photocatalysts for Artificial Photosynthesis and Environmental Remediation: Are We a Step Closer To Achieving Sustainability? Chem. Rev. 2016, 116, 7159–7329. [Google Scholar] [CrossRef]
- He, W.W.; Jia, H.M.; Wamer, W.G.; Zheng, Z.; Li, P.J.; Callahan, J.H.; Yin, J.J. Predicting and identifying reactive oxygen species and electrons for photocatalytic metal sulfide micro-nano structures. J. Catal. 2014, 320, 97–105. [Google Scholar] [CrossRef]
- Vatansever, F.; de Melo, W.C.M.A.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial strategies centered around reactive oxygen species—Bactericidal antibiotics, photodynamic therapy, and beyond. FEMS Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.; Murtaza, M.; Amjad, A.; Zahra, M.; Waseem, A.; Alhodaib, A. NiSe2/Ag3PO4 Nanocomposites for Enhanced Visible Light Photocatalysts for Environmental Remediation Applications. Catalysts 2023, 13, 929. [Google Scholar] [CrossRef]
- Vital-Grappin, A.D.; Ariza-Tarazona, M.C.; Luna-Hernández, V.M.; Villarreal-Chiu, J.F.; Hernández-López, J.M.; Siligardi, C.; Cedillo-González, E.I. The Role of the Reactive Species Involved in the Photocatalytic Degradation of HDPE Microplastics Using C,N-TiO2 Powders. Polymers 2021, 13, 999. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, K.; Shang, R.; Zhang, H.; Shen, J.; Liu, H.; Tressel, J.; Bao, Y.; Hui, A.; Xie, H.; et al. Theory-guided doping of LaCoO3 nanoparticles for enhanced antimicrobial performance. Chem. Eng. J. 2023, 464, 142710. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, W.; Zhang, K.; Liu, H.; Li, J.; Tressel, J.; Chen, S. High-Efficiency Photodynamic Antibacterial Activity of NH2-MIL-101(Fe)@MoS2/ZnO Ternary Composites. ACS Appl. Bio Mater. 2022, 5, 3912–3922. [Google Scholar] [CrossRef]
- Tachikawa, T.; Majima, T. Single-Molecule Fluorescence Imaging of TiO2 Photocatalytic Reactions. Langmuir 2009, 25, 7791–7802. [Google Scholar] [CrossRef]
- Bae, I.K.; Shin, J.-Y.; Son, J.-H.; Wang, K.-K.; Han, W.-S. Antibacterial effect of singlet oxygen depending on bacteria surface charge. Photodiagnosis Photodyn. Ther. 2022, 39, 102975. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DuBois, D.B.; Rivera, I.; Liu, Q.; Yu, B.; Singewald, K.; Millhauser, G.L.; Saltikov, C.; Chen, S. Photocatalytic Generation of Singlet Oxygen by Graphitic Carbon Nitride for Antibacterial Applications. Materials 2024, 17, 3787. https://doi.org/10.3390/ma17153787
DuBois DB, Rivera I, Liu Q, Yu B, Singewald K, Millhauser GL, Saltikov C, Chen S. Photocatalytic Generation of Singlet Oxygen by Graphitic Carbon Nitride for Antibacterial Applications. Materials. 2024; 17(15):3787. https://doi.org/10.3390/ma17153787
Chicago/Turabian StyleDuBois, Davida Briana, Isabelle Rivera, Qiming Liu, Bingzhe Yu, Kevin Singewald, Glenn L. Millhauser, Chad Saltikov, and Shaowei Chen. 2024. "Photocatalytic Generation of Singlet Oxygen by Graphitic Carbon Nitride for Antibacterial Applications" Materials 17, no. 15: 3787. https://doi.org/10.3390/ma17153787





