Comparative Transcriptome Profiling of Resistant and Susceptible Taxodium Trees in Responding to the Infection by Pestalotiopsis maculans
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and P. maculans Inoculation
2.2. RNA Extraction and cDNA Library Construction and Sequencing
2.3. De Novo Assembly and Functional Annotation
2.4. Analysis of Differential Expression Genes and Gene Annotation
2.5. Validation of RNA-Seq Data by Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
3.1. Transcriptome Sequencing and De Novo Assembly
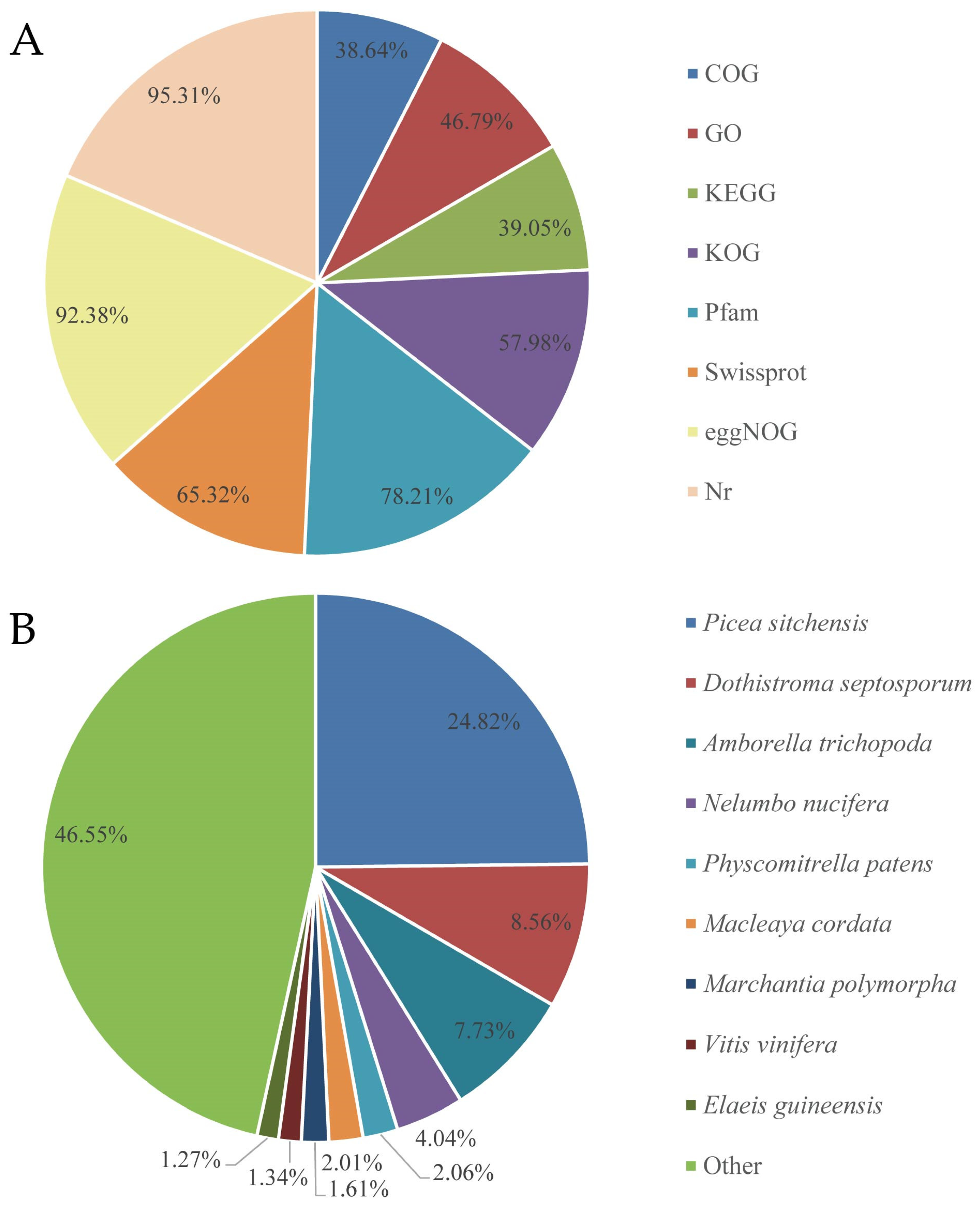
3.2. Gene Annotation of Assembled Transcripts
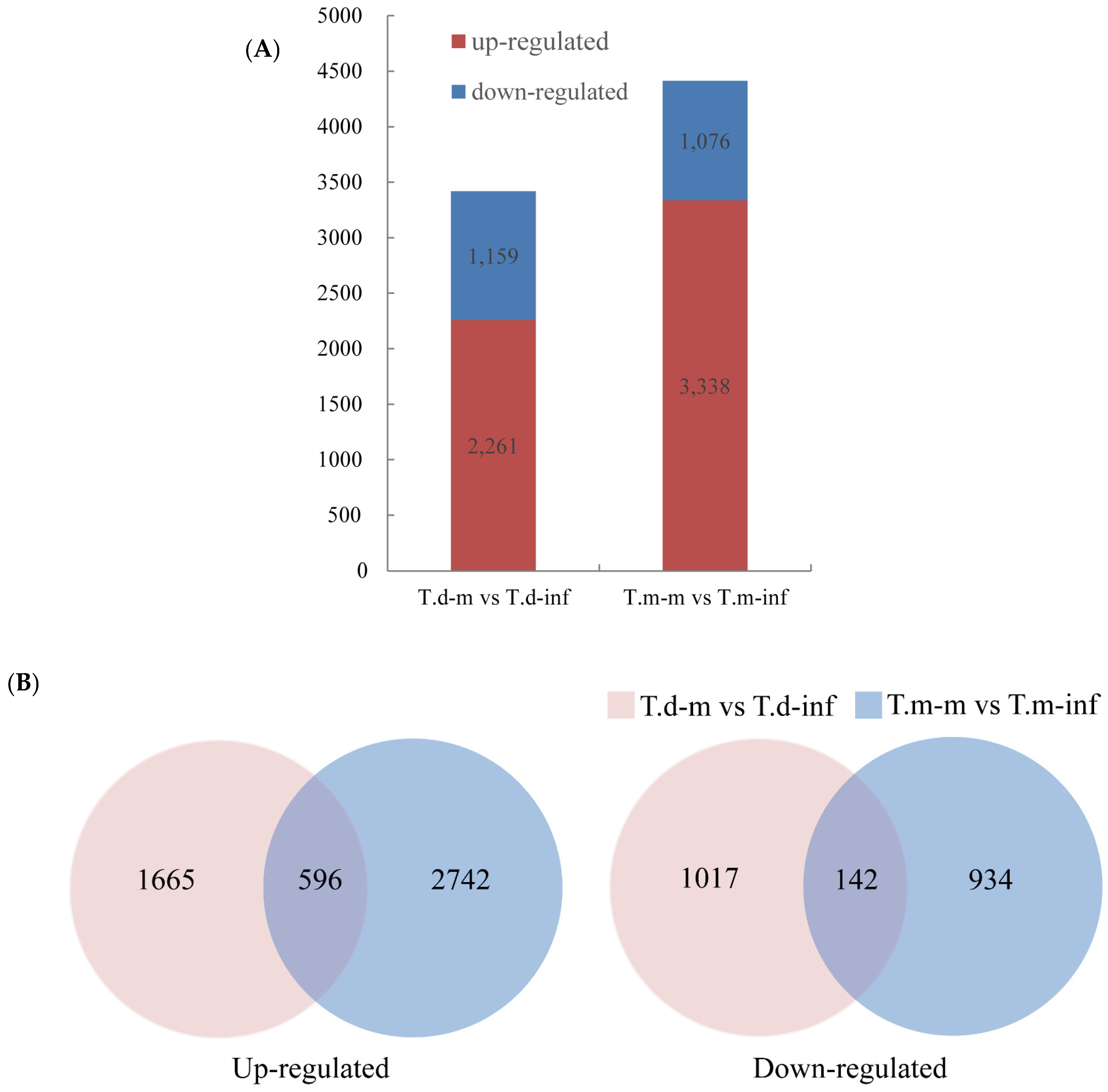
3.3. Different Expressed Genes with and without P. maculans Inoculation
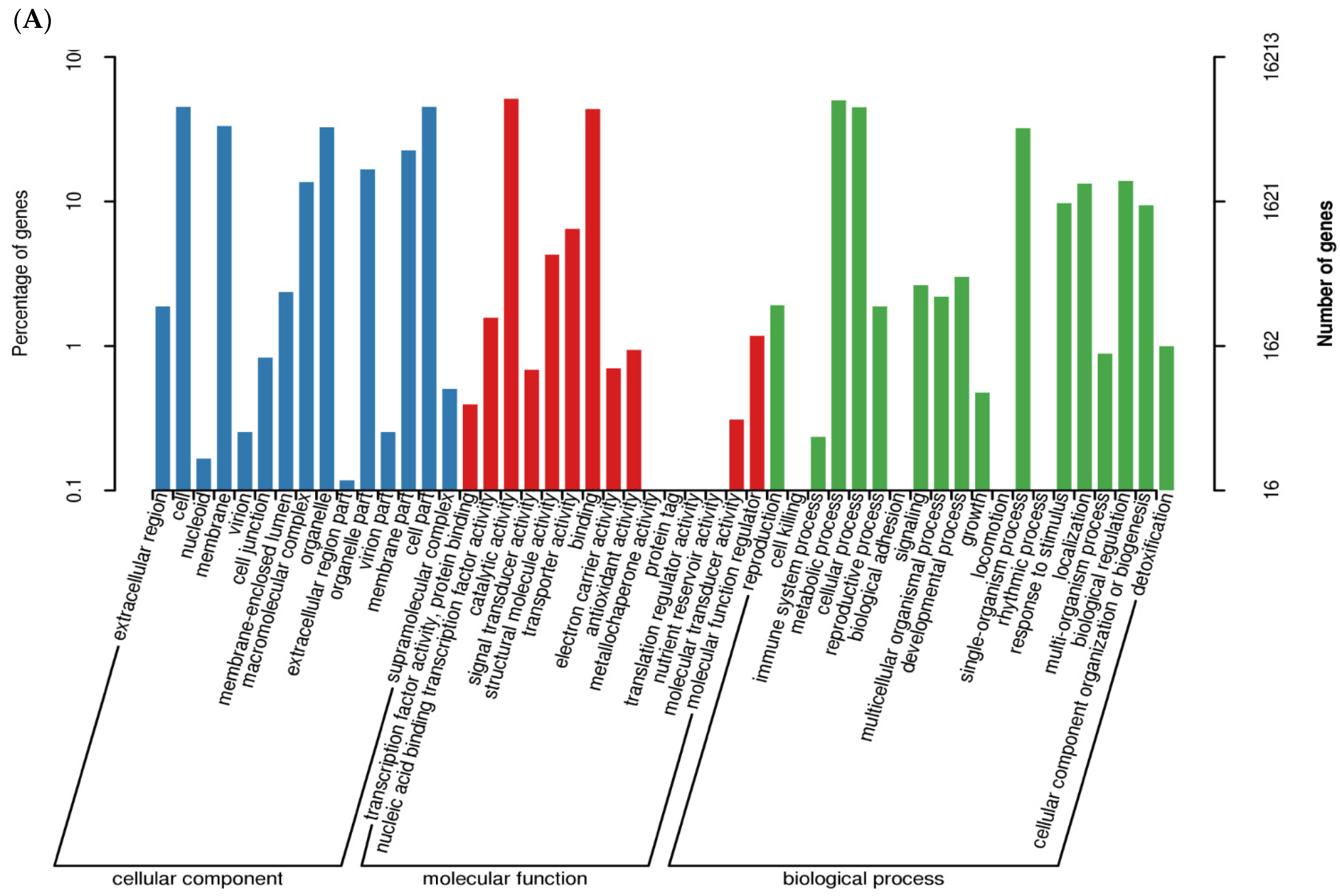
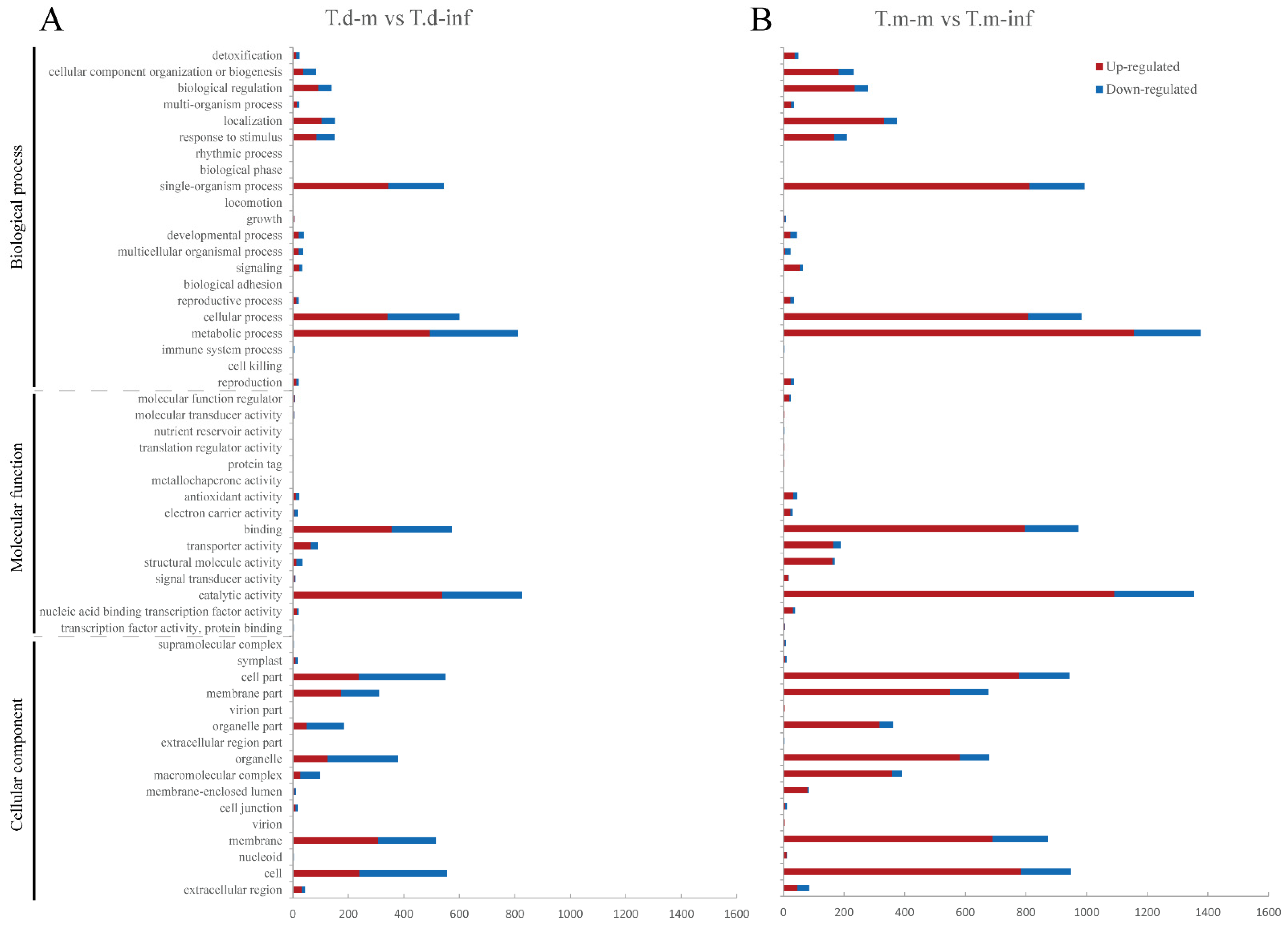
3.4. GO Enrichment Analysis of DEGs after P. maculans Inoculation
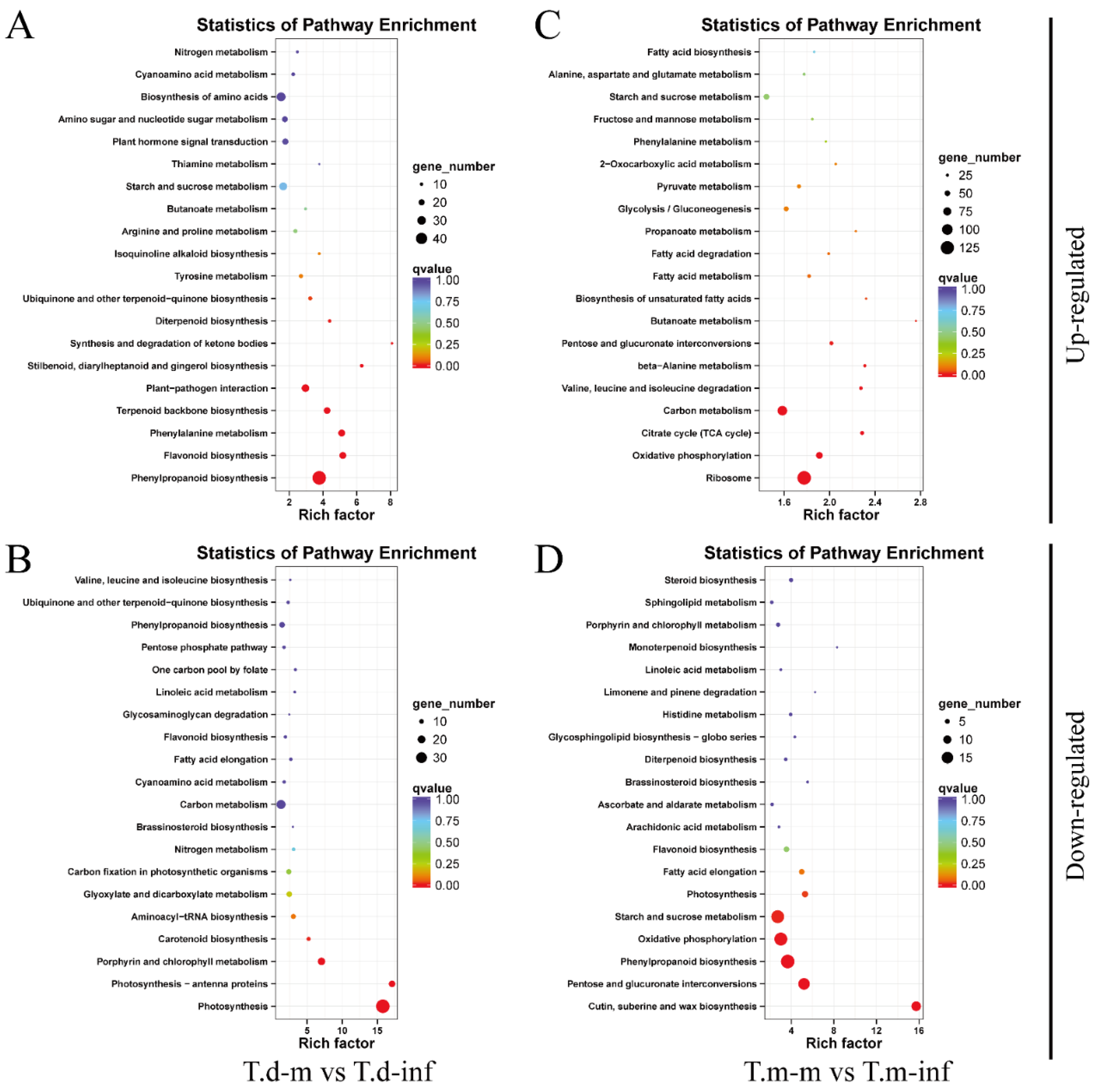
3.5. KEGG Pathway Enrichment Analysis of DEGs after P. maculans Inoculation
3.6. Quantitative Real-Time Reverse Transcription-PCR Validation of Differential Expression
4. Discussion
4.1. Key DEGs in Signal Perception
4.2. Key DEGs in Signal Transduction
4.3. Key DEGs in Phytohormone Metabolism
4.4. Key DEGs in SA-Dependent Pathogen Response
4.5. Key DEGs in Cutin, Suberin, and Wax Modification
4.6. Key DEGs Involved in Cell Wall Modification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duan, H.; Guo, J.; Xuan, L.; Wang, Z.; Li, M.; Yin, Y.; Yang, Y. Comparative chloroplast genomics of the genus Taxodium. BMC Genom. 2020, 21, 114. [Google Scholar] [CrossRef]
- Denny, G.C.; Arnold, M.A. Taxonomy and Nomenclature of Baldcypress, Pondcypress, and Montezuma Cypress: One, Two, or Three Species? Horttechnology 2007, 17, 125–127. [Google Scholar] [CrossRef]
- Creech, D.; Zhou, L.; Yunlong, Y.; Eguiluz-Piedra, T. Can Taxodium Be Improved? Arnoldia 2011, 69, 11–20. [Google Scholar]
- Zhou, L.; Creech, D.L.; Krauss, K.W.; Yunlong, Y.; Kulhavy, D.L. Can We Improve the Salinity Tolerance of Genotypes of Taxodium by Using Varietal and Hybrid Crosses? HortScience 2010, 45, 1773–1778. [Google Scholar] [CrossRef]
- Guo, J.; Duan, H.; Xuan, L.; Wang, Z.; Hua, J.; Yu, C.; Yin, Y.; Li, M.; Yang, Y. Identification and functional analysis of LecRLK genes in Taxodium ‘Zhongshanshan’. PeerJ 2019, 7, e7498. [Google Scholar] [CrossRef]
- Yang, Y.; Xuan, L.; Yu, C.; Wang, Z.; Xu, J.; Fan, W.; Guo, J.; Yin, Y. High-density genetic map construction and quantitative trait loci identification for growth traits in (Taxodium distichum var. distichum × T. mucronatum) × T. mucronatum. BMC Plant Biol. 2018, 18, 263. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, Y.; Yin, Y.; Yu, C.; Yang, Y.; Shi, Q.; Hao, Z.; Li, H. Genetic linkage map construction and QTL mapping of seedling height, basal diameter and crown width of Taxodium ‘Zhongshanshan 302’ × T. mucronatum. SpringerPlus 2016, 5, 1–13. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Han, G. Origin and evolution of the plant immune system. New Phytol. 2018, 222, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xu, S.; Yin, Y. Transcriptome analysis of the Taxodium ‘Zhongshanshan 405’ roots in response to salinity stress. Plant Physiol. Biochem. 2016, 100, 156–165. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhao, M.; Zhong, Q.; Tian, M.; Han, R.; Ren, Y. Comparative transcriptome analysis reveals differentially expressed genes associated with the development of Jerusalem artichoke tuber (Helianthus tuberosus L.). Ind. Crop. Prod. 2020, 151, 112455. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Malukani, K.K.; Chandan, R.K.; Sonti, R.V.; Jha, G. How Plants Respond to Pathogen Attack: Interaction and Communication. In Sensory Biology of Plants; Springer: Berlin/Heidelberg, Germany, 2019; pp. 537–568. [Google Scholar]
- Zhou, J.-M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, J. Regulatory role of receptor-like cytoplasmic kinases in early immune signaling events in plants. FEMS Microbiol. Rev. 2020, 44, 845–856. [Google Scholar] [CrossRef]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef]
- Huaping, H.; Xiaohui, J.; Lunying, W.; Junsheng, H. Chitin elicitor receptor kinase 1 (CERK1) is required for the non-host defense response of Arabidopsis to Fusarium oxysporum f. Sp. cubense. Eur. J. Plant Pathol. 2017, 147, 571–578. [Google Scholar] [CrossRef]
- Erwig, J.; Ghareeb, H.; Kopischke, M.; Hacke, R.; Matei, A.; Petutschnig, E.; Lipka, V. Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 2017, 215, 382–396. [Google Scholar] [CrossRef]
- Geldner, N.; Robatzek, S. Plant Receptors Go Endosomal: A Moving View on Signal Transduction. Plant Physiol. 2008, 147, 1565–1574. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nat. Cell Biol. 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Tena, G.; Asai, T.; Chiu, W.-L.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Wang, C.; Liu, M.; Li, H.; Fu, Y.; Wang, Y.; Nie, Y.; Liu, X.; Ji, W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genom. 2014, 15, 898. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.-Q.; Guo, J.; Zhang, N.; Yao, X.; Wang, H.-B.; Li, J.-F. Cross-Microbial Protection via Priming a Conserved Immune Co-Receptor through Juxtamembrane Phosphorylation in Plants. Cell Host Microbe 2019, 26, 810–822. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, J.; Wang, J.; Chen, S.; Chen, L.; Wang, J.; Wang, H.; Liu, B. The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J. Integr. Plant Biol. 2019, 62, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Loake, G.; Grant, M. Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Hwang, S.-H.; Yie, S.W.; Hwang, D.-J. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011, 181, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Hwang, S.; Fang, I.R.; Kwon, S.I.; Park, S.R.; Ahn, I.; Kim, J.B.; Hwang, D. Molecular characterization of Oryza sativa WRKY 6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015, 208, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, L.; Ma, X.; Zhao, X.; Jiao, B.; Li, C.; Luo, K. The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 2017, 37, 665–675. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2 WRKY13 WRKY42 Transcriptional Regulatory Cascade Is Required for Rice Resistance to Fungal Pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low Oleic Acid-Derived Repression of Jasmonic Acid-Inducible Defense Responses Requires the WRKY50 and WRKY51 Proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca2+/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149.e7. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Kahmann, R. Manipulation of Phytohormone Pathways by Effectors of Filamentous Plant Pathogens. Front. Plant Sci. 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of Hormone Signaling Networks in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef]
- Strawn, M.A.; Marr, S.K.; Inoue, K.; Inada, N.; Zubieta, C.; Wildermuth, M.C. Arabidopsis Isochorismate Synthase Functional in Pathogen-induced Salicylate Biosynthesis Exhibits Properties Consistent with a Role in Diverse Stress Responses. J. Biol. Chem. 2007, 282, 5919–5933. [Google Scholar] [CrossRef]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.-P. Characterization and Biological Function of the ISOCHORISMATE SYNTHASE2 Gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef]
- Nawrath, C.; Métraux, J.-P. Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, W.; Han, X.; Wu, J.; Lyu, B.; Chen, F.; Caplan, A.; Li, C.; Wu, J.; Wang, W.; et al. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018, 19, 1995–2010. [Google Scholar] [CrossRef] [PubMed]
- Shine, M.B.; Yang, J.; El-Habbak, M.; Nagyabhyru, P.; Fu, D.; Navarre, D.; Ghabrial, S.; Kachroo, P.; Kachroo, A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016, 212, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural Basis for Prereceptor Modulation of Plant Hormones by GH3 Proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nat. Cell Biol. 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.D.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Koo, A.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and Function of Arabidopsis JASMONATE ZIM-Domain Genes in Response to Wounding and Herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.-Y.; Hunt, M.D. Systemic Acquired Resistance. Plant Cell 1996, 8, 1809. [Google Scholar] [CrossRef]
- Durrant, W.; Dong, X. Systemic Acquired Resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. Cell 2020, 182, 1093–1108.e18. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Withers, J.; Li, H.; Zwack, P.J.; Rusnac, D.-V.; Shi, H.; Liu, L.; Yan, S.; Hinds, T.R.; Guttman, M.; et al. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature 2020, 586, 311–316. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Backer, R.; Naidoo, S.; van den Berg, N. The nonexpressor of pathogenesis-related genes 1 (NPR1) and related family: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 1996, 143, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Hwang, B.K. Isolation, Partial Sequencing, and Expression of Pathogenesis-Related cDNA Genes from Pepper Leaves Infected by Xanthomonas campestris pv. vesicatoria. Mol. Plant-Microbe Interact. 2000, 13, 136–142. [Google Scholar] [CrossRef]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L. Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci. 2010, 15, 546–553. [Google Scholar] [CrossRef]
- Fich, E.; Segerson, N.A.; Rose, J.K. The Plant Polyester Cutin: Biosynthesis, Structure, and Biological Roles. Annu. Rev. Plant Biol. 2016, 67, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Philippe, G.; Sørensen, I.; Jiao, C.; Sun, X.; Fei, Z.; Domozych, D.S.; Rose, J.K. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef]
- Lim, G.-H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty acid–and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional Roles of Plant Cuticle During Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef]
- Graça, J. Suberin: The biopolyester at the frontier of plants. Front. Chem. 2015, 3, 62. [Google Scholar] [CrossRef]
- Gou, J.-Y.; Yu, X.-H.; Liu, C.-J. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 18855–18860. [Google Scholar] [CrossRef]
- Serra, O.; Hohn, C.; Franke, R.; Prat, S.; Molinas, M.; Figueras, M. A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. Plant J. 2010, 62, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Compagnon, V.; Diehl, P.; Benveniste, I.; Meyer, D.; Schaller, H.; Schreiber, L.; Franke, R.; Pinot, F. CYP86B1 is required for very long chain ω-hydroxyacid and α, ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009, 150, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, A.L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme a: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef]
- Xiao, F.; Goodwin, S.M.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.-M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Mehli, L.; Schaart, J.G.; Kjellsen, T.D.; Tran, D.H.; Salentijn, E.M.J.; Schouten, H.J.; Iversen, T. A gene encoding a polygalacturonase-inhibiting protein (PGIP) shows developmental regulation and pathogen-induced expression in strawberry. New Phytol. 2004, 163, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Vairo, D.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Tandemly Duplicated Arabidopsis Genes That Encode Polygalacturonase-Inhibiting Proteins Are Regulated Coordinately by Different Signal Transduction Pathways in Response to Fungal Infection. Plant Cell 2003, 15, 93–106. [Google Scholar] [CrossRef]
- Oelofse, D.; Dubery, I.A.; Meyer, R.; Arendse, M.S.; Gazendam, I.; Berger, D. Apple polygalacturonase inhibiting protein1 expressed in transgenic tobacco inhibits polygalacturonases from fungal pathogens of apple and the anthracnose pathogen of lupins. Phytochemistry 2006, 67, 255–263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barros-Rios, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef]
- Xie, T.; Liu, Z.; Wang, G. Structural basis for monolignol oxidation by a maize laccase. Nat. Plants 2020, 6, 231–237. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.-Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Soni, N.; Hegde, N.; Dhariwal, A.; Kushalappa, A.C. Role of laccase gene in wheat NILs differing at QTL-Fhb1 for resistance against Fusarium head blight. Plant Sci. 2020, 298, 110574. [Google Scholar] [CrossRef]


| Sample | Clean Reads | Clean Base | Q30 (%) | GC Content (%) |
|---|---|---|---|---|
| T.m-m1 | 23,234,663 | 6,908,744,520 | 44.22% | 94.81% |
| T.m-m2 | 27,191,454 | 8,117,561,816 | 45.78% | 94.34% |
| T.m-m3 | 26,655,673 | 7,963,346,392 | 45.29% | 94.50% |
| T.m-inf1 | 23,359,280 | 6,958,966,304 | 44.08% | 94.59% |
| T.m-inf2 | 25,414,766 | 7,588,528,280 | 44.82% | 93.92% |
| T.m-inf3 | 23,958,561 | 7,165,086,890 | 44.63% | 93.99% |
| T.d-m1 | 20,405,234 | 6,082,914,056 | 44.73% | 94.71% |
| T.d-m2 | 27,830,376 | 8,321,579,488 | 45.46% | 93.84% |
| T.d-m3 | 20,303,095 | 6,062,394,898 | 45.53% | 94.06% |
| T.d-inf1 | 20,896,970 | 6,219,980,106 | 44.79% | 94.50% |
| T.d-inf2 | 24,190,792 | 7,222,586,666 | 44.97% | 93.64% |
| T.d-inf3 | 22,400,353 | 6,685,760,184 | 45.50% | 94.1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Xuan, L.; Chen, H.; Yu, C.; Chong, X.; Yin, Y.; Lu, X. Comparative Transcriptome Profiling of Resistant and Susceptible Taxodium Trees in Responding to the Infection by Pestalotiopsis maculans. Forests 2021, 12, 1090. https://doi.org/10.3390/f12081090
Zhang F, Xuan L, Chen H, Yu C, Chong X, Yin Y, Lu X. Comparative Transcriptome Profiling of Resistant and Susceptible Taxodium Trees in Responding to the Infection by Pestalotiopsis maculans. Forests. 2021; 12(8):1090. https://doi.org/10.3390/f12081090
Chicago/Turabian StyleZhang, Fan, Lei Xuan, Hong Chen, Chaoguang Yu, Xinran Chong, Yunlong Yin, and Xiaoqing Lu. 2021. "Comparative Transcriptome Profiling of Resistant and Susceptible Taxodium Trees in Responding to the Infection by Pestalotiopsis maculans" Forests 12, no. 8: 1090. https://doi.org/10.3390/f12081090
APA StyleZhang, F., Xuan, L., Chen, H., Yu, C., Chong, X., Yin, Y., & Lu, X. (2021). Comparative Transcriptome Profiling of Resistant and Susceptible Taxodium Trees in Responding to the Infection by Pestalotiopsis maculans. Forests, 12(8), 1090. https://doi.org/10.3390/f12081090




