Root Rot Resistance Locus PaLAR3 Is Delivered by Somatic Embryogenesis (SE) Pipeline in Norway Spruce (Picea abies (L.) Karst.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotyping of the PaLAR3 Locus
2.3. SE-Propagation
2.3.1. SE-Initiations
2.3.2. Cryopreservation
2.3.3. Embryo Production Capacity and Embling Viability
2.4. Statistical Analyses
3. Results
3.1. Parent Tree Genotyping
3.2. SE-Initiations
3.3. PaLAR3B Allele Delivery through SE-Pipeline
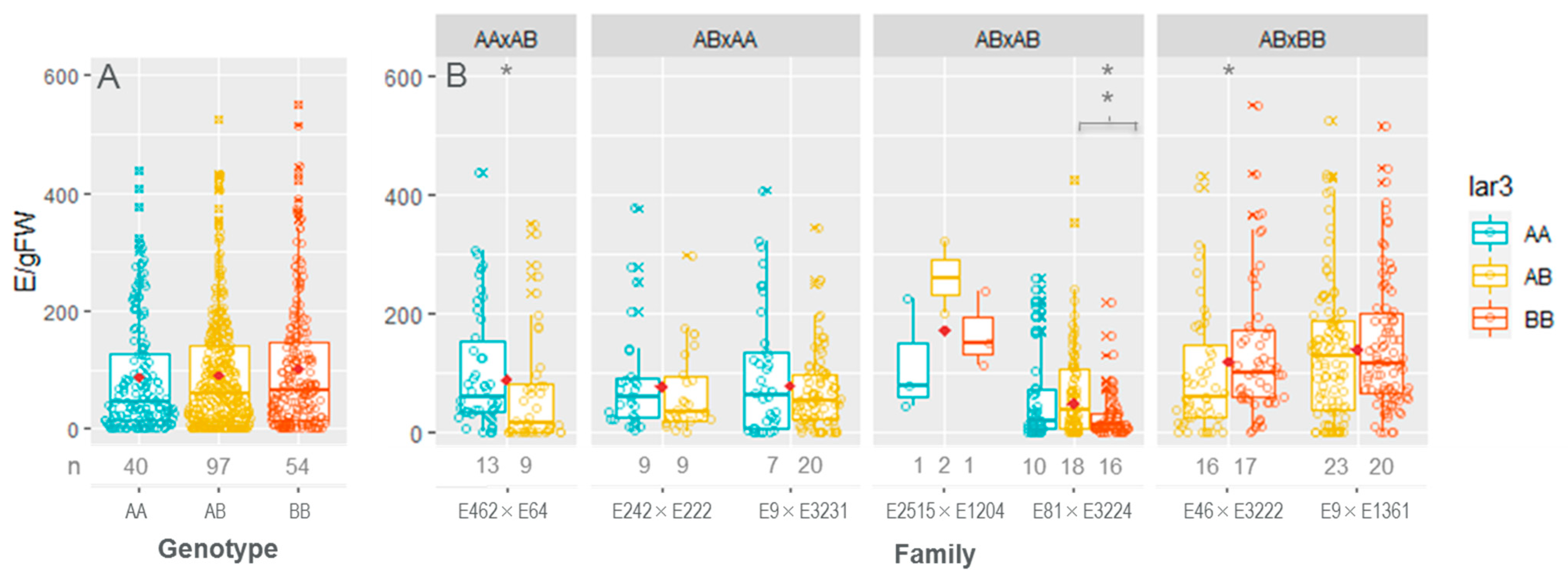
3.4. Embryo Production Ability
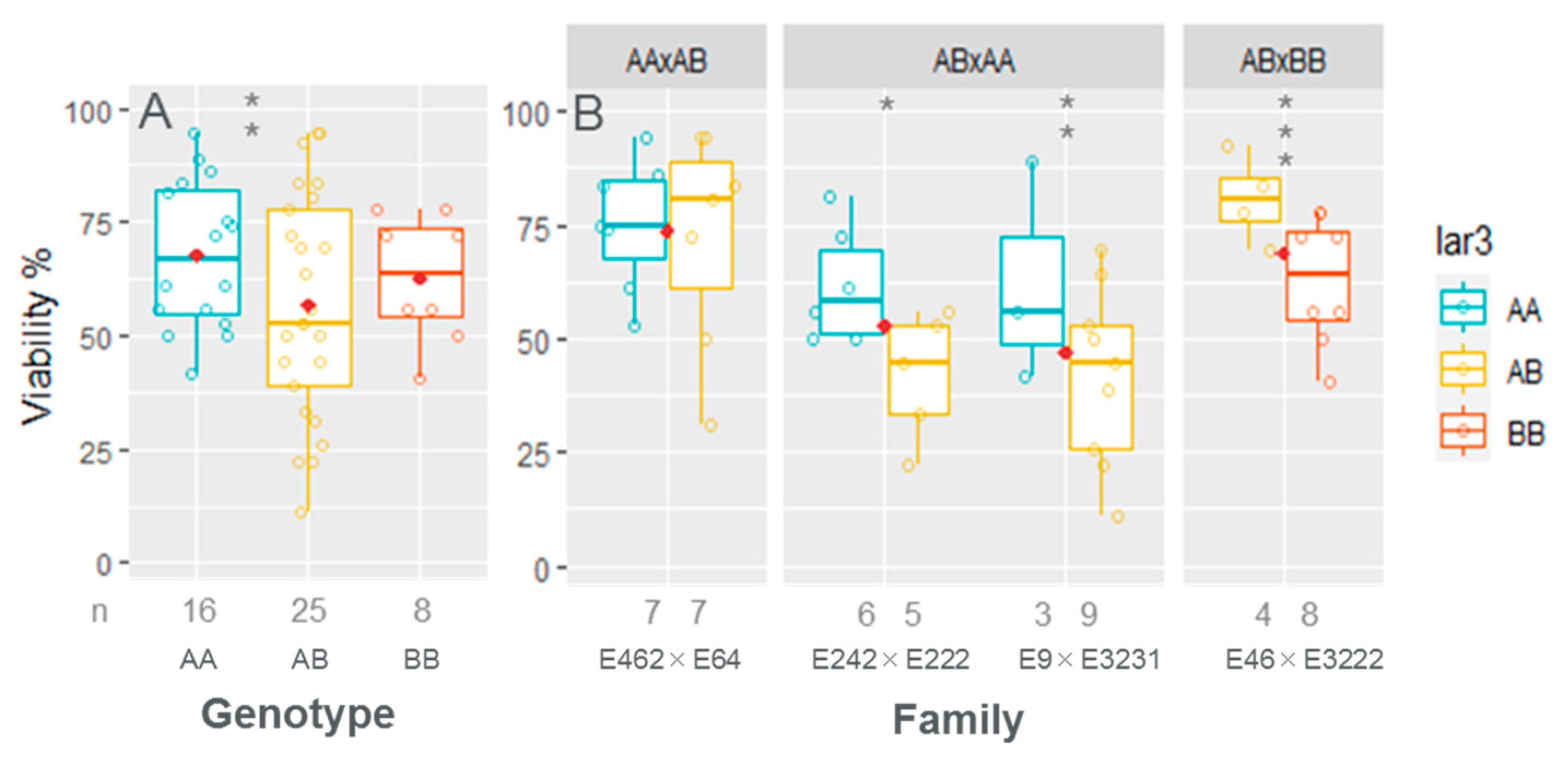
3.5. Embling Viability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jansson, G.; Danusevičius, D.; Grotehusman, H.; Kowalczyk, J.; Krajmerova, D.; Skrøppa, T.; Wolf, H. Norway spruce (Picea Abies (L.) H. Karst). In Forest Tree Breeding in Europe: Current State-of-the-Art and Perspectives; Chambel, M.R., Climent, J., Pichot, C., Ducci, F., Eds.; Springer: Dordrecht, The Netherlands, 2013; Volume 25, pp. 229–265. [Google Scholar] [CrossRef]
- Thor, M.; Möykkynen, T.; Pratt, J.E.; Pukkala, T.; Rönnberg, J.; Shaw, I.G.; Stenlid, J.; Ståhl, G.; Woodward, S. Modeling infection and spread of Heterobasidion annosum in coniferous forests in Europe. USDA For. Serv. Gen. Tech. Rep. PNW 2005, 656, 105–111. [Google Scholar]
- Jansson, G.; Hansen, J.K.; Haapanen, M.; Kvaalen, H.; Steffenrem, A. The genetic and economic gains from forest tree breeding programmes in Scandinavia and Finland. Scand. J. For. Res. 2017, 32, 273–286. [Google Scholar] [CrossRef]
- Chen, Z.; Lunden, K.; Karlsson, B.; Olson, Å.; Vos, I.; Lundqvist, S.-O.; Stenlid, J.; Wu, H.X.; Rosario, M.; Gil, G.; et al. Early selection for resistance to Heterobasidion Parviporum in Norway spruce is not likely to adversely affect growth and wood quality traits in late—Age performance. Eur. J. For. Res. 2018, 137, 517–525. [Google Scholar] [CrossRef]
- Lind, M.; Källman, T.; Chen, J.; Ma, X.F.; Bousquet, J.; Morgante, M.; Zaina, G.; Karlsson, B.; Elfstrand, M.; Lascoux, M.; et al. A Picea abies linkage map based on SNP markers identifies QTLS for four aspects of resistance to Heterobasidion parviporum infection. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Nemesio-Gorriz, M.; Hammerbacher, A.; Ihrmark, K.; Källman, T.; Olson, Å.; Lascoux, M.; Stenlid, J.; Gershenzon, J.; Elfstrand, M. Different alleles of a gene encoding leucoanthocyanidin reductase PaLAR3 influence resistance against the fungus Heterobasidion parviporum in Picea abies. Plant Physiol. 2016, 171, 2671–2681. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, M.; Lundén, K.; Elfstrand, M.; Hu, J.; Zhao, T.; Arnerup, J.; Ihrmark, K.; Swedjemark, G.; Borg-Karlson, A.K.; Stenlid, J. Chemical and transcriptional responses of Norway spruce genotypes with different susceptibility to heterobasidion Spp. infection. BMC Plant Biol. 2011, 11, 1–15. [Google Scholar] [CrossRef]
- Hammerbacher, A.; Paetz, C.; Wright, L.P.; Fischer, T.C.; Bohlmann, J.; Davis, A.J.; Fenning, T.M.; Gershenzon, J.; Schmidt, A. Flavan-3-ols in Norway spruce: Biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol. 2014, 164, 2107–2122. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, V.G.; Lebedeva, T.N.; Chernodubov, A.I.; Shestibratov, K.A. Genomic selection for forest tree improvement: Methods, achievements and perspectives. Forests 2020, 11, 1190. [Google Scholar] [CrossRef]
- Natural Resources Institute Finland–Statistics Service. Available online: https://statdb.luke.fi/PXWeb/pxweb/en/LUKE/ (accessed on 21 December 2020).
- Rosvall, O. Using Norway spruce clones in Swedish forestry: General overview and concepts. Scand. J. For. Res. 2019, 34, 336–341. [Google Scholar] [CrossRef]
- Park, Y.; Bonga, J.; Moon, H. (Eds.) Vegetative Propagation of Forest Trees; National Institute of Forest Science: Seoul, Korea, 2016.
- Adams, G.; Kunze, H.; McCartney, A.; Millican, S.; Park, Y. An industrial perspective on the use of advanced reforestration stock technologies. In Vegetative Propagation of Forest Trees; Park, Y.-S., Bonga, J.M., Moon, H.K., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 323–334. [Google Scholar]
- Lelu-Walter, M.A.; Thompson, D.; Harvengt, L.; Sanchez, L.; Toribio, M.; Pâques, L.E. Somatic embryogenesis in forestry with a focus on Europe: State-of-the-art, benefits, challenges and future direction. Tree Genet. Genomes 2013, 9, 883–899. [Google Scholar] [CrossRef]
- Miguel, C.M.; Rupps, A.; Raschke, J.; Rodrigues, A.; Trotin, J.-F. Impact of molecular studies on somatic embryogenesis development for implementation in conifer multi-varietal forestry. In Vegetative Propagation of Forest Trees; Park, Y., Bonga, J., Moon, H., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 373–421. [Google Scholar]
- Rutledge, R.G.; Stewart, D.; Caron, S.; Overton, C.; Boyle, B.; MacKay, J.; Klimaszewska, K. Potential link between biotic defense activation and recalcitrance to induction of somatic embryogenesis in shoot primordia from adult trees of white spruce (Picea glauca). BMC Plant Biol. 2013, 13, 1–17. [Google Scholar] [CrossRef]
- Businge, E.; Brackmann, K.; Moritz, T.; Egertsdotter, U. Metabolite profiling reveals clear metabolic changes during somatic embryo development of Norway spruce (Picea abies). Tree Physiol. 2012, 32, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Klimaszewska, K.; Park, Y.S.; Overton, C.; Maceacheron, I.; Bonga, J.M. Optimized somatic embryogenesis in Pinus strobus L. In Vitro Cell. Dev. Biol. Plant 2001, 37, 392–399. [Google Scholar] [CrossRef]
- Varis, S.; Ahola, S.; Jaakola, L.; Aronen, T. Reliable and practical methods for cryopreservation of embryogenic cultures and cold storage of somatic embryos of Norway spruce. Cryobiology 2017, 76, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Tikkinen, M.; Varis, S.; Peltola, H.; Aronen, T. Improved germination conditions for Norway spruce somatic cotyledonary embryos increased survival and height growth of emblings. Trees Struct. Funct. 2018, 32, 1489–1504. [Google Scholar] [CrossRef]
- Landis, T.; Dumroese, R.; Haase, D. The Container Tree Nursery Manual: Seedling Processing, Storage, and Outplanting; U.S. Department of Agriculture, Forest Service: Washington, DC, USA, 2010; Volume 7, p. 200.
- Rikala, R. Metsäpuiden Paakkutaimien Kasvatusopas (Manual for Container Seedlings of Forest Trees); Metsäntutkimuslaitos (Finnish Forest Research Institute): Suonenjoki, Finland, 2012.
- Landis, T.; Dumroese, R.; Haase, D. The Container Tree Nursery Manual. Atmospheric Environment; U.S. Department of Agriculture, Forest Service: Washington, DC, USA, 1992; p. 89.
- Meyer, D.; Zeileis, A.; Hornik, K.; Friendly, M. vcd: Visualizing Categorical Data. R Package Version 1.4-8. 2020. Available online: https://cran.r-project.org/web/packages/vcd/vcd.pdf (accessed on 7 February 2021).
- Dunn, O. Multiple comparisons using rank sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Dinno, A. Package ‘Dunn.Test’. Version 1.3.5. 2017. Available online: https://cran.r-project.org/web/packages/dunn.test/dunn.test.pdf (accessed on 7 February 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H. Getting started with Qplot. In Ggplot2; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Pedersen, T.L. Package “ggforce”. Accelerating “Ggplot2.” Version 0.3.2. 2020. Available online: https://cran.r-project.org/web/packages/ggforce/index.html (accessed on 7 February 2021).
- Tikkinen, M.; Varis, S.; Aronen, T. Development of somatic embryo maturation and growing techniques of Norway spruce emblings towards large-scale field testing. Forests 2018, 9, 325. [Google Scholar] [CrossRef]
- Högberg, K.-A.; Varis, S. Vegetative propagation of Norway spruce: Experiences and present situation in Sweden and Finland. In Vegetative Propagation of Forest Trees; Park, Y., Bonga, J., Moon, H., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 538–550. [Google Scholar]
- Dobrowolska, I.; Businge, E.; Abreu, I.N.; Moritz, T.; Egertsdotter, U. Metabolome and transcriptome profiling reveal new insights into somatic embryo germination in Norway spruce (Picea abies). Tree Physiol. 2017, 37, 1752–1766. [Google Scholar] [CrossRef]
- Dhuli, P.; Rohloff, J.; Strimbeck, G.R. Metabolite changes in conifer buds and needles during forced bud break in Norway spruce (Picea abies) and European silver fir (Abies alba). Front. Plant. Sci. 2014, 5, 1–13. [Google Scholar] [CrossRef]
- Puentes, A.; Högberg, K.A.; Björklund, N.; Nordlander, G. Novel avenues for plant protection: Plant propagation by somatic embryogenesis enhances resistance to insect feeding. Front. Plant. Sci. 2018, 871, 1–9. [Google Scholar] [CrossRef]
- Wu, H.X. Benefits and risks of using clones in forestry—A review. Scand. J. For. Res. 2019, 34, 352–359. [Google Scholar] [CrossRef]

| Allele | n | % |
|---|---|---|
| AA | 42 | 52.5 |
| AB | 36 | 45 |
| BB | 2 | 2.50 |
| Family | Parent PaLAR3 Genotype | SE-Progeny PaLAR3 Genotype (n of SE Lines) | |||
|---|---|---|---|---|---|
| AA | AB | BB | Total | ||
| E462 × E64 | AA × AB | 14 | 9 | - | 23 |
| E242 × E222 | AB × AA | 9 | 10 | - | 19 |
| E9 × E3231 | AB × AA | 9 | 21 a | - | 30 |
| E46 × E3222 | AB × BB | - | 17 | 19 | 36 |
| E9 × E1361 | AB × BB | - | 34 | 33 | 67 |
| E2515 × E1204 | AB × AB | 5 | 12 | 5 | 22 |
| E81 × E3224 | AB × AB | 10 | 18 | 16 | 44 |
| Total: | 45 | 122 | 73 | 241 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edesi, J.; Tikkinen, M.; Elfstrand, M.; Olson, Å.; Varis, S.; Egertsdotter, U.; Aronen, T. Root Rot Resistance Locus PaLAR3 Is Delivered by Somatic Embryogenesis (SE) Pipeline in Norway Spruce (Picea abies (L.) Karst.). Forests 2021, 12, 193. https://doi.org/10.3390/f12020193
Edesi J, Tikkinen M, Elfstrand M, Olson Å, Varis S, Egertsdotter U, Aronen T. Root Rot Resistance Locus PaLAR3 Is Delivered by Somatic Embryogenesis (SE) Pipeline in Norway Spruce (Picea abies (L.) Karst.). Forests. 2021; 12(2):193. https://doi.org/10.3390/f12020193
Chicago/Turabian StyleEdesi, Jaanika, Mikko Tikkinen, Malin Elfstrand, Åke Olson, Saila Varis, Ulrika Egertsdotter, and Tuija Aronen. 2021. "Root Rot Resistance Locus PaLAR3 Is Delivered by Somatic Embryogenesis (SE) Pipeline in Norway Spruce (Picea abies (L.) Karst.)" Forests 12, no. 2: 193. https://doi.org/10.3390/f12020193
APA StyleEdesi, J., Tikkinen, M., Elfstrand, M., Olson, Å., Varis, S., Egertsdotter, U., & Aronen, T. (2021). Root Rot Resistance Locus PaLAR3 Is Delivered by Somatic Embryogenesis (SE) Pipeline in Norway Spruce (Picea abies (L.) Karst.). Forests, 12(2), 193. https://doi.org/10.3390/f12020193







