Hybrid and Environmental Effects on Gene Expression in Poplar Clones in Pure and Mixed with Black Locust Stands
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sampling
2.2. RNA Extraction
2.3. RNA Sequencing
2.4. Principal Component Analysis (PCA)
2.5. Identification of Differently Expressed Genes (DEGs) and Sequence Annotation
2.6. Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
3. Results
3.1. RNA Sequencing Data and Transcriptome Annotation
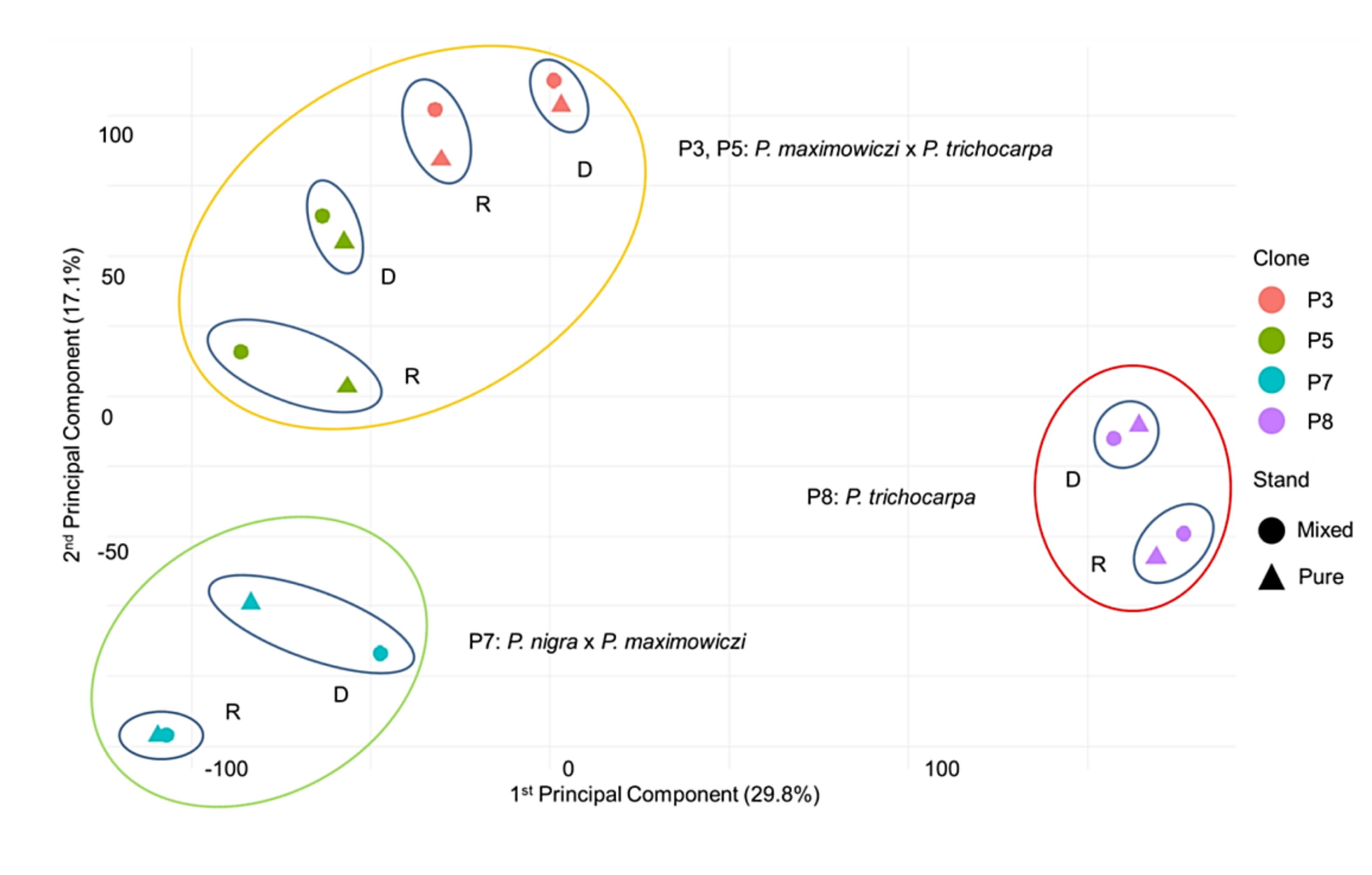
3.2. Principal Component Analysis (PCA)
3.3. Differentially Expressed Genes (DEGs) Analyses
3.4. Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baltodano, J. Monoculture forestry: A critique from an ecological perspective. In Tree Trouble: A Compilation of Testimonies on the Negative Impact of Large-Scale Monoculture Tree Plantations, Proceedings of the 6th COP of the FCCC; Friends of the Earth International, Hague, The Netherlands, 13–15 November 2000; United Nations Climate Change: Bonn, Germany, 2000; pp. 2–10. [Google Scholar]
- Bowyer, J. Forest plantations Threatening or Saving Natural Forests? Arborvitae 2006, 31, 8–9. [Google Scholar]
- Morris, J.; Ningnan, Z.; Zengjiang, Y.; Collopy, J.; Daping, X. Water use by fast-growing Eucalyptus urophylla plantations in southern China. Tree Physiol. 2004, 24, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.C.; Kuchma, O.; Krutovsky, K.V. Mixed-species versus monocultures in plantation forestry: Development, benefits, ecosystem services and perspectives for the future. Glob. Ecol. Conserv. 2018, 15, e00419. [Google Scholar] [CrossRef]
- Benomar, L.; Des Rochers, A.; Larocque, G.R. Comparing growth and fine root distribution in monocultures and mixed plantations of hybrid poplar and spruce. J. For. Res. 2013, 24, 247–254. [Google Scholar] [CrossRef]
- Pretzsch, H.; Block, J.; Dieler, J.; Dong, P.H.; Kohnle, U.; Nagel, J.; Spellmann, H.; Zingg, A. Comparison between the productivity of pure and mixed stands of Norway spruce and European beech along an ecological gradient. Ann. For. Sci. 2010, 67, 712. [Google Scholar] [CrossRef]
- Pretzsch, H.; Dieler, J.; Seifert, T.; Rötzer, T. Climate effects on productivity and resource-use efficiency of Norway spruce (Picea abies [L.] Karst.) and European beech (Fagus sylvatica [L.]) in stands with different spatial mixing patterns. Trees Struct. Funct. 2012, 26, 1343–1360. [Google Scholar] [CrossRef]
- Pretzsch, H.; Schütze, G.; Uhl, E. Resistance of European tree species to drought stress in mixed versus pure forests: Evidence of stress release by inter-specific facilitation. Plant Biol. 2013, 15, 483–495. [Google Scholar] [CrossRef]
- Marron, N.; Epron, D. Are mixed-tree plantations including a nitrogen-fixing species more productive than monocultures? For. Ecol. Manag. 2019, 441, 242–252. [Google Scholar] [CrossRef]
- Sayyad, E.; Hosseini, S.M.; Mokhtari, J.; Mahdavi, R.; Jalali, S.G.; Akbarinia, M.; Tabari, M. Comparison of growth, nutrition and soil properties of pure and mixed stands of Populus deltoides and Alnus subcordata. Silva Fenn. 2006, 40, 27–35. [Google Scholar] [CrossRef]
- Grünewald, H.; Böhm, C.; Quinkenstein, A.; Grundmann, P.; Eberts, J.; von Wühlisch, G. Robinia pseudoacacia L.: A lesser known tree species for biomass production. Bioenergy Res. 2009, 2, 123–133. [Google Scholar] [CrossRef]
- Mantovani, D.; Veste, M.; Freese, D. Effects of Drought Frequency on Growth Performance and Transpiration of Young Black Locust (Robinia pseudoacacia L.). Int. J. For. Res. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Rédei, K.; Veperdi, I.; Meilby, H. Stand structure and growth of mixed white poplar (Populus alba L.) and black locust (Robinia pseudoacacia L.) plantations in Hungary. Acta Silv. Lignaria Hung. 2006, 2, 23–32. [Google Scholar]
- Tanaka-Oda, A.; Kenzo, T.; Koretsune, S.; Sasaki, H.; Fukuda, K. Ontogenetic changes in water-use efficiency (δ13C) and leaf traits differ among tree species growing in a semiarid region of the Loess Plateau, China. For. Ecol. Manag. 2010, 259, 953–957. [Google Scholar] [CrossRef]
- Oliveira, N.; del Río, M.; Forrester, D.I.; Rodríguez-Soalleiro, R.; Pérez-Cruzado, C.; Cañellas, I.; Sixto, H. Mixed short rotation plantations of Populus alba and Robinia pseudoacacia for biomass yield. For. Ecol. Manag. 2018, 410, 48–55. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, M.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Neale, D.B.; Kremer, A. Forest tree genomics: Growing resources and applications. Nat. Rev. Genet. 2011, 12, 111–122. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Plomion, C.; Kirst, M.; Sederoff, R.R. Genomics of growth traits in forest trees. Curr. Opin. Plant Biol. 2009, 12, 148–156. [Google Scholar] [CrossRef]
- Cohen, D.; Bogeat-Triboulot, M.B.; Tisserant, E.; Balzergue, S.; Martin-Magniette, M.L.; Lelandais, G.; Ningre, N.; Renou, J.P.; Tamby, J.P.; Le Thiec, D.; et al. Comparative transcriptomics of drought responses in Populus: A meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genom. 2010, 11, 630. [Google Scholar] [CrossRef]
- Gugger, P.F.; Peñaloza-Ramírez, J.M.; Wright, J.W.; Sork, V.L. Whole-transcriptome response to water stress in a California endemic oak, Quercus lobata. Tree Physiol. 2016, 37, 632–644. [Google Scholar] [CrossRef]
- Hess, M.; Wildhagen, H.; Junker, L.V.; Ensminger, I. Transcriptome responses to temperature, water availability and photoperiod are conserved among mature trees of two divergent Douglas-fir provenances from a coastal and an interior habitat. BMC Genom. 2016, 17, 682. [Google Scholar] [CrossRef]
- Janz, D.; Behnke, K.; Schnitzler, J.P.; Kanawati, B.; Schmitt-Kopplin, P.; Polle, A. Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol. 2010, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.; Best, T.; Zembower, N.; Davitt, J.; Henry, N.; Xu, Y.; Koch, J.; Liang, H.; McGraw, J.; Schuster, S.; et al. The green ash transcriptome and identification of genes responding to abiotic and biotic stresses. BMC Genom. 2016, 17, 702. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Seifert, S.; Lübbe, T.; Leuschner, C.; Finkeldey, R. De novo transcriptome assembly and analysis of differential gene expression in response to drought in European beech. PLoS ONE 2017, 12, e0184167. [Google Scholar] [CrossRef] [PubMed]
- Philippe, R.N.; Ralph, S.G.; Külheim, C.; Jancsik, S.I.; Bohlmann, J. Poplar defense against insects: Genome analysis, full-length cDNA cloning, and transcriptome and protein analysis of the poplar Kunitz-type protease inhibitor family. New Phytol. 2009, 184, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Suontama, M.; Burdon, R.D.; Dungey, H.S. Genotype by environment interactions in forest tree breeding: Review of methodology and perspectives on research and application. Tree Genet. Genomes 2017, 13, 60. [Google Scholar] [CrossRef]
- Rae, A.M.; Pinel, M.P.C.; Bastien, C.; Sabatti, M.; Street, N.R.; Tucker, J.; Dixon, C.; Marron, N.; Dillen, S.Y.; Taylor, G. QTL for yield in bioenergy Populus: Identifying G×E interactions from growth at three contrasting sites. Tree Genet. Genomes 2008, 4, 97–112. [Google Scholar] [CrossRef]
- Stape, J.L.; Binkley, D.; Ryan, M.G.; Fonseca, S.; Loos, R.A.; Takahashi, E.N.; Silva, C.R.; Silva, S.R.; Hakamada, R.E.; de Ferreira, J.M.A.; et al. The Brazil Eucalyptus Potential Productivity Project: Influence of water, nutrients and stand uniformity on wood production. For. Ecol. Manag. 2010, 259, 1684–1694. [Google Scholar] [CrossRef]
- Marron, N.; Priault, P.; Gana, C.; Gérant, D.; Epron, D. Prevalence of interspecific competition in a mixed poplar/black locust plantation under adverse climate conditions. Ann. For. Sci. 2018, 75, 23. [Google Scholar] [CrossRef]
- Izawa, T. Deciphering and prediction of plant dynamics under field conditions. Curr. Opin. Plant Biol. 2015, 24, 87–92. [Google Scholar] [CrossRef]
- Ning, K.; Ding, C.; Huang, Q.; Zhang, W.; Yang, C.; Liang, D.; Fan, R.; Su, X. Transcriptome profiling revealed diverse gene expression patterns in poplar (Populus × euramericana) under different planting densities. PLoS ONE 2019, 14, e0217066. [Google Scholar] [CrossRef]
- Euring, D.; Janz, D.; Polle, A. Wood properties and transcriptional responses of poplar hybrids in mixed cropping with the nitrogen-fixing species Robinia pseudoacacia. Tree Physiol. 2020. in review. [Google Scholar]
- Andersson, A.; Keskitalo, J.; Sjödin, A.; Bhalerao, R.; Sterky, F.; Wissel, K.; Tandre, K.; Aspeborg, H.; Moyle, R.; Ohmiya, Y.; et al. A transcriptional timetable of autumn senescence. Genome Biol. 2004, 5, R24. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Gordon, M.I.; Amarasinghe, V.; Strauss, S.H. Extensive transcriptome changes during seasonal leaf senescence in field-grown black cottonwood (Populus trichocarpa Nisqually-1). Sci. Rep. 2020, 10, 6581. [Google Scholar] [CrossRef] [PubMed]
- Rebola-Lichtenberg, J.; Schall, P.; Annighöfer, P.; Ammer, C.; Leinemann, L.; Polle, A.; Euring, D. Mortality of different Populus genotypes in recently established mixed short rotation coppice with Robinia pseudoacacia L. Forests 2019, 10, 410. [Google Scholar] [CrossRef]
- Schirmer, R.; Haikali, A. Sortenprüfung von Pappelhybriden für Energiewälder. LWF Wissen 2014, 74, 106–118. [Google Scholar]
- Grotehusmann, H.; Janssen, A.; Haikali, A.; Hartmann, K.-U.; Hüller, W.; Karopka, M.; Schildbach, M.; Schirmer, R.; Schuppelius, T.; Töpfner, K. Pappelsortenprüfung im Projekt FastWOOD. Forstarchiv 2015, 86, 67–79. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Urbanek, S.; Bibiko, H.-J.; Stefano, M.L. R: A Language and Environment for Statistical Computing. The R Foundation for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 18 January 2018).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Vu, V.Q. ggbiplot: A Ggplot2 Based Biplot. R Package, Version 0.55. Available online: http://github.com/ (accessed on 18 January 2018).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Huala, E.; Dickerman, A.W.; Garcia-Hernandez, M.; Weems, D.; Reiser, L.; LaFond, F.; Hanley, D.; Kiphart, D.; Zhuang, M.; Huang, W.; et al. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.M.; Yakovlev, I.A.; Strauss, S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Hall, T.A. BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, 43–46. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chhangawala, S.; Rudy, G.; Mason, C.E.; Rosenfeld, J.A. The impact of read length on quantification of differentially expressed genes and splice junction detection. Genome Biol. 2015, 16, 131. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Barghini, E.; Cossu, R.M.; Cavallini, A.; Giordani, T. Transcriptome analysis of response to drought in poplar interspecific hybrids. Genom. Data 2015, 3, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhou, J.; Shi, W.; Cao, X.; Luo, J.; Polle, A.; Luo, Z. Bin Comparative transcriptomic analysis reveals the roles of overlapping heat-/drought-responsive genes in poplars exposed to high temperature and drought. Sci. Rep. 2017, 7, 43215. [Google Scholar] [CrossRef] [PubMed]
- Eckenwalder, J.E. Systematics and evolution of Populus. In Biology of Populus and Its Implications for Management and Conservation; Stettler, R.F., Bradshaw, H.D., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press: Montreal, QC, Canada, 1996; pp. 7–32. [Google Scholar]
- Verlinden, M.S.; Broeckx, L.S.; Van den Bulcke, J.; Van Acker, J.; Ceulemans, R. Comparative study of biomass determinants of 12 poplar (Populus) genotypes in a high-density short-rotation culture. For. Ecol. Manag. 2013, 307, 101–111. [Google Scholar] [CrossRef]
- Hamzeh, M.; Dayanandan, S. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am. J. Bot. 2004, 91, 1398–1408. [Google Scholar] [CrossRef]
- Cervera, M.T.; Storme, V.; Soto, A.; Ivens, B.; Van Montagu, M.; Rajora, O.P.; Boerjan, W. Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theor. Appl. Genet. 2005, 111, 1440–1456. [Google Scholar] [CrossRef]
- Liesebach, H.; Schneck, V.; Ewald, E. Clonal fingerprinting in the genus Populus L. by nuclear microsatellite loci regarding differences between sections, species and hybrids. Tree Genet. Genomes 2010, 6, 259–269. [Google Scholar] [CrossRef]
- Isabel, N.; Lamothe, M.; Thompson, S.L. A second-generation diagnostic single nucleotide polymorphism (SNP)-based assay, optimized to distinguish among eight poplar (Populus L.) species and their early hybrids. Tree Genet. Genomes 2013, 9, 621–626. [Google Scholar] [CrossRef]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef]
- Sham, A.; Moustafa, K.; Al-Ameri, S.; Al-Azzawi, A.; Iratni, R.; AbuQamar, S. Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS ONE 2015, 10, e0125666. [Google Scholar] [CrossRef]
- Wolfe, M.S. The Current Status and Prospects of Multiline Cultivars and Variety Mixtures for Disease Resistance. Annu. Rev. Phytopathol. 1985, 23, 251–273. [Google Scholar] [CrossRef]
- Schäfer, C.; Grams, T.E.E.; Rötzer, T.; Feldermann, A.; Pretzsch, H. Drought stress reaction of growth and δ13C in tree rings of European beech and Norway spruce in monospecific versus mixed stands along a precipitation gradient. Forests 2017, 8, 177. [Google Scholar] [CrossRef]
- Radosevich, S.R.; Hibbs, D.E.; Ghersa, C.M. Effects of species mixtures and growth and stand development of Douglas-fir and red alder. Can. J. For. Res. 2006, 36, 768–782. [Google Scholar] [CrossRef]
- Forrester, D.; Bauhus, J.; Cowie, A. On the success and failure of mixed-species tree plantations: Lessons learned from a model system of Eucalyptus globulus and Acacia mearnsii. For. Ecol. Manag. 2005, 209, 147–155. [Google Scholar] [CrossRef]
| Name | Abbreviation | Species or Hybrid |
|---|---|---|
| Poplar clones | ||
| AF2 | P1 | Populus deltoides × P. nigra |
| Fritzi Pauley | P2 | P. trichocarpa |
| Hybride 275 | P3 | P. maximowiczii × P. trichocarpa |
| I214 | P4 | P. deltoides × P. nigra |
| Matrix 11 | P5 | P. maximowiczii × P. trichocarpa |
| Matrix 49 | P6 | P. maximowiczii × P. trichocarpa |
| Max 1 | P7 | P. nigra × P. maximowiczii |
| Muhle Larsen | P8 | P. trichocarpa |
| Black locust provenances | ||
| HGK 81901, Germany | R1 | Robinia pseudoacacia |
| HGK 81902, Germany | R2 | R. pseudoacacia |
| Nagybudmry, Hungary | R3 | R. pseudoacacia |
| Clone | Gene ID | Description | Biological Process | log2 Fold Change 1 | FDR |
|---|---|---|---|---|---|
| P3 | Potri.001G364300 | agenet domain-containing protein / bromo-adjacent homology (BAH) domain-containing protein | Biological process | 4.78 | 8.97 × 10−5 |
| Potri.001G441400 | S-locus lectin protein kinase family protein | Innate immune response | −5.02 | 8.97 × 10−5 | |
| Potri.T140100 | FAD-binding Berberine family protein | Oxidation-reduction process | −4.61 | 8.97 × 10−5 | |
| Potri.001G067000 | SOUL heme-binding family protein | Red or far-red light signaling pathway | 4.58 | 2.78 × 10−4 | |
| Potri.001G224000 | beta glucosidase 27 | Carbohydrate metabolic process, response to salt stress | 5.07 | 3.83 × 10−4 | |
| Potri.001G430800 | Di-glucose binding protein with Kinesin motor domain | Microtubule-based movement | −4.04 | 8.89 × 10−3 | |
| P5 | Potri.006G160000 | plastidic type i signal peptidase 1 | Protein maturation, proteolysis, signal peptide processing, thylakoid membrane organization | −6.42 | 1.73 × 10−10 |
| Potri.005G154600 | ATP synthase subunit alpha | ATP synthesis coupled proton transport, dATP biosynthetic process from ADP, defense response to bacterium | −5.63 | 2.93 × 10−3 | |
| Potri.013G073000 | mechanosensitive channel of small conductance-like 10 | Anion transport, leaf senescence, programmed cell death in response to reactive oxygen species | 4.49 | 7.07 × 10−3 | |
| Potri.003G001500 | chloride channel C | Chloride transmembrane transport | −4.04 | 3.18 × 10−2 | |
| Potri.018G126000 | bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | Lipid transport | −2.81 | 4.06 × 10−2 | |
| Potri.011G124600 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Methylation, nicotinate metabolic process | 3.17 | 6.98 × 10−2 | |
| Potri.013G051700 | RmlC-like cupins superfamily protein | Nutrient reservoir activity, response to abscisic acid | −4.09 | 6.98 × 10−2 | |
| P7 | Potri.008G162100 | zinc knuckle (CCHC-type) family protein | Regulation of morning-specific hypocotyl growth, response to blue light, regulation of transcription, DNA-templated | 6.43 | 3.47 × 10−10 |
| Potri.005G133700 | methyl esterase 1 | Defense response to fungus, jasmonic acid metabolic process, salicylic acid metabolic process | −5.17 | 4.65 × 10−6 | |
| Potri.013G137100 | photosystem II reaction center protein B | Photosynthesis, light reaction, photosynthetic electron transport in photosystem II, photosystem II assembly | 5.19 | 1.17 × 10−4 | |
| Potri.017G025900 | GRAS family transcription factor | Leaf development, negative gravitropism | −4.46 | 2.09 × 10−3 | |
| Potri.001G441400 | S-locus lectin protein kinase family protein | Innate immune response, protein phosphorylation, | −4.09 | 8.47 × 10−3 | |
| Potri.017G138800 | glutathione S-transferase phi 12 | Anthocyanin-containing compound metabolic process, regulation of flavonol biosynthetic process, | 4.03 | 2.24 × 10−2 | |
| Potri.013G142900 | Unknown | 8.78 | 4.87 × 10−2 | ||
| P8 | Potri.017G025900 | GRAS family transcription factor | Leaf development, negative gravitropism | 5.71 | 1.02 × 10−7 |
| Potri.002G221000 | DNAJ heat shock N-terminal domain-containing protein | Cellular response to misfolded protein | −5.69 | 1.46 × 10−7 | |
| Potri.T131400 | pathogenesis-related gene 1 | Defense response, response to water deprivation, systemic acquired resistance | −2.09 | 2.74 × 10−5 | |
| Potri.T120200 | ZPR1 zinc-finger domain protein | Biological process | 4.52 | 3.92 × 10−4 | |
| Potri.T019900 | polyketide cyclase/dehydrase and lipid transport superfamily protein | Defense response, response to biotic stimulus | 4.09 | 6.93 × 10−3 | |
| Potri.010G018600 | receptor kinase 3 | Protein phosphorylation | 3.95 | 7.84 × 10−3 | |
| Potri.001G226200 | beta glucosidase 32 | Carbohydrate metabolic process, glucosinolate catabolic process, response to other organism, response to salt stress | −4.81 | 1.36 × 10−2 | |
| Potri.010G219000 | Unknown | Biological process | −4.88 | 1.81 × 10−2 | |
| Potri.013G082700 | Phosphorylase superfamily protein | Nucleoside metabolic process | −2.24 | 3.22 × 10−2 | |
| Potri.016G008400 | Biological process | 4.04 | 3.33 × 10−2 | ||
| Potri.004G146000 | 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein | Many biosynthetic and catabolic reactions | −2.61 | 6.82 × 10−2 | |
| Potri.018G119600 | RING/U-box superfamily protein | Protein ubiquitination | 3.64 | 6.82 × 10−2 |
| Clone | Deppoldshausen | Reinshof | Total |
|---|---|---|---|
| P3 | 522 | 685 | 1207 |
| P5 | 793 | 339 | 1132 |
| P7 | 1186 | 520 | 1706 |
| P8 | 835 | 442 | 1277 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuchma, O.; Janz, D.; Leinemann, L.; Polle, A.; Krutovsky, K.V.; Gailing, O. Hybrid and Environmental Effects on Gene Expression in Poplar Clones in Pure and Mixed with Black Locust Stands. Forests 2020, 11, 1075. https://doi.org/10.3390/f11101075
Kuchma O, Janz D, Leinemann L, Polle A, Krutovsky KV, Gailing O. Hybrid and Environmental Effects on Gene Expression in Poplar Clones in Pure and Mixed with Black Locust Stands. Forests. 2020; 11(10):1075. https://doi.org/10.3390/f11101075
Chicago/Turabian StyleKuchma, Oleksandra, Dennis Janz, Ludger Leinemann, Andrea Polle, Konstantin V. Krutovsky, and Oliver Gailing. 2020. "Hybrid and Environmental Effects on Gene Expression in Poplar Clones in Pure and Mixed with Black Locust Stands" Forests 11, no. 10: 1075. https://doi.org/10.3390/f11101075
APA StyleKuchma, O., Janz, D., Leinemann, L., Polle, A., Krutovsky, K. V., & Gailing, O. (2020). Hybrid and Environmental Effects on Gene Expression in Poplar Clones in Pure and Mixed with Black Locust Stands. Forests, 11(10), 1075. https://doi.org/10.3390/f11101075








