Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection
Abstract
:1. Introduction
2. Metal Nanomaterials
2.1. Silver Nanoclusters


2.2. Copper Nanoclusters

2.3. Gold Nanopartilces

3. Quantum Dots



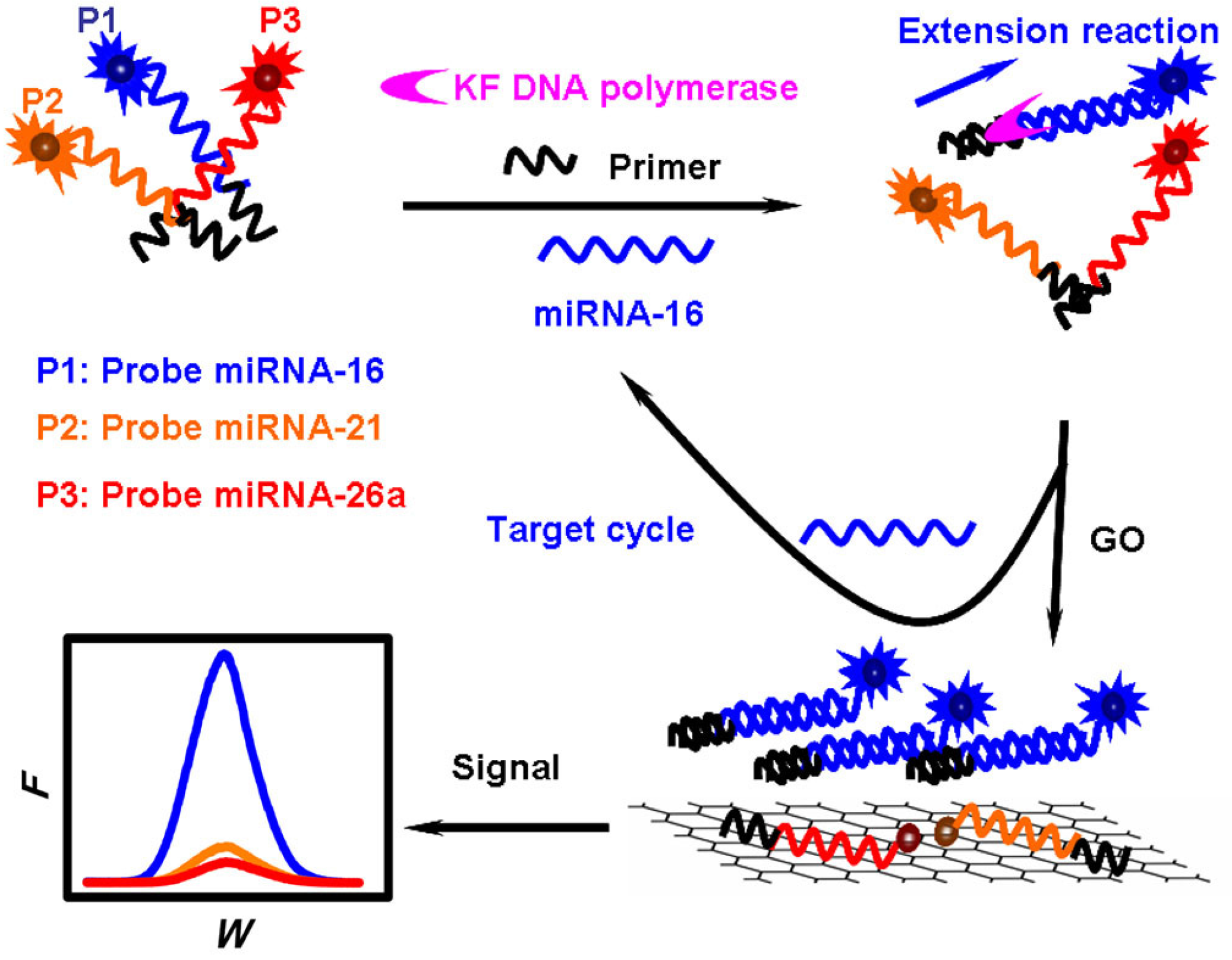
4. Graphene Oxide




5. Silicon


6. Conclusions
| Nanomaterials | Signal Amplification | Detection ranges | Detection limits | References |
|---|---|---|---|---|
| AgNCs | - | 0~1.5 μM | <0.25 μM | [18] |
| AgNCs | - | 5~125 nM | 1.7 nM | [22] |
| AgNCs | HCR | 1.56~400 nM | 0.78 nM | [24] |
| AgNCs | Target recycling | 0.5~50 nM | 0.16 nM | [23] |
| AgNCs | TAIEA | 10 aM~1 nM | 2 aM | [21] |
| CuNCs | RCR | 10~400 nM | 10 pM | [29] |
| CuNCs | TAIEA | 1 pM~10 nM | 1 pM | [28] |
| AuNPs | - | 0.05~50 pM | 0.01 pM | [33] |
| AuNPs | DSN | - | <25 pM | [34] |
| CdSe/ZnS | DSN | 0.53 pM~3.9 nM | 0.28 pM | [47] |
| CdTe/CdS | - | 10 fM~10 nM | 10 fM | [45] |
| CdSe nanocrystals | Cation-Exchange | 0.1 pM~5 μM | 35 fM | [49] |
| 488QDs | PG-RCA | 0.1 fM~1 nM | 50.9 aM | [46] |
| 605QDs | EXPAR | 0.1 aM–10 fM | 0.1 aM | [44] |
| GO | - | 50~400 nM | - | [56] |
| GO | Endonuclease | 20 pM~1 nM | 9 pM | [63] |
| GO | HCR | 1 pM~5 nM | - | [59] |
| GO | DSN | 0.5 pM~1 nM | 160 fM | [61] |
| GO | CEA | 0.06~12 pM | 10.8 fM | [64] |
| GO | Endonuclease | 0.02~100 pM | 3 fM | [60] |
| GO | ISDPR | 5 fM~5 pM | 2.1 fM | [58] |
| Nano GO | - | 0~100 nM | 2 nM | [57] |
| Nano GO | - | 0~1 μM | 1 pM | [67] |
| SiO2 | - | 0.5~40 nM | 0.18 nM | [74] |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Grasso, M.; Piscopo, P.; Confaloni, A.; Denti, M.A. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules 2014, 19, 6891–6910. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. microRNAs in cancer management. Lancet Oncol. 2013, 13, e249–258. [Google Scholar] [CrossRef]
- Ajit, S.K. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors 2012, 12, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, detection, and bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-D.; La, M.; Zhou, B.-B. Strategies for designing of electrochemical microRNA genesensors based on the difference in the structure of RNA and DNA. Int. J. Electrochem. Sci. 2014, 9, 7228–7238. [Google Scholar]
- Jamali, A.A.; Pourhassan-Moghaddam, M.; Dolatabadi, J.E.N.; Omidi, Y. Nanomaterials on the road to microRNA detection with optical and electrochemical nanobiosensors. TrAC Trends Anal. Chem. 2014, 55, 24–42. [Google Scholar] [CrossRef]
- Tian, K.; He, Z.; Wang, Y.; Chen, S.-J.; Gu, L.-Q. Designing a polycationic probe for simultaneous enrichment and detection of microRNAs in a nanopore. ACS Nano 2013, 7, 3962–3969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.Q. Nanopore–based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M.; Dadosh, T.; Ray, V.; Jin, J.; McReynolds, L.; Drndić, M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010, 5, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Fricke, B.L.; Gu, L.-Q. Programming nanopore ion flow for encoded multiplex microRNA detection. ACS Nano 2014, 8, 3444–3450. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.Q.; Wanunu, M.; Wang, M.X.; McReynolds, L.; Wang, Y. Detection of miRNAs with a nanopore single-molecule counter. Expert Rev. Mol. Diagn. 2012, 12, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Zhang, L. Nanomaterials-based sensing strategies for electrochemical detection of microRNAs. Materials 2014, 7, 5366–5384. [Google Scholar] [CrossRef]
- Guo, W.W.; Yuan, J.P.; Dong, Q.Z.; Wang, E.K. Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J. Am. Chem. Soc. 2010, 132, 932–934. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.T.; Lan, G.Y.; Chen, W.Y.; Chang, H.T. Detection of copper ions through recovery of the fluorescence of DNA-templated copper/silver nanoclusters in the presence of mercaptopropionic acid. Anal. Chem. 2010, 82, 8566–8572. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Choi, S.; Dickson, R.M. Shuttle-based fluorogenic silver-cluster biolabels. Angew. Chem. Int. Ed. 2009, 48, 318–320. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.-Q.; Yu, C.-Y.; Yin, B.-C.; Ye, B.-C. Multiplexed detection of microRNAs by tuning DNA-scaffolded silver nanoclusters. Analyst 2013, 138, 4812–4817. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Rørvig-Lund, A.; Chaabane, S.B.; Thulstrup, P.W.; Kjaergaard, H.G.; Fron, E.; Hofkens, J.; Yang, S.W.; Vosch, T. Design aspects of bright red emissive silver nanoclusters/DNA probes for microRNA detection. ACS Nano 2012, 6, 8803–8814. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Vosch, T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal. Chem. 2011, 83, 6935–6939. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Cho, S.K.; Thulstrup, P.W.; Bhang, Y.-J.; Ahn, J.C.; Choi, S.W.; Rørvig-Lund, A.; Yang, S.W. Effect of salts, solvents and buffer on miRNA detection using DNA silver nanocluster (DNA/AgNCs) probes. Nanotechnology 2014, 25, 045101. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Thulstrup, P.W.; Cho, S.K.; Bhang, Y.-J.; Ahn, J.C.; Choi, S.W.; Bjerrum, M.J.; Yang, S.W. In-solution multiplex miRNA detection using DNA-templated silver nanocluster probes. Analyst 2014, 139, 2158–2166. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Q.; Zhang, M.; Yin, B.-C.; Ye, B.-C. Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal. Chem. 2012, 84, 5165–5169. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Ha, Y.; Hu, S.; Wang, J. Hairpin DNA probe with 5′-TCC/CCC-3′ overhangsfor the creation of silver nanoclusters and miRNA assay. Biosens. Bioelectron. 2014, 51, 36–39. [Google Scholar] [CrossRef]
- Dong, H.; Hao, K.; Tian, Y.; Jin, S.; Lu, H.; Zhou, S.-F.; Zhang, X. Label-free and ultrasensitive microRNA detection based on novel molecular beacon binding readout and target recycling amplification. Biosens. Bioelectron. 2014, 53, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Wang, P.; Cao, Z. Hybridization chain reaction modulated DNA-hosted silver nanoclusters for fluorescent identification of single nucleotide polymorphisms in the let-7 miRNA family. Biosens. Bioelectron. 2014, 60, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rotaru, A.; Dutta, S.; Jentzsch, E.; Gothelf, K.; Mokhir, A. Selective dsDNA-templated formation of copper nanoparticles in solution. Angew. Chem. Int. Ed. 2010, 49, 5665–5667. [Google Scholar] [CrossRef]
- Qing, Z.H.; He, X.X.; He, D.G.; Wang, K.M.; Xu, F.Z.; Qing, T.P.; Yang, X. Poly(thymine)-templated selective formation of fluorescent copper nanoparticles. Angew. Chem. Int. Ed. 2013, 52, 9719–9722. [Google Scholar] [CrossRef]
- Jia, X.; Li, J.; Han, L.; Ren, J.; Yang, X.; Wang, E. DNA-hosted copper nanoclusters for fluorescent identification of single nucleotide polymorphisms. ACS Nano 2012, 6, 3311–3317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-P.; Yin, B.-C.; Ye, B.-C. A novel fluorescence probe of dsDNA-templated copper nanoclusters for quantitative detection of microRNAs. RSC Adv. 2013, 3, 8633–8636. [Google Scholar] [CrossRef]
- Xu, F.; Shi, H.; He, X.; Wang, K.; He, D.; Guo, Q.; Qing, Z.; Yan, L.; Ye, X.; Li, D.; Tang, J. Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal. Chem. 2014, 86, 6976–6982. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Zhang, L.; Wang, G.; Feng, Q.; Liu, L. Label-free and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles. Biosens. Bioelectron. 2013, 47, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Balcioglu, M.; Rana, M.; Robertson, N.; Yigit, M.V. DNA-length-dependent quenching of fluorescently labeled iron oxide nanoparticles with gold, graphene oxide and MoS2 nanostructures. ACS Appl. Mater. Interfaces 2014, 6, 12100–12110. [Google Scholar] [CrossRef] [PubMed]
- Swierczewska, M.; Lee, S.; Chen, X. The design and application of fluorophore-gold nanoparticle activatable probes. Phys. Chem. Chem. Phys. 2011, 13, 9929–9941. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Wu, P.; Zhang, H.; Cai, C. Fluorescence quenching of gold nanoparticles integrating with a conformation–switched hairpin oligonucleotide probe for microRNA detection. Chem. Commun. 2012, 48, 10718–10720. [Google Scholar] [CrossRef]
- Degliangeli, F.; Kshirsagar, P.; Brunetti, V.; Pompa, P.P.; Fiammengo, R. Absolute and direct microRNA quantification using DNA−gold nanoparticle probes. J. Am. Chem. Soc. 2014, 136, 2264–2267. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gao, Y.; Liu, H.; Xia, N. An ultrasensitive electrochemical miRNAs sensor based on miRNAs-initiated cleavage of DNA by duplex-specific nuclease and signal amplification of enzyme plus redox cycling reaction. Sens. Actuators B Chem. 2015, 208, 137–142. [Google Scholar] [CrossRef]
- Rosa, J.; Conde, J.; de la Fuente, J.M.; Lima, J.C.; Baptista, P.V. Gold-nanobeacons for real-time monitoring of RNA synthesis. Biosens. Bioelectron. 2012, 36, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Rosa, J.; de la Fuente, J.M.; Baptista, P.V. Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events. Biomaterials 2013, 34, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Miao, Q.Q.; Liang, G.L. Quantum dots as multifunctional materials for tumor imaging and therapy. Materials 2013, 6, 483–499. [Google Scholar] [CrossRef]
- Härmä, H.; Pihlasalo, S.; Cywinski, P.J.; Mikkonen, P.; Hammann, T.; Lohmannsröben, H.-G.; Hänninen, P. Protein quantification using resonance energy transfer between donor nanoparticles and acceptor quantum dots. Anal. Chem. 2013, 85, 2921–2926. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.-Q.; Li, W.; Li, Y.; Tan, C.-Y.; Li, J.-X.; Jin, Y.-X.; Ruan, K.-C. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005, 33, e17. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, J.; Liang, G.L.; Yang, X.R. Rapid fluorescent detection of neurogenin3 by CdTe quantum dot aggregation. Analyst 2012, 137, 1775–1778. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Zhang, L.-F.; Zhang, Y.; Zhang, C.-Y. Single quantum dot based nanosensor for renin assay. Anal. Chem. 2012, 84, 8846–8852. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Granek, J.; Deschamps, J.R.; Boeneman, K.; Blanco-Canosa, J.B.; Dawson, P.E.; Susumu, K.; Stewart, M.H.; Medintz, I.L. Monitoring botulinum neurotoxin a activity with peptide-functionalized quantum dot resonance energy transfer sensors. ACS Nano 2011, 5, 2687–2699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, C.-Y. Sensitive detection of microRNA with isothermal amplification and a single-quantum-dot-based nanosensor. Anal. Chem. 2012, 84, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Fan, J.; Xue, B.; Yuwen, L.; Liu, X.; Pan, D.; Fan, C.; Wang, L. DNA-conjugated quantum dot nanoprobe for high-sensitivity fluorescent detection of DNA and microRNA. ACS Appl. Mater. Interfaces 2014, 6, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.-P.; Zhu, G.; Yang, X.-Y.; Cao, J.; Jing, Z.-L.; Zhang, C.-Y. A quantum dot-based microRNA nanosensor for point mutation assays. Chem. Commun. 2014, 50, 7160–7162. [Google Scholar] [CrossRef]
- Jou, A.F.; Lu, C.-H.; Ou, Y.-C.; Wang, S.-S.; Hsu, S.-L.; Willner, I.; Ho, J.A. Diagnosing the miR-141 prostate cancer biomarker using nucleic acid-functionalized CdSe/ZnS QDs and telomerase. Chem. Sci. 2015, 6, 659–665. [Google Scholar] [CrossRef]
- Kucur, E.; Boldt, F.M.; Cavaliere-Jaricot, S.; Ziegler, J.; Nann, T. Quantitative analysis of cadmium selenide nanocrystal concentration by comparative techniques. Anal. Chem. 2007, 79, 8987–8993. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Schachermeyer, S.; Wang, Y.; Yin, Y.; Zhong, W. Detection of microRNA by fluorescence amplification based on cation-exchange in nanocrystals. Anal. Chem. 2009, 81, 9723–9729. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Adsorption of DNA onto gold nanoparticles and graphene oxide: surface science and applications. Phys. Chem. Chem. Phys. 2012, 14, 10485–10496. [Google Scholar] [CrossRef] [PubMed]
- Manohar, S.; Mantz, A.R.; Bancro, K.E.; Hui, C.Y.; Jagota, A.; Vezenov, D.V. Peeling single-stranded DNA from graphite surface to determine oligonucleotide binding energy by force spectroscopy. Nano Lett. 2008, 8, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Yang, H.H.; Zhu, C.L.; Chen, X.; Chen, G.N. A graphene platform for sensing biomolecules. Angew. Chem. Int. Ed. 2009, 48, 4785–4787. [Google Scholar] [CrossRef]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Chang, H.; Tang, L.; Wang, Y.; Jiang, J.; Li, J. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal. Chem. 2010, 82, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Kim, Y.K.; Kwon, H.M.; Yeo, W.S.; Kim, D.E.; Min, D.H. A grapheme-based platform for the assay by helicase. Angew. Chem. Int. Ed. 2010, 49, 5703–5707. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, L.; Deng, Y.; Li, S.; He, N. Graphene oxide for rapid microRNA detection. Nanoscale 2012, 4, 5840–5842. [Google Scholar] [CrossRef] [PubMed]
- Hizir, M.S.; Balcioglu, M.; Rana, M.; Robertson, N.M.; Yigit, M.V. Simultaneous detection of circulating oncomiRs from body fluids for prostate cancer staging using nanographene oxide. ACS Appl. Mater. Interfaces 2014, 6, 14772–14778. [Google Scholar] [PubMed]
- Dong, H.; Zhang, J.; Ju, H.; Lu, H.; Wang, S.; Jin, S.; Hao, K.; Du, H.; Zhang, X. Highly sensitive multiple microRNA detection based on fluorescence quenching of graphene oxide and isothermal strand-displacement polymerase reaction. Anal. Chem. 2012, 84, 4587–4593. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, C.; Ren, W.; Li, Z. Graphene surface-anchored fluorescence sensor for sensitive detection of microRNA coupled with enzyme-free signal amplification of hybridization chain reaction. ACS Appl. Mater. Interfaces 2012, 4, 6450–6453. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Li, W.; Wu, P.; Zhang, H.; Cai, C. Fluorescence quenching of graphene oxide integrating with the site-specific cleavage of the endonuclease for sensitive and selective microRNA detection. Anal. Chem. 2013, 85, 2536–2542. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Yang, F.; Zhang, Y.; Ning, Y.; Yao, Q.; Zhang, G.-J. Amplified fluorescence sensing of miRNA by combination of graphene oxide with duplexspecific nuclease. Anal. Methods 2014, 6, 3598–3603. [Google Scholar] [CrossRef]
- Tang, Z.; Wu, H.; Cort, J.R.; Buchko, G.W.; Zhang, Y.; Shao, Y.; Aksay, I.A.; Liu, J.; Lin, Y. Constraint of DNA on functionalized graphene improves its biostability and specificity. Small 2010, 6, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lin, X.; Lin, N.; Song, Y.; Zhu, Z.; Chen, X.; Yang, C.J. Graphene oxide-protected DNA probes for multiplex microRNA analysis in complex biological samples based on a cyclic enzymatic amplification method. Chem. Commun. 2012, 48, 194–196. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.; Wang, Q.; Duan, L.; Tang, B. Graphene fluorescence switch-based cooperative amplification: A sensitive and accurate method to detection microRNA. Anal. Chem. 2014, 86, 5487–5493. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Hasan, A.; Kindi, H.A.; Gaharwar, A.K.; Rao, V.T.S.; Nikkhah, M.; Shin, S.R.; Krafft, D.; Dokmeci, M.R.; Shum-Tim, D.; et al. Injectable graphene oxide/hydrogel-based angiogenic gene delivery system for vasculogenesis and cardiac repair. ACS Nano 2014, 8, 8050–8062. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, W.J. Photothermally controlled gene delivery by reduced graphene oxide-polyethylenimine nanocomposite. Small 2014, 10, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.-R.; Lee, J.; Yeo, J.; Na, H.-K.; Kim, Y.-K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.; Orbay, H.; Cai, W. Molecular imaging strategies for in Vivo tracking of microRNAs: A comprehensive review. Curr. Med. Chem. 2013, 20, 3594–3603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Cheng, F.; Zhou, R.; Cao, J.; Li, J.; Burda, C.; Min, Q.; Zhu, J.-J. DNA-hybrid-gated nultifunctional mesoporous silica nanocarriers for dual-targeted and microRNA-responsive controlled drug delivery. Angew. Chem. Int. Ed. 2014, 53, 2371–2375. [Google Scholar] [CrossRef]
- Rivera-Gil, P.; de Aberasturi, D.J.; Wulf, V.; Pelaz, B.; Del Pino, P.; Zhao, Y.Y.; de la Fuente, J.M.; de Larramendi, I.R.; Rojo, T.; Liang, X.J.; et al. The challenge to relate the physicochemical properties of colloidal nanoparticles to their cytotoxicity. Acc. Chem. Res. 2013, 46, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Kuang, L.; Battle, C.; Shaner, T.; Mitchell, B.S.; Fink, M.J.; Jayawickramarajah, J. Mild two-step method to construct DNA-conjugated silicon nanoparticles: Scaffolds for the detection of microRNA-21. Bioconjug. Chem. 2014, 25, 1739–1743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fu, Y.; Mei, Y.; Jiang, F.; Lakowicz, J.R. Fluorescent metal nanoshell probe to detect single miRNA in lung cancer cell. Anal. Chem. 2010, 82, 4464–4471. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Lei, J.; Ju, H.; Zhi, F.; Wang, H.; Guo, W.; Zhu, Z.; Yan, F. Target-cell-specific delivery, imaging, and detection of intracellular microRNA with a multifunctional SnO2 nanoprobe. Angew. Chem. 2012, 124, 4685–4690. [Google Scholar] [CrossRef]
- Li, H.; Mu, Y.; Lu, J.; Wei, W.; Wan, Y.; Liu, S. Target-cell-specific fluorescence silica nanoprobes for imaging and theranostics of cancer cells. Anal. Chem. 2014, 86, 3602–3609. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La, M.; Liu, L.; Zhou, B.-B. Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection. Materials 2015, 8, 2809-2829. https://doi.org/10.3390/ma8052809
La M, Liu L, Zhou B-B. Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection. Materials. 2015; 8(5):2809-2829. https://doi.org/10.3390/ma8052809
Chicago/Turabian StyleLa, Ming, Lin Liu, and Bin-Bin Zhou. 2015. "Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection" Materials 8, no. 5: 2809-2829. https://doi.org/10.3390/ma8052809
APA StyleLa, M., Liu, L., & Zhou, B.-B. (2015). Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection. Materials, 8(5), 2809-2829. https://doi.org/10.3390/ma8052809






