Annual Shoot Segmentation and Physiological Age Classification from TLS Data in Trees with Acrotonic Growth
Abstract
1. Introduction
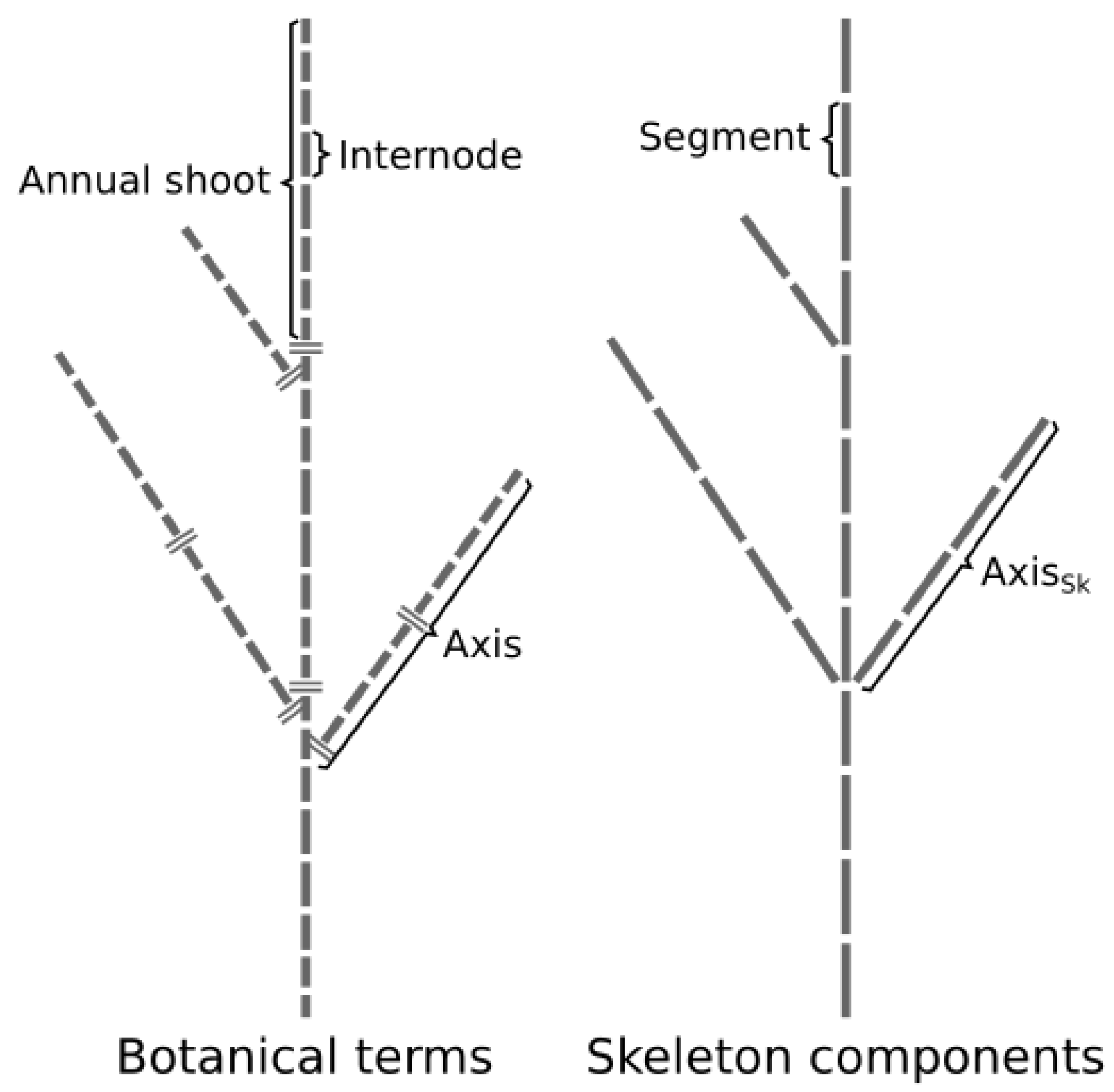
2. Description of the Annual Shoot Segmentation Model
- The segment that is the elementary element of a skeleton defined by 3D coordinates of its starting and ending points and by its unique identifier.
- The skeleton’s axes (axes) are a linear assemblage of connected segments starting from a branching point (or at the tree base for the trunk) and ending at a tip of the skeleton (i.e., a segment without a child segment).
- data preparation prior to annual shoot segmentation (steps 1 to 4)
- annual shoot segmentation (steps 5 and 6)
- annual shoot classification into physiological ages (step 7)
2.1. Data Preparation Prior to Annual Shoot Segmentation (Steps 1 to 4)



2.2. Annual Shoot Segmentation (Steps 5 and 6)
- acrotony occurs at least on the main axes of the tree structure
- -
- the longest lateral axes are close to the AS tip,
- -
- an AS ends immediately or shortly after the last branching point, and
- -
- annual shoots are composed of only one growth unit (i.e., there is no polycyclism) so that branching occurs close to the tip of the AS.
- an AS produced at year n can only bear an AS produced at year
- The AS are correctly segmented. This is usually the case for axes of small order (i.e., main branches and trunk). This is because large axes usually bear many child axes, which are usually well reconstructed by skeletonization methods and because branching accidents are relatively rare in axes of lower order.
- The AS are not segmented. This is typically the case for axes of higher order (i.e., short axes) that are usually not branched or poorly branched. This results in an AS longer than it should be (compare steps 6.4 to 6.3 in Figure 2).
- Oversegmentation (i.e., the addition of an AS that does not exists in reality) occurs. This results in an AS shorter than it should be, usually one segment long and mostly occurring at the tip of an annual shoot.
- if , the annual shoots of the axis are correctly segmented (case 1)
- if , some annual shoots are not segmented (case 2)
- if , some supernumerary segmentation occurred (case 3)
2.3. Classification of Physiological Ages
3. Material and Methods
- at the tree and AS level using “perfect data”
- at the tree level using simulated TLS data
3.1. Tree Sampling, Modeling, and Physiological Age Classification
3.1.1. Sampled Trees and Architectural Measurements
3.1.2. Physiological Age Classification from Manual Measurements
- mean value of the distribution: mu = 2 cm, 10 cm, 30 cm, and 50 cm
- standard deviation: sigma = 10
- the (optional) proportion of the data contained in each distribution (pi) is not provided
3.1.3. Architectural Model Calibration
3.2. TLS Simulations
3.3. Simulated TLS Data Skeletonization
3.4. Comparisons of Modeled vs. Simulated Annual Shoots and Physiological Ages
3.5. Improving the Reconstruction through Non-Reconstructed Axes Modeling
- Markov chains were used to determine branching probabilities in order to estimate pPA4 and position modeled PA4 axes along an AS (using parameters shown in Supplementary Materials Figure S1b)
- binomial distributions were used to generate random PA4 ASs (using parameters shown in Supplementary Materials Figure S1c)
3.6. An Example from the Real World
3.6.1. Tree Sampling and QSM Reconstructions
3.6.2. A New Functionality of the Annual Shoot Segmentation Model
- Near the trimming point (i.e., at the crown center), which results in highly heliotrope branches (i.e., nearly vertical) borne via old branches.
- On the trunk bellow the crown, which results in a less vertical TR orientation and TRs that emerge on older branches compared to case one.
4. Results
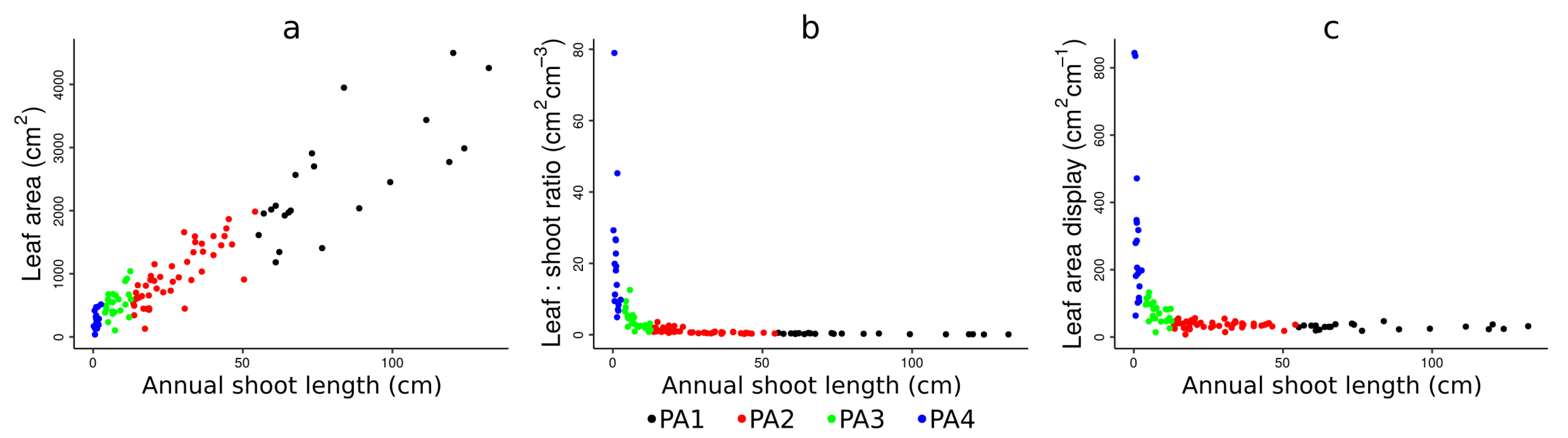
4.1. Physiological Ages Partitioning and Functional Attributes
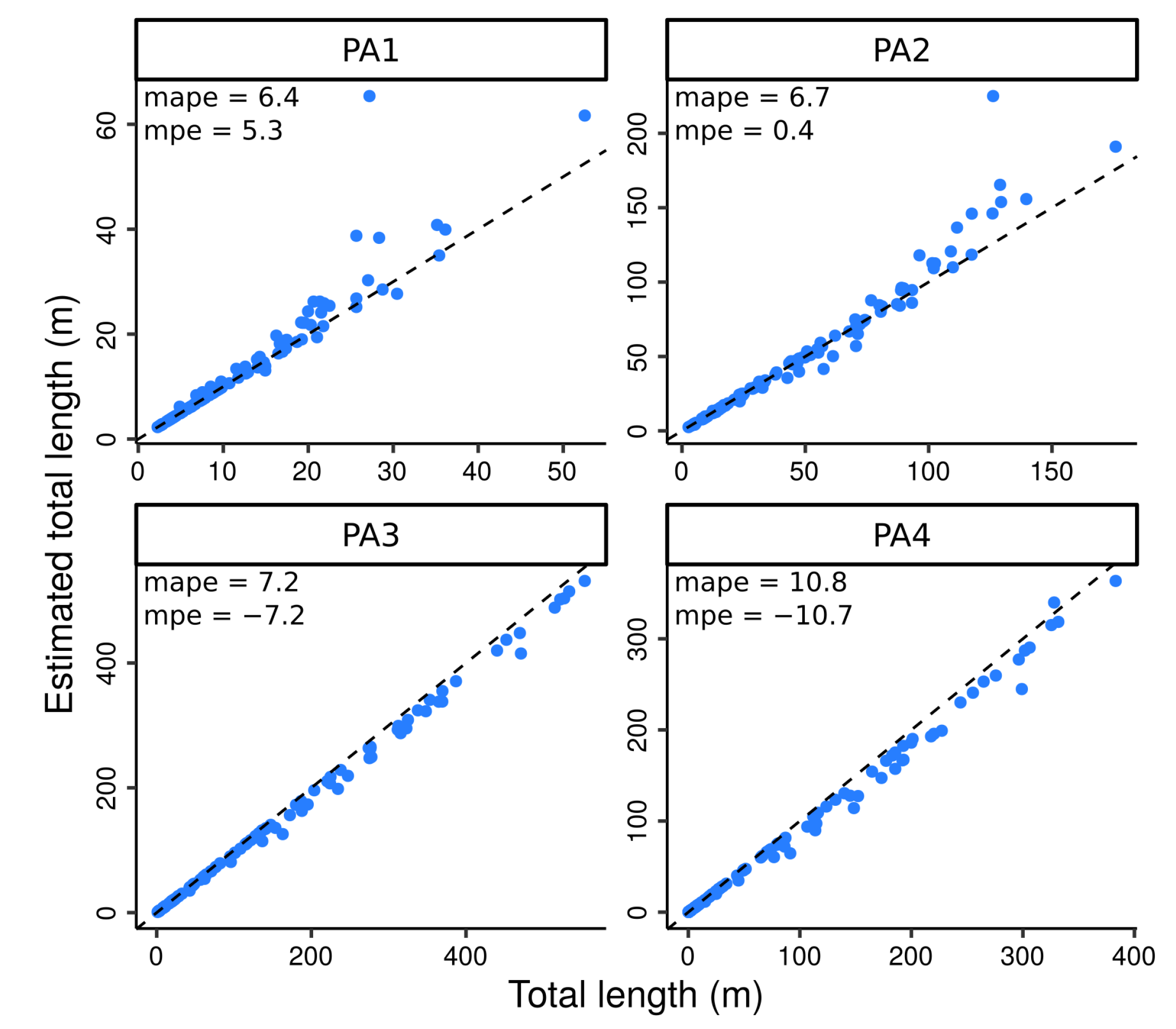
4.2. Testing the Algorithm against Perfect Data
4.3. Testing the Algorithm against Skeletons Obtained from Simulated TLS Data
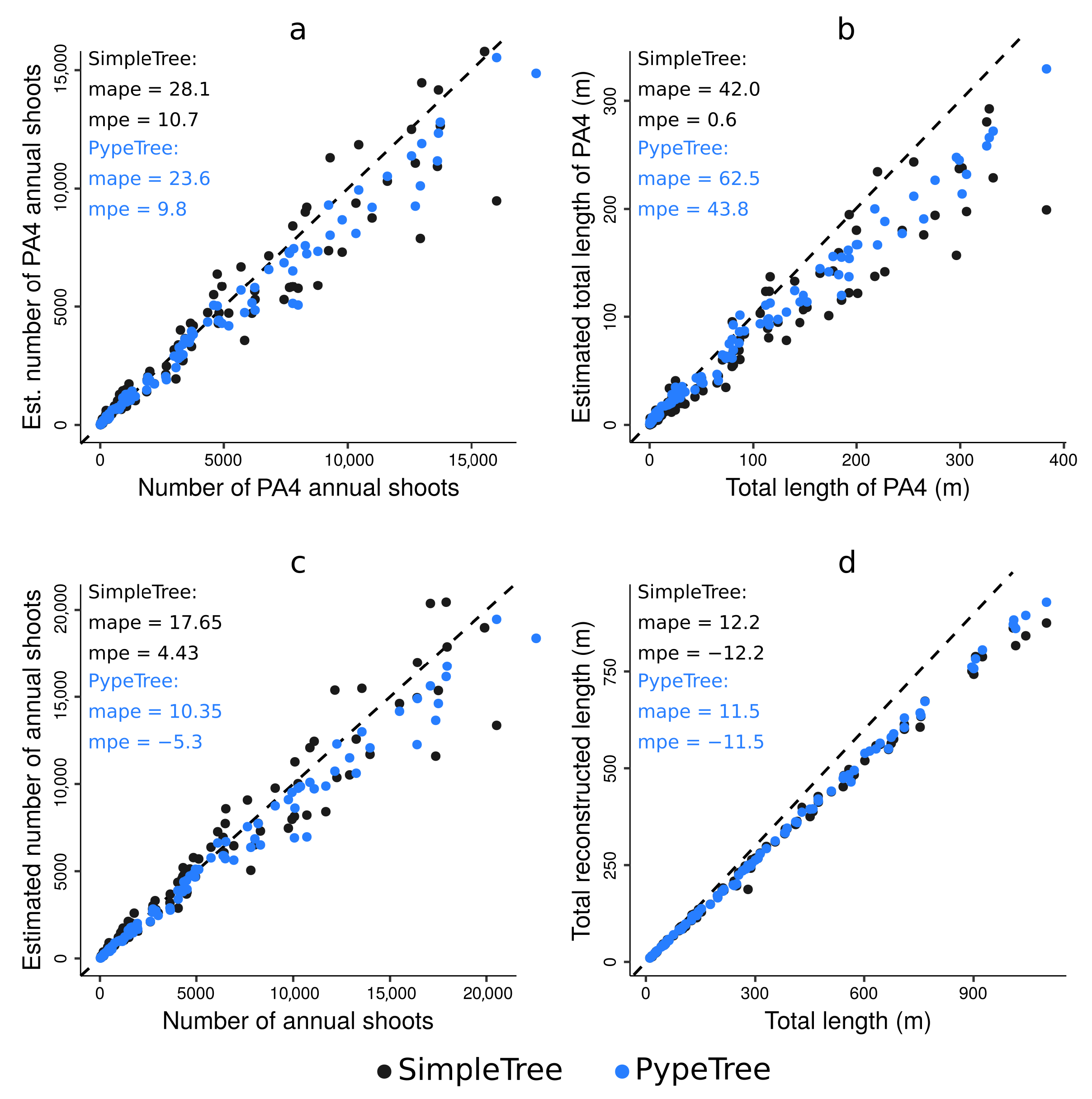
4.4. Real-Life Example
5. Discussion
5.1. Model Accuracy, Possible Applications, and Limitations
- the capacity of the QSM and skeletonization methods to capture all the finest details of the point cloud; and
- the stage of development that influences the branching pattern, especially due to the drift effect [1]. This would limit the applicability of this method to large trees that do not express acrotony anymore.
5.2. On the Use of Distribution Mixture Models to Retrieve Annual Shoot Physiological Ages
5.3. Toward a More Complex Model
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barthélémy, D.; Caraglio, Y. Plant Architecture: A Dynamic, Multilevel and Comprehensive Approach to Plant Form, Structure and Ontogeny. Ann. Bot. 2007, 99, 375–407. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Kikuzawa, K. Patterns in Size-Related Variations in Current-Year Shoot Structure in Eight Deciduous Tree Species. J. Plant Res. 1999, 112, 343–352. [Google Scholar] [CrossRef]
- Yagi, T. Morphology and Biomass Allocation of Current-Year Shoots of Ten Tall Tree Species in Cool Temperate Japan. J. Plant Res. 2000, 113, 171–183. [Google Scholar] [CrossRef]
- Puntieri, J.; Torres, C.; Magnin, A.; Stecconi, M.; Grosfeld, J. Structural Differentiation among Annual Shoots as Related to Growth Dynamics in Luma Apiculata Trees (Myrtaceae). Flora 2018, 249, 86–96. [Google Scholar] [CrossRef]
- Taugourdeau, O.; Delagrange, S.; Lecigne, B.; Sousa-Silva, R.; Messier, C. Sugar Maple (Acer Saccharum Marsh.) Shoot Architecture Reveals Coordinated Ontogenetic Changes between Shoot Specialization and Branching Pattern. Trees 2019, 33, 1615–1625. [Google Scholar] [CrossRef]
- Fernández-Sarría, A.; Martínez, L.; Velázquez-Martí, B.; Sajdak, M.; Estornell, J.; Recio, J.A. Different Methodologies for Calculating Crown Volumes of Platanus Hispanica Trees Using Terrestrial Laser Scanner and a Comparison with Classical Dendrometric Measurements. Comput. Electron. Agric. 2013, 90, 176–185. [Google Scholar] [CrossRef]
- Bayer, D.; Reischl, A.; Rötzer, T.; Pretzsch, H. Structural Response of Black Locust (Robinia Pseudoacacia L.) and Small-Leaved Lime (Tilia Cordata Mill.) to Varying Urban Environments Analyzed by Terrestrial Laser Scanning: Implications for Ecological Functions and Services. Urban For. Urban Green. 2018, 35, 129–138. [Google Scholar] [CrossRef]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non Destructive Method for Biomass Prediction Combining TLS Derived Tree Volume and Wood Density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- Paynter, I.; Genest, D.; Peri, F.; Schaaf, C. Bounding Uncertainty in Volumetric Geometric Models for Terrestrial Lidar Observations of Ecosystems. Interface Focus 2018, 8, 20170043. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.; Herold, M. Quantifying Branch Architecture of Tropical Trees Using Terrestrial LiDAR and 3D Modelling. Trees 2018, 32, 1219–1231. [Google Scholar] [CrossRef]
- Owers, C.J.; Rogers, K.; Woodroffe, C.D. Terrestrial Laser Scanning to Quantify Above-Ground Biomass of Structurally Complex Coastal Wetland Vegetation. Estuar. Coast. Shelf Sci. 2018, 204, 164–176. [Google Scholar] [CrossRef]
- Stovall, A.E.L.; Anderson-Teixeira, K.J.; Shugart, H.H. Assessing Terrestrial Laser Scanning for Developing Non-Destructive Biomass Allometry. For. Ecol. Manag. 2018, 427, 217–229. [Google Scholar] [CrossRef]
- Fan, G.; Nan, L.; Dong, Y.; Su, X.; Chen, F. AdQSM: A New Method for Estimating Above-Ground Biomass from TLS Point Clouds. Remote Sens. 2020, 12, 3089. [Google Scholar] [CrossRef]
- Nock, C.A.; Lecigne, B.; Taugourdeau, O.; Greene, D.F.; Dauzat, J.; Delagrange, S.; Messier, C. Linking Ice Accretion and Crown Structure: Towards a Model of the Effect of Freezing Rain on Tree Canopies. Ann. Bot. 2016, 117, 1163–1173. [Google Scholar] [CrossRef]
- Martin-Ducup, O.; Robert, S.; Fournier, R.A. Response of Sugar Maple (Acer Saccharum, Marsh.) Tree Crown Structure to Competition in Pure versus Mixed Stands. For. Ecol. Manag. 2016, 374, 20–32. [Google Scholar] [CrossRef]
- Lecigne, B.; Delagrange, S.; Messier, C. Exploring Trees in Three Dimensions: VoxR, a Novel Voxel-Based R Package Dedicated to Analysing the Complex Arrangement of Tree Crowns. Ann. Bot. 2018, 121, 589–601. [Google Scholar] [CrossRef]
- Malhi, Y.; Jackson, T.; Patrick Bentley, L.; Lau, A.; Shenkin, A.; Herold, M.; Calders, K.; Bartholomeus, H.; Disney, M.I. New Perspectives on the Ecology of Tree Structure and Tree Communities through Terrestrial Laser Scanning. Interface Focus 2018, 8, 20170052. [Google Scholar] [CrossRef]
- Hosoi, F.; Omasa, K. Voxel-Based 3-D Modeling of Individual Trees for Estimating Leaf Area Density Using High-Resolution Portable Scanning Lidar. IEEE Trans. Geosci. Remote Sens. 2006, 44, 3610–3618. [Google Scholar] [CrossRef]
- Béland, M.; Widlowski, J.-L.; Fournier, R.A.; Côté, J.-F.; Verstraete, M.M. Estimating Leaf Area Distribution in Savanna Trees from Terrestrial LiDAR Measurements. Agric. For. Meteorol. 2011, 151, 1252–1266. [Google Scholar] [CrossRef]
- Béland, M.; Widlowski, J.-L.; Fournier, R.A. A Model for Deriving Voxel-Level Tree Leaf Area Density Estimates from Ground-Based LiDAR. Environ. Model. Softw. 2014, 51, 184–189. [Google Scholar] [CrossRef]
- Li, S.; Dai, L.; Wang, H.; Wang, Y.; He, Z.; Lin, S. Estimating Leaf Area Density of Individual Trees Using the Point Cloud Segmentation of Terrestrial LiDAR Data and a Voxel-Based Model. Remote Sens. 2017, 9, 1202. [Google Scholar] [CrossRef]
- Xie, D.; Wang, X.; Qi, J.; Chen, Y.; Mu, X.; Zhang, W.; Yan, G. Reconstruction of Single Tree with Leaves Based on Terrestrial LiDAR Point Cloud Data. Remote Sens. 2018, 10, 686. [Google Scholar] [CrossRef]
- Magney, T.S.; Eitel, J.U.H.; Griffin, K.L.; Boelman, N.T.; Greaves, H.E.; Prager, C.M.; Logan, B.A.; Zheng, G.; Ma, L.; Fortin, E.A.; et al. LiDAR Canopy Radiation Model Reveals Patterns of Photosynthetic Partitioning in an Arctic Shrub. Agric. For. Meteorol. 2016, 221, 78–93. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Tao, S.; Su, Y. VBRT: A Novel Voxel-Based Radiative Transfer Model for Heterogeneous Three-Dimensional Forest Scenes. Remote Sens. Environ. 2018, 206, 318–335. [Google Scholar] [CrossRef]
- Seidel, D. A Holistic Approach to Determine Tree Structural Complexity Based on Laser Scanning Data and Fractal Analysis. Ecol. Evol. 2018, 8, 128–134. [Google Scholar] [CrossRef]
- Martin-Ducup, O.; Ploton, P.; Barbier, N.; Momo Takoudjou, S.; Mofack, G.; Kamdem, N.G.; Fourcaud, T.; Sonké, B.; Couteron, P.; Pélissier, R. Terrestrial Laser Scanning Reveals Convergence of Tree Architecture with Increasingly Dominant Crown Canopy Position. Funct. Ecol. 2020, 34, 2442–2452. [Google Scholar] [CrossRef]
- Verroust, A.; Lazarus, F. Extracting Skeletal Curves from 3D Scattered Data. In Proceedings of the Shape Modeling International ’99. International Conference on Shape Modeling and Applications, Aizu-Wakamatsu, Japan, 1–4 March 1999. [Google Scholar]
- Bucksch, A.; Lindenbergh, R.; Menenti, M. SkelTre: Robust Skeleton Extraction from Imperfect Point Clouds. Vis. Comput. 2010, 26, 1283–1300. [Google Scholar] [CrossRef]
- Bucksch, A.; Lindenbergh, R. CAMPINO—A Skeletonization Method for Point Cloud Processing. ISPRS J. Photogramm. Remote Sens. 2008, 63, 115–127. [Google Scholar] [CrossRef]
- Li, R.; Bu, G.; Wang, P. An Automatic Tree Skeleton Extracting Method Based on Point Cloud of Terrestrial Laser Scanner. Int. J. Opt. 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Delagrange, S.; Jauvin, C.; Rochon, P. PypeTree: A Tool for Reconstructing Tree Perennial Tissues from Point Clouds. Sensors 2014, 14, 4271–4289. [Google Scholar] [CrossRef]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree—An Efficient Open Source Tool to Build Tree Models from TLS Clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Du, S.; Lindenbergh, R.; Ledoux, H.; Stoter, J.; Nan, L. AdTree: Accurate, Detailed, and Automatic Modelling of Laser-Scanned Trees. Remote Sens. 2019, 11, 2074. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive Estimates of Above-Ground Biomass Using Terrestrial Laser Scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Stovall, A.E.L.; Shugart, H.H. Improved Biomass Calibration and Validation with Terrestrial LiDAR: Implications for Future LiDAR and SAR Missions. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3527–3537. [Google Scholar] [CrossRef]
- Disney, M.I.; Boni Vicari, M.; Burt, A.; Calders, K.; Lewis, S.L.; Raumonen, P.; Wilkes, P. Weighing Trees with Lasers: Advances, Challenges and Opportunities. Interface Focus 2018, 8, 20170048. [Google Scholar] [CrossRef] [PubMed]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Bartholomeus, H.M.; Herold, M.; Kooistra, L. Non-Destructive Tree Volume Estimation through Quantitative Structure Modelling: Comparing UAV Laser Scanning with Terrestrial LIDAR. Remote Sens. Environ. 2019, 233, 111355. [Google Scholar] [CrossRef]
- Delagrange, S.; Rochon, P. Reconstruction and Analysis of a Deciduous Sapling Using Digital Photographs or Terrestrial-LiDAR Technology. Ann. Bot. 2011, 108, 991–1000. [Google Scholar] [CrossRef]
- Qi, J.; Xie, D.; Zhang, W. Reconstruction of Individual Trees Based on LiDAR and in Situ Data. In Proceedings of the 2nd ISPRS International Conference on Computer Vision in Remote Sensing, Xiamen, China, 2 March 2016; Wang, C., Ji, R., Wen, C., Eds.; International Society for Optics and Photonics: Xiamen, China, 2016; p. 99011E. [Google Scholar]
- Åkerblom, M.; Raumonen, P.; Casella, E.; Disney, M.I.; Danson, F.M.; Gaulton, R.; Schofield, L.A.; Kaasalainen, M. Non-Intersecting Leaf Insertion Algorithm for Tree Structure Models. Interface Focus 2018, 8, 20170045. [Google Scholar] [CrossRef]
- Calders, K.; Origo, N.; Burt, A.; Disney, M.; Nightingale, J.; Raumonen, P.; Åkerblom, M.; Malhi, Y.; Lewis, P. Realistic Forest Stand Reconstruction from Terrestrial LiDAR for Radiative Transfer Modelling. Remote Sens. 2018, 10, 933. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Clustering Method: Clustering Criterion and Agglomerative Algorithm. arXiv 2011, arXiv:1111.6285. [Google Scholar]
- R Core Team. Available online: https://www.r-project.org/ (accessed on 14 January 2021).
- Millet, J. L’architecture des Arbres des réGions Tempérées, 1st ed.; MultiMondes: Montréal, QC, Canada, 2012; p. 397. [Google Scholar]
- Perrette, G.; Delagrange, S.; Messier, C. Optimizing Reduction Pruning of Trees Under Electrical Lines: The Influence of Intensity and Season of Pruning on Epicormic Branch Growth and Wound Compartmentalization. Arboric. Urban For. 2020, 46, 432–449. [Google Scholar] [CrossRef]
- Macdonald, P.; Du, J. mixdist: Finite Mixture Distribution Models. R Package. Available online: Https://CRAN.R-project.org/package=mixdist (accessed on 14 January 2021).
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Researché. Available online: Https://CRAN.R-project.org/package=agricolae (accessed on 14 January 2021).
- Barczi, J.-F.; Rey, H.; Caraglio, Y.; de Reffye, P.; Barthelemy, D.; Dong, Q.X.; Fourcaud, T. AmapSim: A Structural Whole-Plant Simulator Based on Botanical Knowledge and Designed to Host External Functional Models. Ann. Bot. 2007, 101, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, S.; Höfle, B. Helios: A multi-purpose LiDAR simulation framework for research, planning and training of laser scanning operations with airborne, ground-based mobile and stationary platforms. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 3, 161–168. [Google Scholar] [CrossRef]
- Adler, D.; Murdoch, D.; Nenadic, O.; Urbanek, S.; Chen, M.; Gebhardt, A.; Bolker, B.; Csardi, G.; Strzelecki, A.; Senger, A.; et al. RGL: A R-Library for 3D Visualization with OpenGL. 11. R Package. Available online: Https://CRAN.R-project.org/package=rgl (accessed on 14 January 2021).
- CloudCompare. Available online: https://www.danielgm.net/cc/ (accessed on 14 January 2021).
- Hackenberg, J.; Morhart, C.; Sheppard, J.; Spiecker, H.; Disney, M. Highly Accurate Tree Models Derived from Terrestrial Laser Scan Data: A Method Description. Forests 2014, 5, 1069–1105. [Google Scholar] [CrossRef]
- Computree. Available online: http://computree.onf.fr/?lang=en (accessed on 14 January 2021).
- Faro. Available online: https://www.faro.com/ (accessed on 14 January 2021).
- Deal, R.L. Development of Epicormic Sprouts in Sitka Spruce Following Thinning and Pruning in South-East Alaska. Forestry 2003, 76, 401–412. [Google Scholar] [CrossRef]
- O’Hara, K.L.; York, R.A.; Heald, R.C. Effect of Pruning Severity and Timing of Treatment on Epicormic Sprout Development in Giant Sequoia. Forestry 2008, 81, 103–110. [Google Scholar] [CrossRef]
- Attocchi, G. Effects of Pruning and Stand Density on the Productionof New Epicormic Shoots in Young Stands of Pedunculate Oak (Quercus robur L.). Ann. For. Sci. 2013, 70, 663–673. [Google Scholar] [CrossRef]
- Maurin, V.; DesRochers, A. Physiological and Growth Responses to Pruning Season and Intensity of Hybrid Poplar. For. Ecol. Manag. 2013, 304, 399–406. [Google Scholar] [CrossRef]
- Desrochers, A.; Maurin, V.; Tarroux, E. Production and Role of Epicormic Shoots in Pruned Hybrid Poplar: Effects of Clone, Pruning Season and Intensity. Ann. For. Sci. 2015, 72, 425–434. [Google Scholar] [CrossRef]
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A Review of Computer Vision Technologies for Plant Phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Lauri, P.-E.; Kelner, J.-J. Shoot Type Demography and Dry Matter Partitioning: A Morphometric Approach in Apple (Malus x Domestica). Can. J. Bot. 2001, 79, 1270–1273. [Google Scholar] [CrossRef]
- Stephan, J.; Sinoquet, H.; Dones, N.; Haddad, N.; Talhouk, S.; Lauri, P.-E. Light Interception and Partitioning between Shoots in Apple Cultivars Influenced by Training. Tree Physiol. 2008, 28, 331–342. [Google Scholar] [CrossRef]
- Sinoquet, H.; Godin, C.; Rivet, P. Assessment of the Three-Dimensional Architecture of Walnut Trees Using Digitising. Silva Fenn. 1997, 31, 265–273. [Google Scholar] [CrossRef][Green Version]
- Colaço, A.F.; Molin, J.P.; Rosell-Polo, J.R.; Escolà, A. Application of Light Detection and Ranging and Ultrasonic Sensors to High-Throughput Phenotyping and Precision Horticulture: Current Status and Challenges. Hortic. Res. 2018, 5, 35. [Google Scholar] [CrossRef]
- Cournède, P.-H.; Kang, M.-Z.; Mathieu, A.; Barczi, J.-F.; Yan, H.-P.; Hu, B.-G.; de Reffye, P. Structural Factorization of Plants to Compute Their Functional and Architectural Growth. Simulation 2006, 82, 427–438. [Google Scholar] [CrossRef]
- Abegg, M.; Boesch, R.; Schaepman, M.E.; Morsdorf, F. Impact of Beam Diameter and Scanning Approach on Point Cloud Quality of Terrestrial Laser Scanning in Forests. IEEE Trans. Geosci. Remote Sens. 2020, 1–15. [Google Scholar] [CrossRef]
- Taugourdeau, O.; Caraglio, Y.; Sabatier, S.; Guédon, Y. Characterizing the Respective Importance of Ontogeny and Environmental Constraints in Forest Tree Development Using Growth Phase Duration Distributions. Ecol. Model. 2015, 300, 61–72. [Google Scholar] [CrossRef]
- GuéDon, Y.; BarthéLéMy, D.; Caraglio, Y.; Costes, E. Pattern Analysis in Branching and Axillary Flowering Sequences. J. Theor. Biol. 2001, 212, 481–520. [Google Scholar] [CrossRef]
- Guédon, Y.; Caraglio, Y.; Heuret, P.; Lebarbier, E.; Meredieu, C. Analyzing Growth Components in Trees. J. Theor. Biol. 2007, 248, 418–447. [Google Scholar] [CrossRef] [PubMed]








| Average | se | % Data | Range | |
|---|---|---|---|---|
| PA1 | 58.06 | 2.09 | 1.47 | 54.51+ |
| PA2 | 20.76 | 14.42 | 34.31 | 13.24–54.51 |
| PA3 | 6.96 | 3.38 | 38.11 | 3.7–13.24 |
| PA4 | 1.8 | 1.24 | 26.1 | 0–3.7 |
| LA (cm) | LAD (cm.cm) | L:S (cm.cm) | ||||
|---|---|---|---|---|---|---|
| Mean (±sd) | gr | Mean (±sd) | gr | Mean (±sd) | gr | |
| PA1 | 2478 (±933.25) | a | 30.39 (±7.34) | d | 0.28 (±0.14) | d |
| PA2 | 973.76 (±453.39) | b | 37.55 (±12.46) | c | 1.02 (±0.73) | c |
| PA3 | 537.26 (±222.44) | c | 73.85 (±30.95) | b | 3.99 (±2.95) | b |
| PA4 | 271.76 (±136.70) | d | 296.17 (±230.60) | a | 21.06 (±18.22) | a |
| Annual Shoot Length | ||||||
| Total (m) | PA1 (m-%) | PA2 (m-%) | PA3 (m-%) | PA4 (m-%) | TR (m-%) | |
| Heavy 1 | 42.02 | 3.72–8.89 | 17.26–40.08 | 6.78–16.14 | 4.19–9.97 | 10.07–23.96 |
| Heavy 2 | 43.61 | 1.39–3.19 | 17.52–40.17 | 10.44–23.94 | 4.75–10.89 | 9.51–21.81 |
| Medium 1 | 132.19 | 6.5–4.92 | 70.15–53.07 | 34.14–25.83 | 14.31–10.83 | 7.09–5.36 |
| Medium 2 | 76.41 | 3.75–4.91 | 44.09–57.70 | 17.68–23.14 | 5.88–7.70 | 5.01–6.56 |
| Control 1 | 153.76 | 18.73–12.18 | 76.28–49.61 | 45.06–29.31 | 13.69–8.90 | - |
| Control 2 | 215.18 | 39.77–18.48 | 110.02–51.13 | 51.88–24.11 | 13.51–6.28 | - |
| Number of Annual Shoot | ||||||
| Total (n) | PA1 (n-%) | PA2 (n-%) | PA3 (n-%) | PA4 (n-%) | TR (n-%) | |
| Heavy 1 | 243 | 8–3.29 | 62–25.51 | 70–28.81 | 83–34.15 | 20–8.23 |
| Heavy 2 | 288 | 1–0.35 | 74–25.69 | 94–32.64 | 94–32.64 | 25–8.68 |
| Medium 1 | 1098 | 9–0.81 | 280–25.50 | 366–33.33 | 426–38.80 | 17–1.54 |
| Medium 2 | 455 | 5–1.20 | 151–33.20 | 160–35.16 | 130–28.57 | 9–1.98 |
| Control 1 | 1021 | 29–2.84 | 262–25.66 | 435–42.61 | 295–28.89 | - |
| Control 2 | 1349 | 66–4.89 | 412–30.54 | 535–39.65 | 336–24.91 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lecigne, B.; Delagrange, S.; Taugourdeau, O. Annual Shoot Segmentation and Physiological Age Classification from TLS Data in Trees with Acrotonic Growth. Forests 2021, 12, 391. https://doi.org/10.3390/f12040391
Lecigne B, Delagrange S, Taugourdeau O. Annual Shoot Segmentation and Physiological Age Classification from TLS Data in Trees with Acrotonic Growth. Forests. 2021; 12(4):391. https://doi.org/10.3390/f12040391
Chicago/Turabian StyleLecigne, Bastien, Sylvain Delagrange, and Olivier Taugourdeau. 2021. "Annual Shoot Segmentation and Physiological Age Classification from TLS Data in Trees with Acrotonic Growth" Forests 12, no. 4: 391. https://doi.org/10.3390/f12040391
APA StyleLecigne, B., Delagrange, S., & Taugourdeau, O. (2021). Annual Shoot Segmentation and Physiological Age Classification from TLS Data in Trees with Acrotonic Growth. Forests, 12(4), 391. https://doi.org/10.3390/f12040391






