Abstract
The Acer L. (Sapindaceae) is one of the most diverse and widespread genera in the Northern Hemisphere. Section Platanoidea harbours high genetic and morphological diversity and shows the phylogenetic conflict between A. catalpifolium and A. amplum. Chloroplast (cp) genome sequencing is efficient for the enhancement of the understanding of phylogenetic relationships and taxonomic revision. Here, we report complete cp genomes of five species of Acer sect. Platanoidea. The length of Acer sect. Platanoidea cp genomes ranged from 156,262 bp to 157,349 bp and detected the structural variation in the inverted repeats (IRs) boundaries. By conducting a sliding window analysis, we found that five relatively high variable regions (trnH-psbA, psbN-trnD, psaA-ycf3, petA-psbJ and ndhA intron) had a high potential for developing effective genetic markers. Moreover, with an addition of eight plastomes collected from GenBank, we displayed a robust phylogenetic tree of the Acer sect. Platanoidea, with high resolutions for nearly all identified nodes, suggests a promising opportunity to resolve infrasectional relationships of the most species-rich section Platanoidea of Acer.
1. Introduction
Chloroplasts (cp) are essential organelles in plant cells for the processes of photosynthesis and carbon fixation [1]. They possess uniparental inheritance and their genome has a high conservation structure in most land plants [2]. Generally, cp genomes in most angiosperms are circular DNA molecules composed of four parts, namely two inverted repeats (IRs) at approximately 20–28 kb, a vast single-copy region (LSC) at 80–90 kb and a small single-copy region (SSC) at 16–27 kb [3]. The composition of angiosperm cp genomes is relatively conserved and encodes four ribosomal RNAs (rRNAs), roughly 30 transfer RNAs (tRNAs) and approximately 80 single-copy genes [4]. With the rapid development of next-generation sequencing (NGS) and other methods for obtaining the cp genome sequences, the availability of cp genome sequences has increased dramatically for land plants, offering opportunities for the comprehensive structure comparison, improvement of horticultural plant breeding [5,6] and reconstruction of evolutionary relationships [7,8]. In most angiosperms, the cp genome is inherited from the patrilineal lineage and exhibits little or no recombination [9]. Due to its relatively conserved features, cp sequences are commonly used as DNA barcodes for genetic identification, plant systematic studies, and research into plant biodiversity, biogeography, adaptation, etc. [10,11].
Acer L. (Maple), a diverse genus of family Sapindaceae L., contains more than 124 species [12]. Most extant species of the genus are native to Asia, whereas others occur in North America, Europe and North Africa [12,13,14]. Most Acer species are famous ornamental plants [13], and also another usage for pharmaceutical and chemical products [15]. To date, 17 species belonging to the Acer section Platanoidea have been recognised in China, in which the section shows high genetics and morphology diversity. [12]. Among them, four species are widespread in various vegetation regions (A. amplum, A. longipes, A. mono and A. truncatum) [16], and some are endangered, such as A. catalpifolium, A. miaotaiense, and A. yangjuechi.
However, some species with diversified leaf morphology have taxonomic controversy due to unresolved phylogenetic relationships (for example, A. longipes and A. amplum) and require further studies and clarification [17]. The comparative plastome analysis allows detailed insights to affirm the phylogenetic placement of these plants and will be useful for species identification, to verify taxonomic levels and identify phylogenetic relationships [8,18]. Recently, cp genomes of A. miaotaiense and A. truncatum have been reported, but merely the sequence information was provided without further analyses [19,20]. Thus, a comparative study among these two published cp genomes and five newly generated plastomes of sect. Platanoidea is conducted and applied to address phylogenetic and taxonomic validity.
Firstly, we reported newly completed cp genomes of five species of the Acer sect. Platanoidea (A. catalpifolium, A. amplum, A. longipes, A. yanjuechi and A. mono). Then, we compared the gene contents and the plastomic organisation with two published cp genomes in the sect. Platanoidea to identify variable loci that can apply to the species or population-level studies on Acer. The aims of this study are: (i) to deepen our understanding of the genetic and structural diversity within the sect. Platanoidea, (ii) to increase our understanding of phylogenetic relationships of species within the sect. Platanoidea, and (iii) to reconstruct a phylogenetic tree based on these plastomes. Our study also provides genetic resources for future research in this genus.
2. Materials and Methods
2.1. Sampling and DNA Extraction
Young leaves of five Acer species (A. catalpifolium, A. amplum, A. longipes, A. yanjuechi and A. mono) were collected and dried immediately with silica gel for preservation, for each species we collected the leaves from one healthy plant. Collection information of the plant materials is showed in Table S1. Vouchers taxonomical determination was by the Beijing Forestry University herbarium and deposited at the College of Forestry, Beijing Forestry University, China. We isolated total genomic DNA from silica gel-dried leaves according to the modified CTAB method [21].
2.2. Chloroplast Genome Sequencing, Assembling and Annotation
Purified genomic DNA was sequenced using an Illumina MiSeq sequencer at Shanghai OE Biotech. Co., Ltd. A paired-end library was constructed with an insert size of 300 bp, yielding at least 8 GB of 150 bp paired-end reads for each species. Clean reads were obtained with NGS QC Toolkit v2.3.3 (cut-off read length for HQ = 70%, cut-off quality score = 20, trim reads from 5′ = 3, trim reads from 3′ = 7) [22]. We used MITObim v. 1.8 [23] to assemble the five new Acer cp sequences with the reference cp genomes, A. miaotaiense (KX098452) [19], A. davidii (KU977442) [24] and A. morrisonense (KT970611) [25]. Gene functions were annotated using DOGMA [26], and the protein-encoding genes (PCG), tRNAs and rRNA were determined and verified using the BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Divergence Hotspot Identification
In order to determine the divergence level, a MAFFT: multiple sequence alignment program [27] was used to align cp sequences of seven Acer sect. Platanoidea species, and then sliding windows of the nucleotide variability (pi) was conducted using DnaSP 5.0 with 600-bp window length and 200-bp step size [28].
2.4. Phylogenomic Reconstruction
To determine the phylogenetic relationship of the Acer sect. Platanoidea, we performed phylogenetic analyses using 13 cp genome sequences, which comprised five plastome sequences generated in this study, six plastomes of the Acer species collected from GenBank, and two of the Dipteronia species as an outgroup (Table S2). Consequently, a total length of 160,886 bp was aligned using MAFFT [27]. The best-fitting substitution model (GTR + I + G) was inferred using Modeltest 3.7 [29]. Finally, phylogenomic relationships were reconstructed with Bayesian Inference (BI) and Maximum Likelihood (ML) using MrBayes 3.2 [30] and phyML v3.0 [31], respectively. For the BI tree, 10 million generations were simulated using two parallel Markov Chain Monte Carlo (MCMC) simulations, sampled every 1000 generations. The first 25% of the simulations were discarded (burn-in) to generate a consensus tree. For the ML tree, 1000 bootstrap replicates were conducted to evaluate the supporting values of each node.
3. Results and Discussion
3.1. Chloroplast Genome Organisation of the Acer sect. Platanoidea
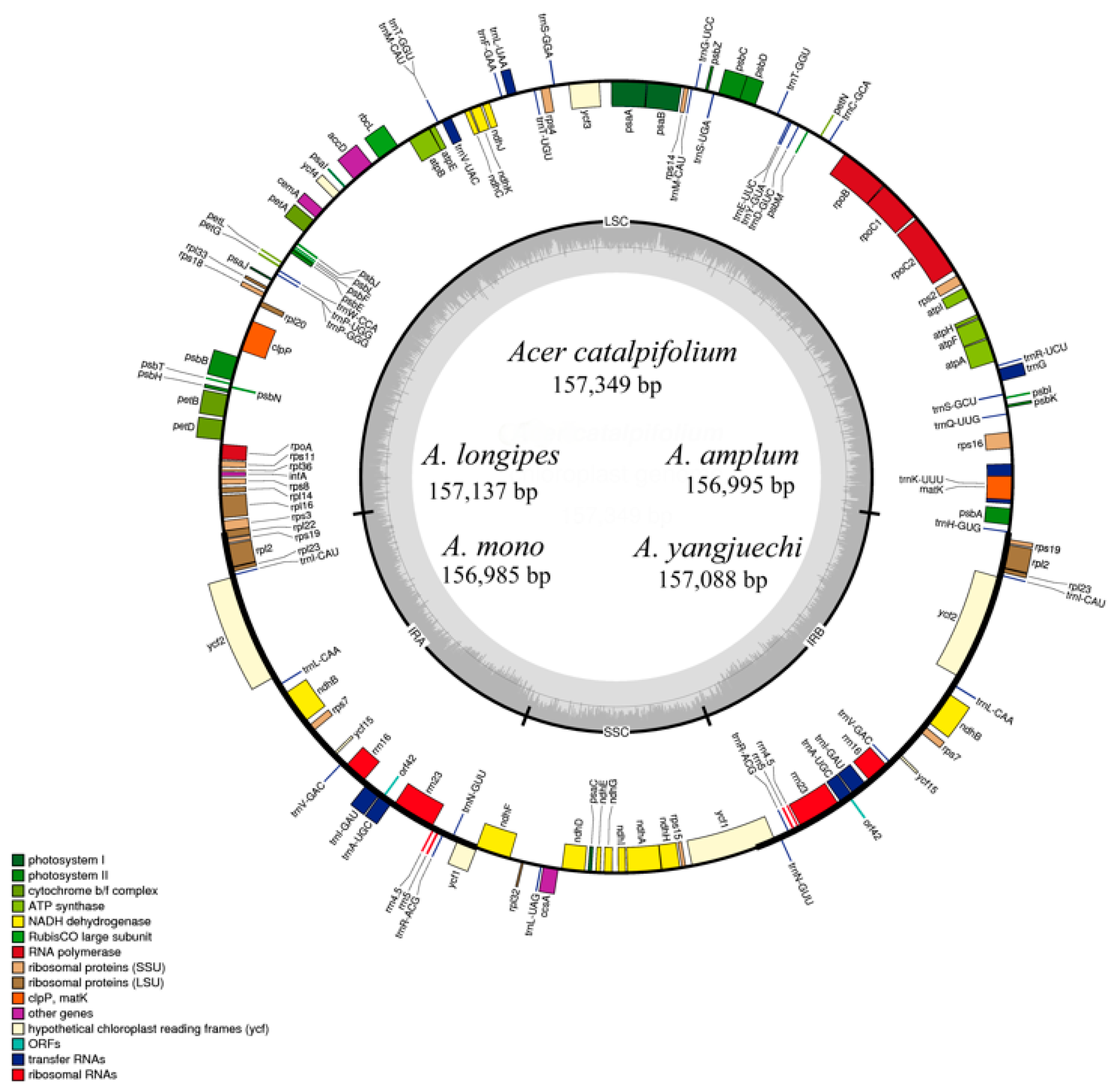
The nucleotide sequences of the seven Acer sect. Platanoidea cp genomes ranged from 156,262 bp to 157,349 bp in length (Figure 1, Table 1), which are similar to other reported cp genomes of Acer [19,25]. The variation of chloroplast genome length is mainly caused by the change of LSC region length. The quadripartite structures of these cp genomes were identical to most angiosperms containing a LSC region, SSC region and two inverted repeat regions (IRa and IRb) [32]. The overall GC content of these cp genomes accounts for 37.9% and the GC content of IR regions accounts for 42.8% higher than the LSC (35.9%) and the SSC (32.3%). The new sequences possess 117 genes, including four unique rRNAs, 31 tRNAs, and 82 PCGs, respectively (Table 2). Among them, most cp genes were single copy, while 23 genes exhibited double copies, including four rRNA (4.5S, 5S, 16S and 23S rRNA), nine tRNA (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnT-GGU and trnV-GAC) and 10 PCGs (ndhB, rpl2, rps12, rpl23, rps19, rps7, ycf1, orf42, ycf2 and ycf15). Additionally, a total of 18 genes harboured introns, and three genes (ycf3, clpP and rps12) contained two introns.
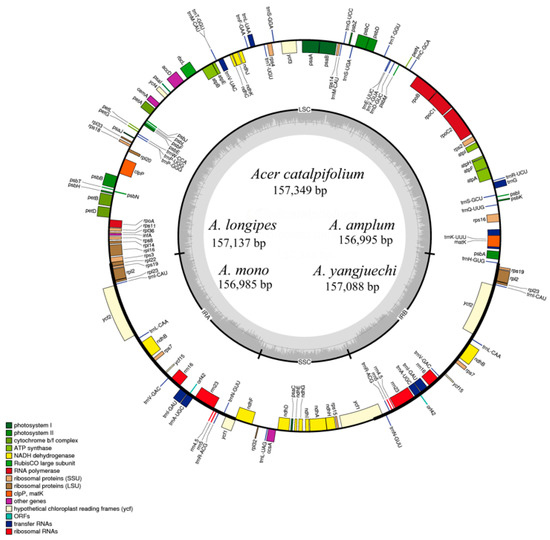
Figure 1.
Merged gene map of the complete chloroplast genomes of five Acer sect. Platanoidea species. Genes belonging to different functional groups are colour-coded. The genes drawn inside the circle are transcribed clockwise, while those outside are transcribed counter-clockwise. Darker grey in the inner circle corresponds to the GC content of the chloroplast genome.

Table 1.
General features of the Acer sect. Platanoidea chloroplast genomes compared in this study.

Table 2.
Genes presented in the Acer sect. Platanoidea chloroplast genome.
3.2. Comparative Analysis of the Genomic Structure
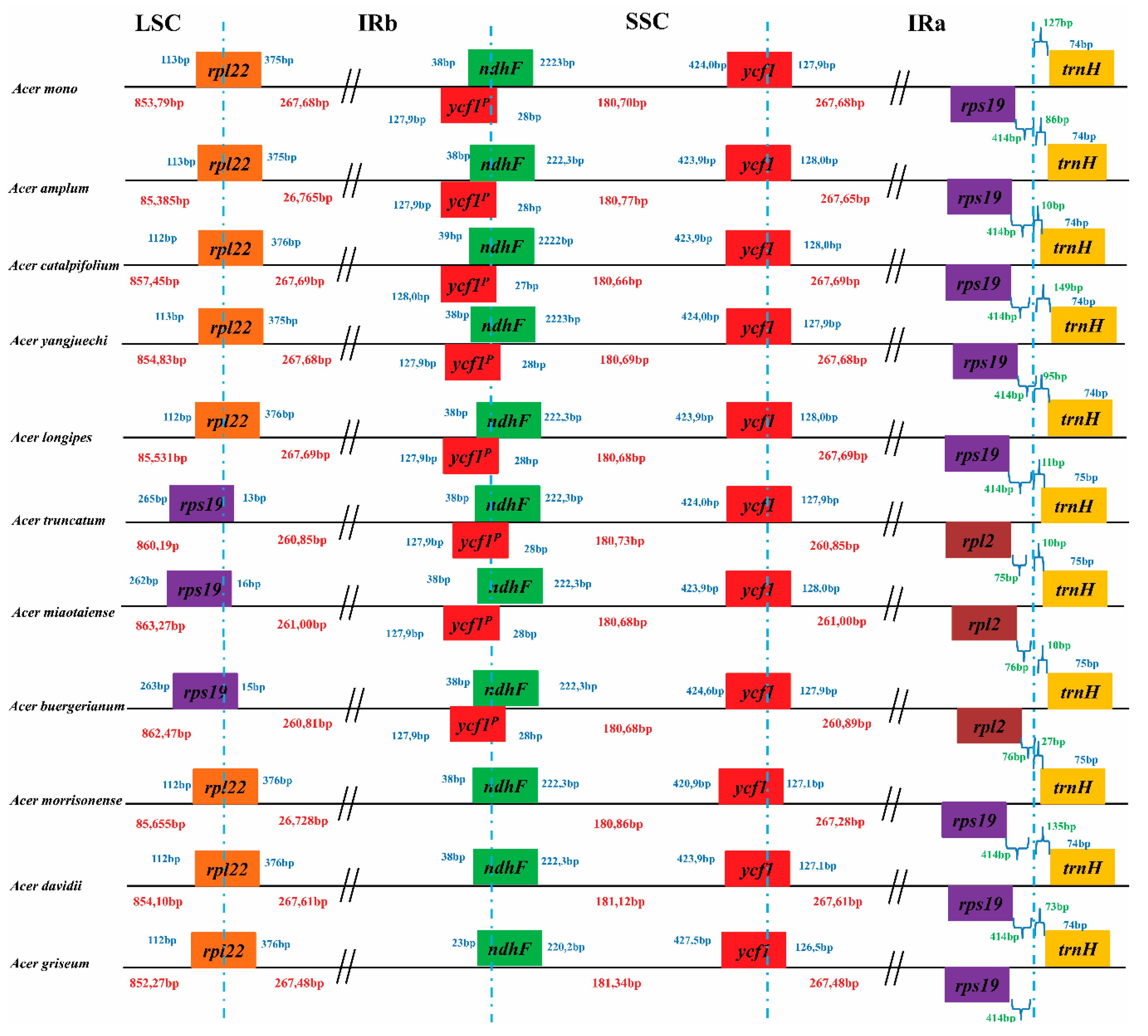
Contraction and expansion of the IRs, LSC and SSC are important to the evolution of cp genomes [33,34], which is the leading cause of gene order and the size changes of the cp genome [35]. Detailed structure comparisons among the 13 cp genomes of the Acer species were presented in Figure 2. rpl22 in the LSC/IR boundary and rps19 was the last gene in most Acer sect. Platanoidea cp genomes included A. mono, A. amplum, A. catalpifolium, A. yangjuechi, A. longipes, A. morrisonense, A. davidii and A. griseum. However, the different structures of rps19 in the LSC/IR boundary and the last gene rpl2 were found in A. truncatum, A. miaotaiense and A. buergerianum. Among the Arecoideae species, rpl22 and rps19 gene order changes in the IRA/LSC borders were also observed and has become the most varied rearrangement in this section [36]. Similarly, the rearrangement has been reported in the Apiales species, which also has two structure types of rpl23 and rps19 in the LSC/IR boundary [37]. Length variations of the Acer cp genomes were found by contraction and expansion of the LSC and IRs, the length of the LSC varies from 85,379 bp to 86,327 bp, and the length of the IR varies from 26,085 bp to 26,769 bp (Figure 2). Moreover, the type of unique structural borders of the cp genome (A. truncatum, A. miaotaiense, and A. buergerianum) also show a contraction of IRs, which indicates variations in boundary genes may be caused by variations in length.
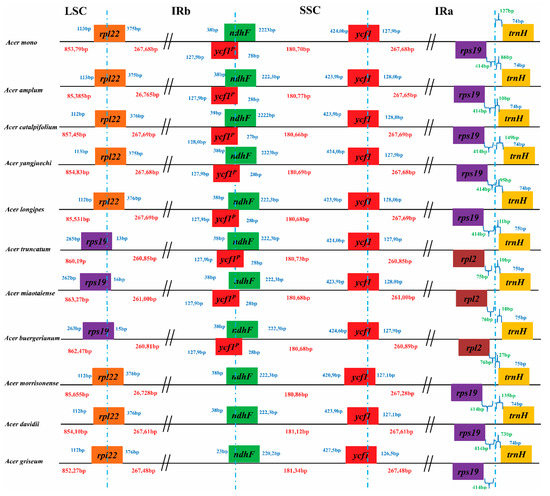
Figure 2.
Comparison of the junction sites of the LSC, IRs and SSC regions among 13 Acer sect. Platanoidea species chloroplast genomes. Different colour boxes indicate specific genes and those above the genome lines indicate their transcriptions in a forward direction, while the under-line boxes are in the reverse direction. The length of the LSC, SSC, and IR regions is shown in red, the length of the gene distance from the boundary is shown in green, the length of the gene is shown in blue, and the P represents the pseudogene.
3.3. Divergence Hotspot of the Acer sect. Platanoidea Species
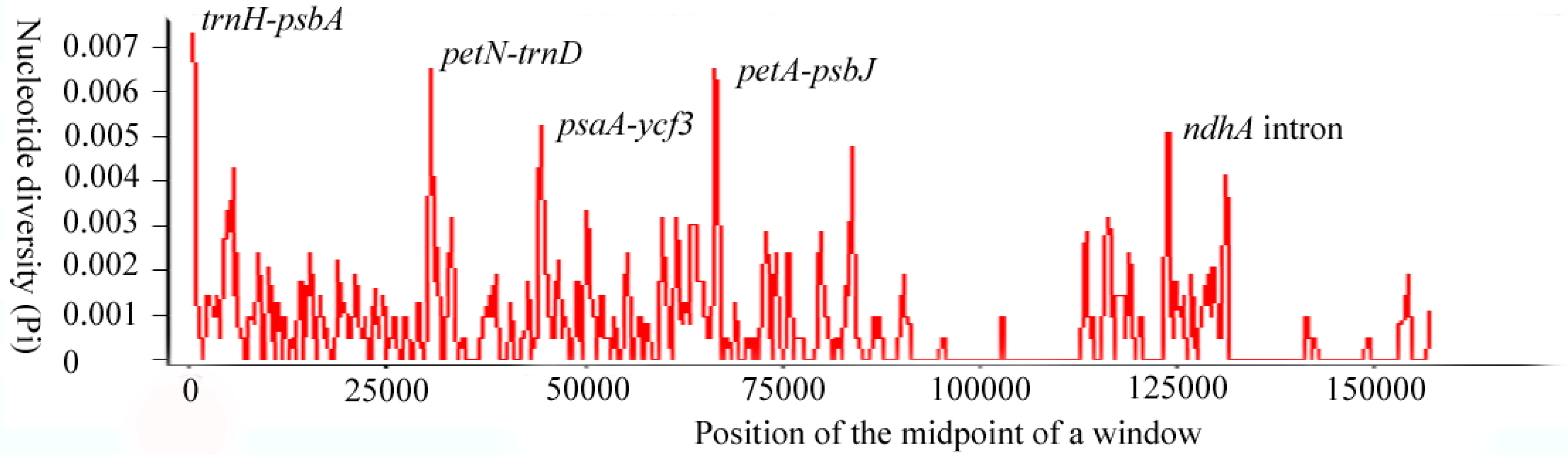
Sliding window analysis of the whole cp genome was performed to identify hotspots of the Acer sect. Platanoidea species. In Figure 3, it is apparent that the trnH-psbA, psbN-trnD, psaA-ycf3, petA-psbJ and ndhA intron nucleotide variability was higher than other regions. Most divergent hotspot loci are located in the LSC region, which allows for the proper design of the genetic markers. Only one hotspot ndhA intron was located in the SSC region. The IR regions were much more conserved. This result was similar to other cp genomes [37,38]. The general barcode trnH-psbA has demonstrated extreme variation in plant groups [39,40]. Thus, the highest variation trnH-psbA has the potential to be used in DNA barcoding in the Acer sect. Platanoidea. Additionally, the evolutionary history of A. mono has been inferred by using psbA-trnH, trnL-trnF and an intron of rpl16 [16]. The regions of the psaA-ycf3, petA-psbJ and ndhA intron have been indicated as high variations in previous studies. In witch-hazel (genus Hamamelis L., Hamamelidaceae), appropriate variations of psaA-ycf3 were used to reconstruct phylogenetic relationships [41]. In Scutellaria, petA-psbJ was one of six fast-evolving DNA sequences in the cp genome [42], while a systematic study shows that Muhlenbergiinae has high variation at the ndhA intron [43].
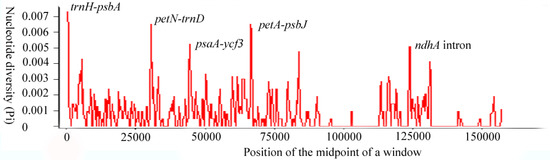
Figure 3.
Sliding-window analysis on the cp genomes for seven Acer sect. Platanoidea species.
The endangered plants, A. catalpifolium, A. yangjuechi and A. miaotaiense in the Acer sect. Platanoidea, have small population sizes [44]. Population genetics studies of these species are relatively weak, and with limited conservation goals [45,46]. These high variability regions can provide alternative sites for subsequent studies and will contribute to the conservation of endangered plants.
3.4. Phylogenetic Analysis
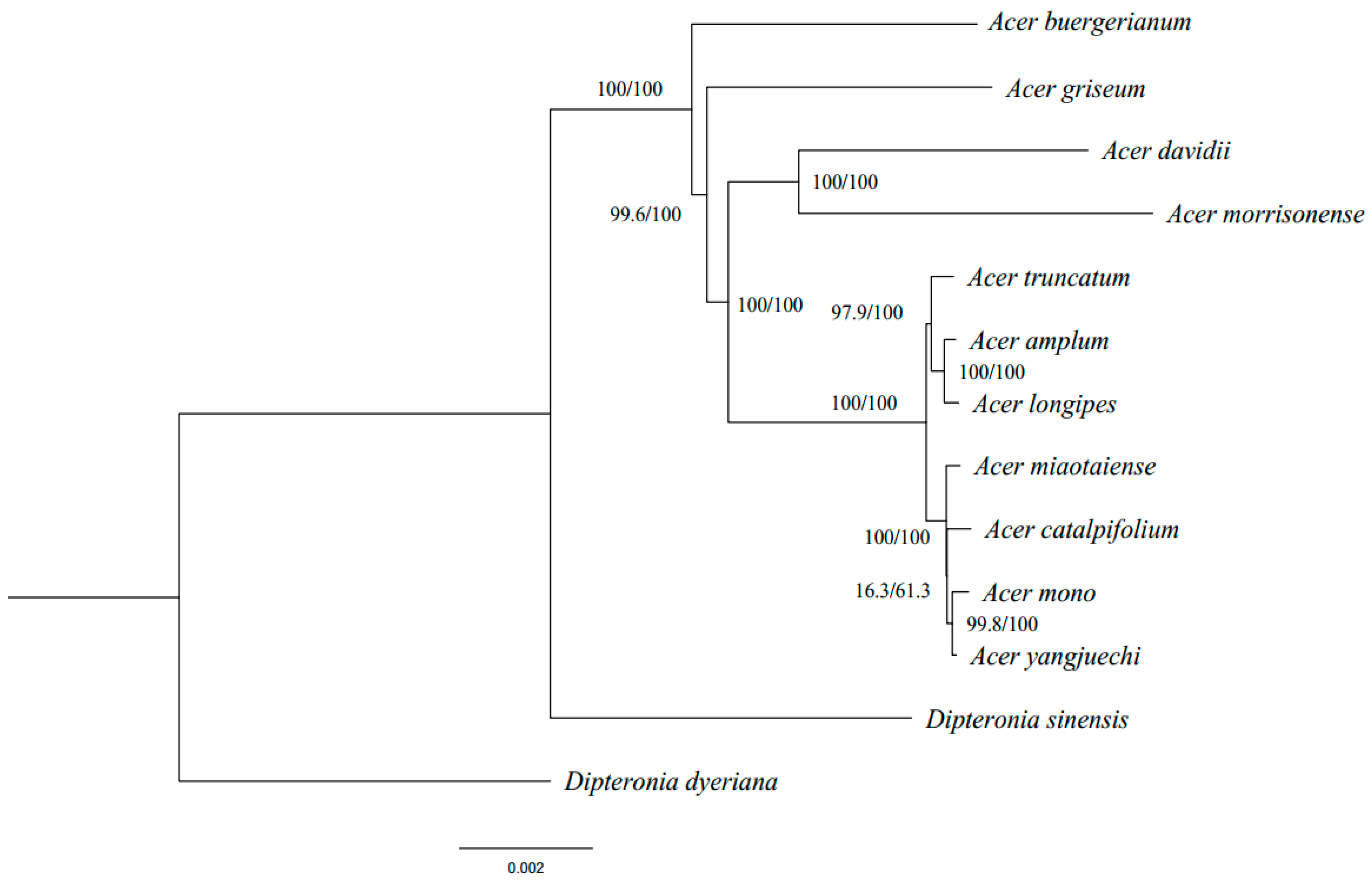
The phylogenomic inference based on cp genomes shows high bootstrap supports in most nodes that provided robust evolutionary placement and relationship of the Acer species (Figure 4). The results showed that seven sampled species of the Acer sect. Platanoidea formed a single clade, which is consistent with previous studies of the Acer phylogeny [14,47]. In this clade, A. catalpifolium, A. mono, A. miaotaiense and A. yangjuechi had the closest phylogenetic relationship and formed a subclade, while A. truncatum, A. longipes, A. amplum formed another one. Previous phylogenetic inference of the Acer did not contain species with a small population size in the sect. Platanoidea (such as A. catalpifolium and A. yangjuechi) [14,47]. Phylogenetic analysis exhibited in this study for the issue of A. longipes A. amplum complex in the sect. Platanoidea raised earlier [17], and the phylogenetic position of A. catalpifolium was also redefined.
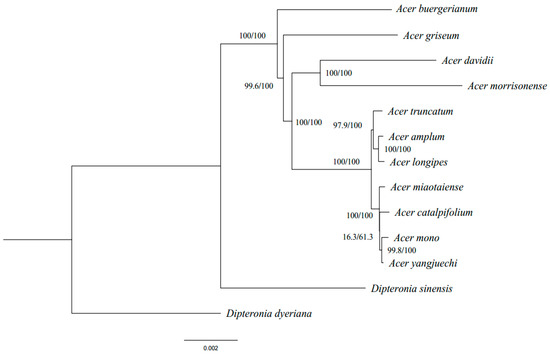
Figure 4.
Phylogenetic tree of 13 Acer species inferred by Maximum Likelihood (ML) and Bayesian Inference (BI) methods, based on the whole cp genome sequences. The numbers above the branches are the bootstrap values of ML methods and the posterior probabilities of BI.
It is somewhat surprising that A. mono is not a sister with A. truncatum but with A. yangjuechi, which differs from some published studies [13]. Since A. mono is widespread in Asia and comprised of multiple local varieties, we cannot rule out that the possibility of the grouping of A. mono and A. yangjuechi due to adjacent sampling localities of A. mono and A. yangjuechi in Lin’an, Zhejiang province of China, the only extant habitat of A. yangjuechi [48]. Liu et al. [49] showed that the population composition of the Lin’an population is significantly different from the neighbouring populations of A. mono. The clustering of A. mono and A. yangjuechi inferred in this study may not only reflect the truth of geographic divergence in the genetics of A. mono but also implies the chloroplast capture by ancient hybridisation events between the two species in Lin’an.
4. Conclusions
In this study, we firstly reported complete cp genomes of five Acer sect. Platanoidea species (A. catalpifolium, A. amplum, A. longipes, A. yanjuechi and A. mono) using the NGS technology. In comparison with other published Acer species from NCBI, we found that the Acer species have similar cp genome structure and gene content. The divergence hotspots identified in the cp genome of the Acer sect. Platanoidea could be applied to develop molecular markers for further population genetics studies. The high variation at the IR/LSC and IR/SSC boundaries were also reported. The phylogenetic analysis strongly supported that A. catalpifolium has the closest relationship with A. miaotaiense, followed by A. mono, and A. yanjuechi, which confirms the species-complex relationship of A. longipes and A. amplum. The available genomic data presented in this paper provides a basis for further research on the evolutionary history and conservation genetics of endangered species of genus Acer.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/4/462/s1, Table S1: General features of the Acer sect. Platanoidea chloroplast genomes compared in this study. Table S2: Acer taxa sampled in this study.
Author Contributions
T.Y. and J.G. conceived and designed the work; J.G. and Y.-Y.Z. collected samples; T.Y., J.G. and B.-H.H. performed the experiments and analysed the data; T.Y. and J.G. wrote the manuscript; P.-C.L., W.-B.M., B.D. and J.-Q.L. critically reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the program ‘‘Reintroduction Technologies and Demonstration of Extremely Rare Wild Plant Population’’ of National Key Research and Development Program (2016YFC0503106) to J.Q.L., the Ministry of Science and Technology of Taiwan (grant number: 108-2628-B-003-001) and National Taiwan Normal University (NTNU) to P.C.L. and “The biogeographical feature and competitive hybridization of Maple (Acer L.) in East Asia” of National Natural Science Foundation of China (41901063) to J.G.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V. Uniparental inheritance of chloroplast genomes. Adv. Photosynth. Respir. 1998, 7, 93–113. [Google Scholar]
- Choi, K.S.; Chung, M.G.; Park, S.J. The complete chloroplast genome sequences of three veroniceae species (plantaginaceae): Comparative analysis and highly divergent regions. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Lv, J.; Li, J.; Du, F.K.; Yin, K. The complete chloroplast genome of the dove tree Davidia involucrata (Nyssaceae), a relict species endemic to China. Conserv. Genet. Resour. 2016, 8, 1–4. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Singh, V.P.; Gupta, D.K.; Singh, N.K.; Sharma, T.R. Genomic resources in horticultural crops: Status, utility and challenges. Biotechnol. Adv. 2011, 29, 199–209. [Google Scholar] [CrossRef]
- Xiong, J.S.; Ding, J.; Li, Y. Genome-editing technologies and their potential application in horticultural crop breeding. Hortic. Res. 2015, 2. [Google Scholar] [CrossRef]
- Cai, J.; Ma, P.F.; Li, H.T.; Li, D.Z. Complete plastid genome sequencing of fourtiliaspecies (malvaceae): A comparative analysis and phylogenetic implications. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Ruhsam, M.; Rai, H.S.; Mathews, S.; Ross, T.G.; Graham, S.W.; Raubeson, L.A.; Mei, W.; Thomas, P.I.; Gardner, M.F.; Ennos, R.A.; et al. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour. 2015, 15, 1067–1078. [Google Scholar] [CrossRef]
- Yi, X.; Gao, L.; Wang, B.; Su, Y.J.; Wang, T. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol. Evol. 2013, 5, 688–698. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.L.; Henry, R.J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.Z.; Chen, Y.S.; de Jong, P.C.; Oterdoom, H.J. Aceraceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science press: Beijing, China, 2008; Volume 11, pp. 515–553. [Google Scholar]
- Van Gelderen, D.M.; De Jong, P.C.; Oterdoom, H.J. Maples of the World; Timber Press: Portland, OR, USA, 1994; ISBN 0881920002. [Google Scholar]
- Renner, S.S.; Grimm, G.W.; Schneeweiss, G.M.; Stuessy, T.F.; Ricklefs, R.E. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for north American/Asian disjunctions. Syst. Biol. 2008, 57, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Rosado, A.; Vera-Vélez, R.; Cota-Sánchez, J.H. Floral morphology and reproductive biology in selected maple ( Acer, L.) species (Sapindaceae). Braz. J. Bot. 2018, 41, 1–14. [Google Scholar] [CrossRef]
- Guo, X.; Wang, H.; Bao, L.; Wang, T.; Bai, W.; Ye, J.; Ge, J. Evolutionary history of a widespread tree species Acer mono in East Asia. Ecol. Evol. 2014, 4, 4332–4345. [Google Scholar] [PubMed]
- Grimm, G.W.; Denk, T. The Colchic region as refuge for relict tree lineages: Cryptic speciation in field maples. Turk. J. Bot. 2014, 38, 1050–1066. [Google Scholar] [CrossRef]
- Kumar, S.; Hahn, F.M.; McMahan, C.M.; Cornish, K.; Whalen, M.C. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009, 9, 131. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Chen, H.; Wang, Y. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conserv. Genet. Resour. 2016, 8, 1–3. [Google Scholar] [CrossRef]
- Wang, R.; Fan, J.; Chang, P.; Zhu, L.; Zhao, M.; Li, L. Genome survey sequencing of acer truncatum bunge to identify genomic information, simple sequence repeat (ssr) markers and complete chloroplast genome. Forests 2019, 10, 87. [Google Scholar] [CrossRef]
- Doyle, J.J. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Dai, M.; Thompson, R.C.; Maher, C.; Contrerasgalindo, R.; Kaplan, M.H.; Markovitz, D.M.; Omenn, G.; Meng, F. NGSQC: Cross-platform quality analysis pipeline for deep sequencing data. BMC Genom. 2010, 11. [Google Scholar] [CrossRef]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, J.; He, Y.L.; He, Y.; Niu, C.; Gong, L.L.; Li, Z.H. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conserv. Genet. Resour. 2016, 8, 1–3. [Google Scholar] [CrossRef]
- Li, Z.-H.; Xie, Y.-S.; Zhou, T.; Jia, Y.; He, Y.-L.; Yang, J. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). Mitochondrial DNA Part A 2015, 28, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PHYML Online–A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33, 557–559. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Guo, H.; Liul, J.; Luo, L.; Wei, X.; Zhang, J.; Qi, Y.; Zhang, B.; Liu, H.; Xiao, P. Complete chloroplast genome sequences of Schisandra chinensis: Genome structure, comparative analysis, and phylogenetic relationship of basal angiosperms. Sci. China Life Sci. 2017, 60, 1286–1290. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; Paola, D.D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other asteraceae. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H. The complete chloroplast genome sequence of the medicinal plant salvia miltiorrhiza. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Han, Y.W.; Duan, D.; Ma, X.F.; Jia, Y.; Liu, Z.L.; Zhao, G.F.; Li, Z.H. Efficient identification of the forest tree species in aceraceae using DNA barcodes. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2. [Google Scholar] [CrossRef]
- Pang, X.; Chang, L.; Shi, L.; Rui, L.; Dong, L.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnh–Psba intergenic spacer region and its combinations as plant dna barcodes: a meta-analysis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Lahaye, R.R.Y.; Savolainen, V.; Duthoit, S.; Maurin, O.; Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef]
- Falchi, A.; Paolini, J.; Desjobert, J.M.; Melis, A.; Costa, J.; Varesi, L. Phylogeography of Cistus creticus L. on Corsica and Sardinia inferred by the TRNL-F and RPL32-TRNL sequences of cpDNA. Mol. Phylogenetics Evol. 2009, 52, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Baillie, J.E.M.; Hilton, C.; Stuart, S.N. IUCN Red List of Threatened Species: A Global Species Assessment; IUCN-The World Conservation Union: Grand, Switzerland, 2004. [Google Scholar]
- Zhang, Y.; Yu, T.; Ma, W.; Tian, C.; Sha, Z.; Li, J. Morphological and physiological response of Acer catalpifolium Rehd. Seedlings to water and light stresses. Glob. Ecol. Conserv. 2019, 19, 10. [Google Scholar] [CrossRef]
- Zhao, J.; Yao, X.; Xi, L.; Yang, J.; Chen, H.; Zhang, J. Characterization of the chloroplast genome sequence of acer miaotaiense: comparative and phylogenetic analyses. Molecules 2018, 23, 1740. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Yue, J.P.; Shoup, S. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harv. Pap. Bot. 2006, 11, 101–115. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Su, J.; Cong, Y. Extraction of genomic DNA from Acer yangjuechi and system optimization of SRAP-PCR. Jiangsu J. Agric. Sci. 2009, 25, 538–541. [Google Scholar]
- Liu, C.; Tsuda, Y.; Shen, H.; Hu, L.; Saito, Y.; Ide, Y. Genetic structure and hierarchical population divergence history of Acer mono var. mono in South and Northeast China. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).