Machine Learning-Based Species Classification Methods Using DART-TOF-MS Data for Five Coniferous Wood Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Wood Materials
2.2. DART-TOF-MS Conditions
2.3. Data Preprocessing
2.4. Modeling for Classification
- “n_estimators” means the number of trees in the forest;
- “max_depth” means the maximum depth of the trees;
- “min_samples_split” means the minimum number of samples required to split an internal node;
- “min_samples_leaf” means the minimum number of samples required to be at a leaf node;
- “max_features” means the number of features to consider when looking for the best split;
- “max_leaf_nodes” means the number of groups to be classified;
- “max_samples” means the number of samples to draw from the total data to train each base estimator.
- We optimized n_estimators as 1000 and max_depth as 10 and allowed the rest of the parameters to be set to default values by Scikit-learn (min_samples_split, 2; min_samples_leaf, 1; max_features, auto; max_leaf_nodes, none; max_samples, none).
3. Results
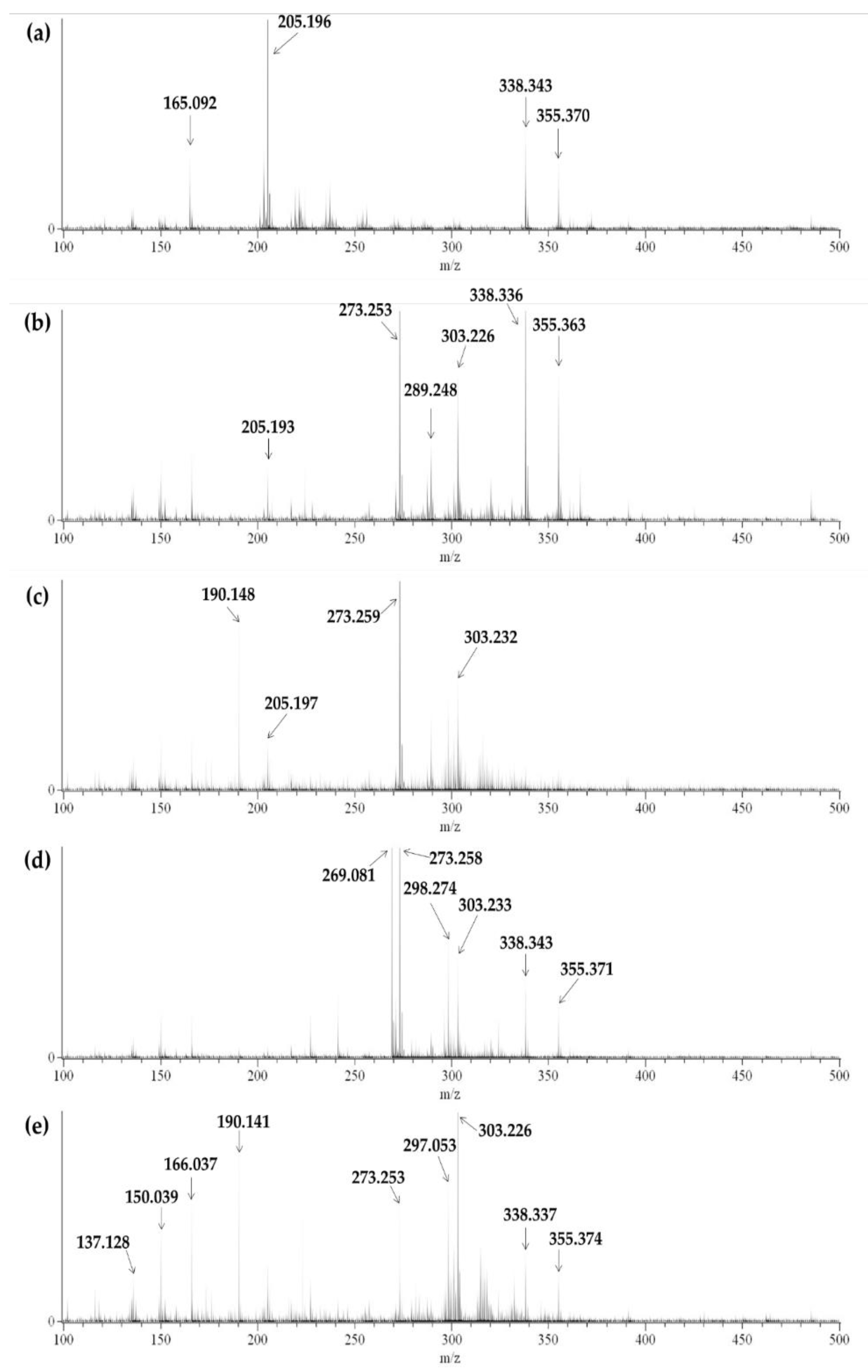
3.1. Figures, Tables, and Schemes
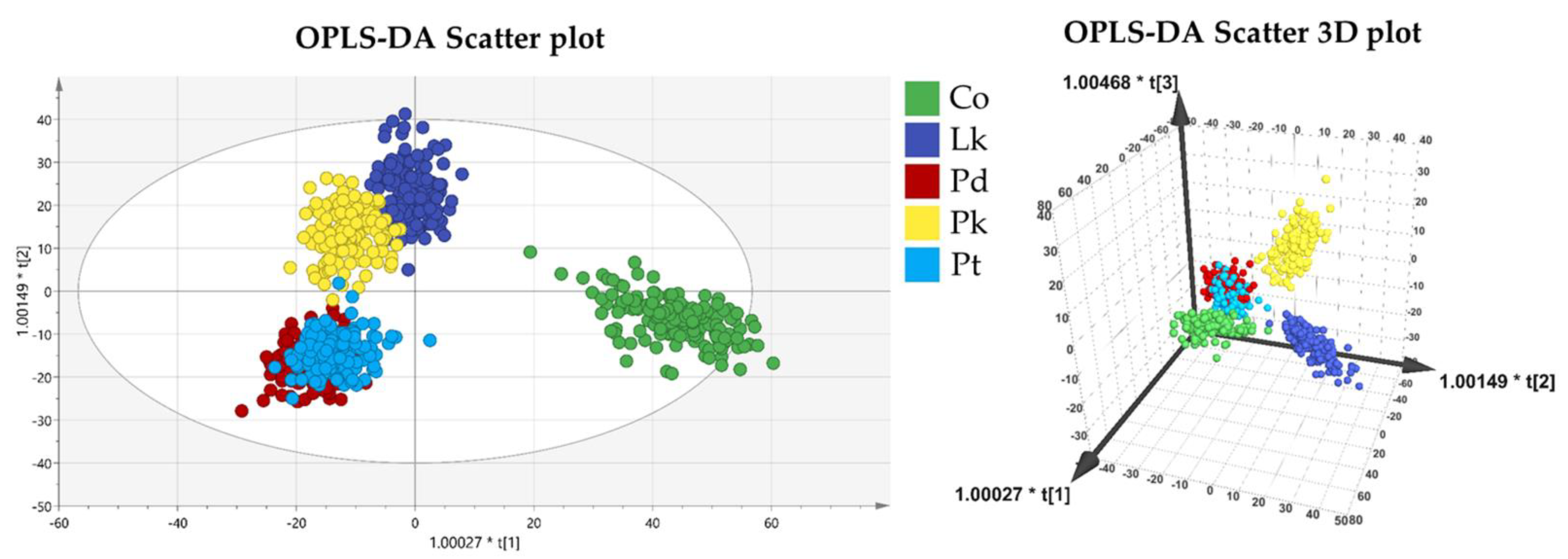
3.2. Multivariate Analysis
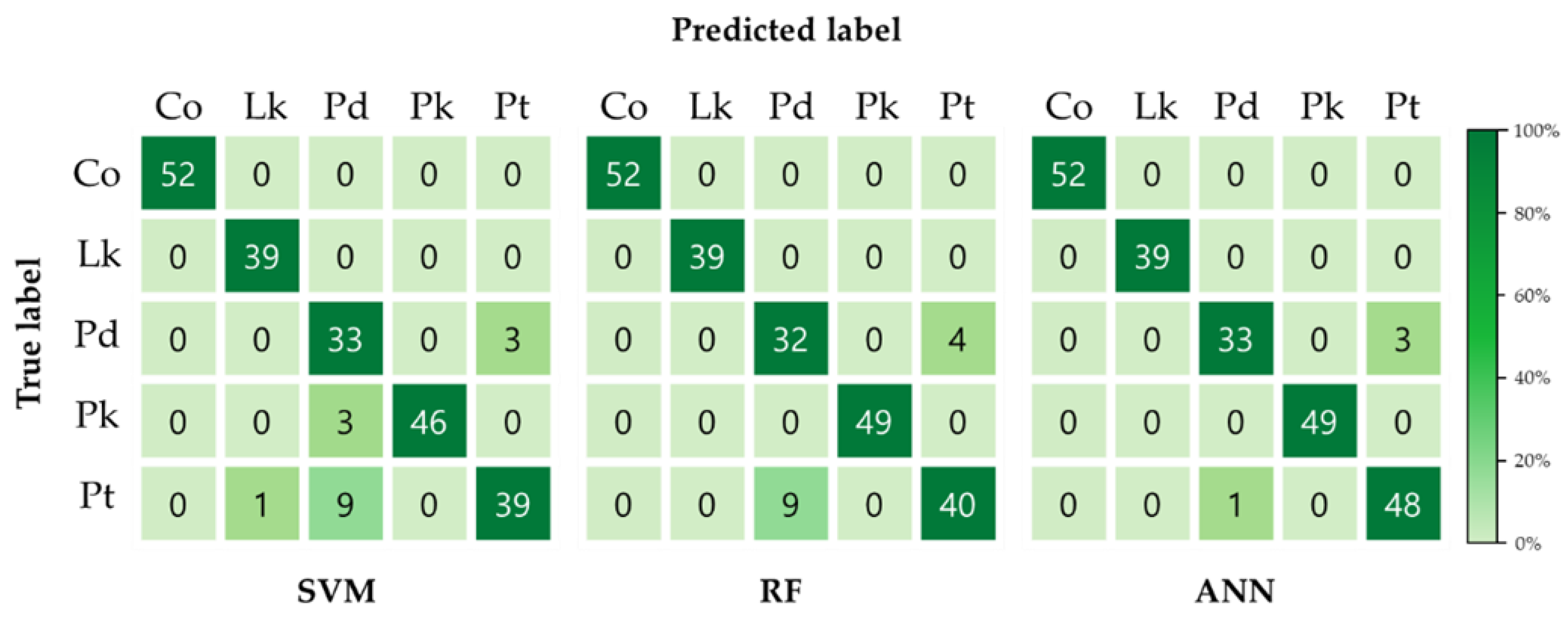
3.3. Classification Model Performance
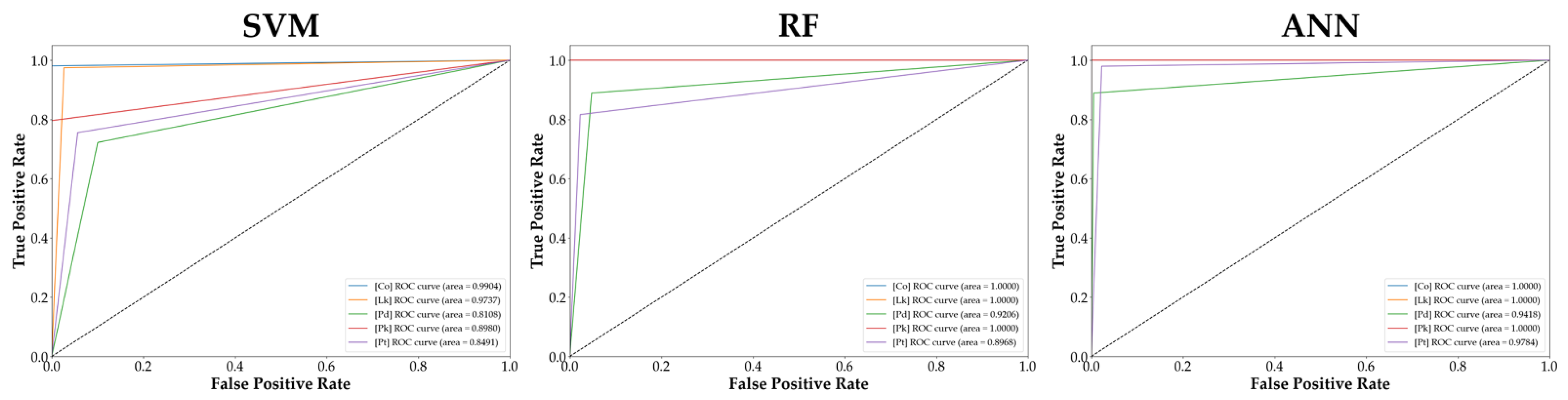
3.4. Evaluation of Model Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reboredo, F. Socio-economic, environmental, and governance impacts of illegal logging. Environ. Syst. Decis. 2013, 33, 295–304. [Google Scholar] [CrossRef]
- Dormontt, E.E.; Boner, M.; Braun, B.; Breulmann, G.; Degen, B.; Espinoza, E.; Gardner, S.; Guillery, P.; Hermanson, J.C.; Koch, G.; et al. Forensic timber identification: It’s time to integrate disciplines to combat illegal logging. Biol. Conserv. 2015, 191, 790–798. [Google Scholar] [CrossRef]
- Schmitz, N.; Boner, M.; Cervera, M.T.; Chavesta, M.; Cronn, R.; Degen, B.; Deklerck, V.; Diaz-Sala, C.; Dormontt, E.; Ekué, M.; et al. General sampling guide for timber tracking: How to collect reference samples for timber identification. General sampling guide for timber tracking: How to collect reference samples for timber identification. Glob. Timber Track. Netw. GTTN Secr. Eur. For. Inst. Thuenen Inst. 2019, 43, 1–43. [Google Scholar]
- Schmitz, N.; Beeckman, H.; Blanc-Jolivet, C.; Boeschoten, L.; Braga, J.; Cabezas, J.A.; Chaix, G.; Crameri, S.; Deklerck, V.; Degen, B.; et al. Overview of current practices in data analysis for wood identification. A guide for the different timber tracking methods. Glob. Timber Track. Netw. GTTN Secr. Eur. For. Inst. Thuenen Inst. 2020. [Google Scholar]
- Jozsa, L.A.; Middleton, G.R. A Discussion of Wood Quality Attributes and Their Practical Implications; Forintek Canada Corporation Vancouver: Vancouver, BC, Canada, 1994. [Google Scholar]
- Schweingruber, F.H. Trees and Wood in Dendrochronology: Morphological, Anatomical, and Tree-Ring Analytical Characteristics of Trees Frequently Used in Dendrochronology; Springer Science & Business Media: Berlin/Heidelberg, Germary, 2012. [Google Scholar]
- Lowe, A.J.; Cross, H.B. The Applicat ion of DNA methods to Timber Tracking and Origin Verificat ion. IAWA J. 2011, 32, 251–262. [Google Scholar] [CrossRef]
- Wu, F.; Gazo, R.; Haviarova, E.; Benes, B. Wood identification based on longitudinal section images by using deep learning. Wood Sci. Technol. 2021, 55, 553–563. [Google Scholar] [CrossRef]
- Höltken, A.M.; Schröder, H.; Wischnewski, N.; Degen, B.; Magel, E.; Fladung, M. Development of DNA-based methods to identify CITES-protected timber species: A case study in the Meliaceae family. Holzforschung 2012, 66, 97–104. [Google Scholar] [CrossRef]
- Bächle, H.; Zimmer, B.; Wegener, G. Classification of thermally modified wood by FT-NIR spectroscopy and SIMCA. Wood Sci. Technol. 2012, 46, 1181–1192. [Google Scholar] [CrossRef]
- Nisgoski, S.; de Oliveira, A.A.; de Muñiz, G.I.B. Artificial neural network and SIMCA classification in some wood discrimination based on near-infrared spectra. Wood Sci. Technol. 2017, 51, 929–942. [Google Scholar] [CrossRef]
- Sohn, S.-I.; Oh, Y.-J.; Pandian, S.; Lee, Y.-H.; Zaukuu, J.-L.Z.; Kang, H.-J.; Ryu, T.-H.; Cho, W.-S.; Cho, Y.-S.; Shin, E.-K. Identification of Amaranthus Species Using Visible-Near-Infrared (Vis-NIR) Spectroscopy and Machine Learning Methods. Remote Sens. 2021, 13, 4149. [Google Scholar] [CrossRef]
- Cody, R.B.; Dane, A.J.; Dawson-Andoh, B.; Adedipe, E.O.; Nkansah, K. Rapid classification of White Oak (Quercus alba) and Northern Red Oak (Quercus rubra) by using pyrolysis direct analysis in real time (DART™) and time-of-flight mass spectrometry. J. Anal. Appl. Pyrolysis 2012, 95, 134–137. [Google Scholar] [CrossRef]
- Evans, P.D.; Mundo, I.A.; Wiemann, M.C.; Chavarria, G.D.; McClure, P.J.; Voin, D.; Espinoza, E.O. Identification of selected CITES-protected Araucariaceae using DART TOFMS. IAWA J. 2017, 38, 266–S3. [Google Scholar] [CrossRef]
- Espinoza, E.O.; Lancaster, C.A.; Kreitals, N.M.; Hata, M.; Cody, R.B.; Blanchette, R.A. Distinguishing wild from cultivated agarwood (Aquilaria spp.) using direct analysis in real time and time of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2014, 28, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, G.J.; Liu, B.; He, T.; Guo, J.; Jiang, X.; Yin, Y. Wood discrimination analyses of Pterocarpus tinctorius and endangered Pterocarpus santalinus using DART-FTICR-MS coupled with multivariate statistics. IAWA J. 2019, 40, 58–74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhao, G.; Guo, J.; Wiedenhoeft, A.C.; Liu, C.C.; Yin, Y. Timber species identification from chemical fingerprints using direct analysis in real time (DART) coupled to Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS): Comparison of wood samples subjected to different treatments. Holzforschung 2019, 73, 975–985. [Google Scholar] [CrossRef]
- Pierce, C.Y.; Barr, J.R.; Cody, R.B.; Massung, R.F.; Woolfitt, A.R.; Moura, H.; Thompson, H.A.; Fernandez, F.M. Ambient generation of fatty acid methyl ester ions from bacterial whole cells by direct analysis in real time (DART) mass spectrometry. Chem. Commun. 2006, 8, 807–809. [Google Scholar] [CrossRef]
- Kim, H.J.; Seo, Y.T.; Park, S.-I.; Jeong, S.H.; Kim, M.K.; Jang, Y.P. DART–TOF–MS based metabolomics study for the discrimination analysis of geographical origin of Angelica gigas roots collected from Korea and China. Metabolomics 2014, 11, 64–70. [Google Scholar] [CrossRef]
- Sisco, E.; Forbes, T.P. Forensic applications of DART-MS: A review of recent literature. Forensic Chem. 2020, 22, 100294. [Google Scholar] [CrossRef]
- Arora, M.; Zambrzycki, S.C.; Levy, J.M.; Esper, A.; Frediani, J.K.; Quave, C.L.; Fernández, F.M.; Kamaleswaran, R. Machine Learning Approaches to Identify Discriminative Signatures of Volatile Organic Compounds (VOCs) from Bacteria and Fungi Using SPME-DART-MS. Metabolites 2022, 12, 232. [Google Scholar] [CrossRef]
- Deklerck, V.; Finch, K.; Gasson, P.; Bulcke, J.V.D.; Van Acker, J.; Beeckman, H.; Espinoza, E. Comparison of species classification models of mass spectrometry data: Kernel Discriminant Analysis vs Random Forest; A case study of Afrormosia (Pericopsis elata(Harms) Meeuwen). Rapid Commun. Mass Spectrom. 2017, 31, 1582–1588. [Google Scholar] [CrossRef] [Green Version]
- Deklerck, V.; Mortier, T.; Goeders, N.; Cody, R.B.; Waegeman, W.; Espinoza, E.; Van Acker, J.; Bulcke, J.V.D.; Beeckman, H. A protocol for automated timber species identification using metabolome profiling. Wood Sci. Technol. 2019, 53, 953–965. [Google Scholar] [CrossRef]
- Finch, K.; Espinoza, E.; Jones, F.A.; Cronn, R. Source Identification of Western Oregon Douglas-Fir Wood Cores Using Mass Spectrometry and Random Forest Classification. Appl. Plant Sci. 2017, 5, 1600158. [Google Scholar] [CrossRef] [PubMed]
- Pavlovich, M.J.; Dunn, E.E.; Hall, A.B. Chemometric brand differentiation of commercial spices using direct analysis in real time mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 1123–1130. [Google Scholar] [CrossRef]
- Samuel, A.L. Some Studies in Machine Learning Using the Game of Checkers. II-Recent Progress. In Computer Games I; Levi, D.N.L., Ed.; Springer: New York, NY, USA, 1988; pp. 366–400. [Google Scholar] [CrossRef]
- Salem, H.; Kabeel, A.; El-Said, E.M.; Elzeki, O.M. Predictive modelling for solar power-driven hybrid desalination system using artificial neural network regression with Adam optimization. Desalination 2021, 522, 115411. [Google Scholar] [CrossRef]
- Kowsher, M.; Hossen, I.; Tahabilder, A.; Prottasha, N.J.; Habib, K.; Azmi, Z.R.M. Support Directional Shifting Vector: A Direction Based Machine Learning Classifier. Emerg. Sci. J. 2021, 5, 700–713. [Google Scholar] [CrossRef]
- Cord, M.; Cunningham, P. Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Barlow, H.B. Unsupervised learning. Neural Comput. 1989, 1, 295–311. [Google Scholar] [CrossRef]
- Wongpoo, T.; Sriwan, W.; Titijaroonroj, T.; Jamsri, P. Chertify: Wood Identification-Based Mobile Cross-platform by Deep Learning Technique. In International Conference on Computing and Information Technology; Springer: Cham, Switzerland, 2022; pp. 77–87. [Google Scholar] [CrossRef]
- Liu, S.; He, T.; Wang, J.; Chen, J.; Guo, J.; Jiang, X.; Wiedenhoeft, A.C.; Yin, Y. Can quantitative wood anatomy data coupled with machine learning analysis discriminate CITES species from their look-alikes? Wood Sci. Technol. 2022, 56, 1567–1583. [Google Scholar] [CrossRef]
- Nag, A.; Gerritsen, A.; Doeppke, C.; Harman-Ware, A. Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data. Int. J. Mol. Sci. 2021, 22, 4107. [Google Scholar] [CrossRef]
- Silvello, G.C.; Bortoletto, A.M.; de Castro, M.C.; Alcarde, A.R. New approach for barrel-aged distillates classification based on maturation level and machine learning: A study of cachaça. LWT 2021, 140, 110836. [Google Scholar] [CrossRef]
- He, T.; Jiao, L.; Wiedenhoeft, A.C.; Yin, Y. Machine learning approaches outperform distance- and tree-based methods for DNA barcoding of Pterocarpus wood. Planta 2019, 249, 1617–1625. [Google Scholar] [CrossRef]
- He, T.; Jiao, L.; Yu, M.; Guo, J.; Jiang, X.; Yin, Y. DNA barcoding authentication for the wood of eight endangered Dalbergia timber species using machine learning approaches. Holzforschung 2018, 73, 277–285. [Google Scholar] [CrossRef]
- Esteban, L.G.; De Palacios, P.; Conde, M.; Fernández, F.G.; Garcia, M.C.; González-Alonso, M. Application of artificial neural networks as a predictive method to differentiate the wood of Pinus sylvestris L. and Pinus nigra Arn subsp. salzmannii (Dunal) Franco. Wood Sci. Technol. 2017, 51, 1249–1258. [Google Scholar] [CrossRef]
- Chen, J.; Li, G. Prediction of moisture content of wood using Modified Random Frog and Vis-NIR hyperspectral imaging. Infrared Phys. Technol. 2020, 105, 103225. [Google Scholar] [CrossRef]
- Ozsahin, S.; Murat, M. Prediction of equilibrium moisture content and specific gravity of heat treated wood by artificial neural networks. Eur. J. Wood Wood Prod. 2017, 76, 563–572. [Google Scholar] [CrossRef]
- Xi, B.; Gu, H.; Baniasadi, H.; Raftery, D. Statistical Analysis and Modeling of Mass Spectrometry-Based Metabolomics Data. In Mass Spectrometry in Metabolomics; Humana Press: New York, NY, USA, 2014; Volume 1198, pp. 333–353. [Google Scholar] [CrossRef] [Green Version]
- El Margae, S.; Sanae, B.; Mounir, A.K.; Youssef, F. Traffic sign recognition based on multi-block LBP features using SVM with normalization. In Proceedings of the 2014 9th International Conference on Intelligent Systems: Theories and Applications (SITA-14), Rabat, Morocco, 7–8 May 2014. [Google Scholar]
- Amarappa, S.; Sathyanarayana, S. Data classification using Support Vector Machine (SVM), a simplified approach. Int. J. Electron. Comput. Sci. Eng. 2014, 3, 435–445. [Google Scholar]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hsu, C.-W.; Lin, C.-J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Karatzoglou, A.; Meyer, D.; Hornik, K. Support vector machines in R. J. Stat. Softw. 2006, 15, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rumelhart, E.D.; McClelland, J.L. PDP Research Group. In Parallel Distributed Processing; IEEE: New York, NY, USA, 1998; Volume 1. [Google Scholar]
- Svozil, D.; Kvasnicka, V.; Pospichal, J. Introduction to multi-layer feed-forward neural networks. Chemom. Intell. Lab. Syst. 1997, 39, 43–62. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corsaro, C.; Vasi, S.; Neri, F.; Mezzasalma, A.M.; Neri, G.; Fazio, E. NMR in Metabolomics: From Conventional Statistics to Machine Learning and Neural Network Approaches. Appl. Sci. 2022, 12, 2824. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Cha, J. Partial least squares. Adv. Methods Mark. Res. 1994, 407, 52–78. [Google Scholar]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Mahadevan, S.; Shah, S.L.; Marrie, T.J.; Slupsky, C.M. Analysis of Metabolomic Data Using Support Vector Machines. Anal. Chem. 2008, 80, 7562–7570. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ling, C. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar] [CrossRef]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for multi-class classification: An overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Ranawana, R.; Palade, V. Optimized Precision—A New Measure for Classifier Performance Evaluation. In Proceedings of the 2006 IEEE International Conference on Evolutionary Computation, Vancouver, BC, Canada, 16–21 July 2006; pp. 2254–2261. [Google Scholar] [CrossRef]
- Hanley, J.A. Receiver operating characteristic (ROC) methodology: The state of the art. Crit. Rev. Comput. Tomogr. 1989, 29, 307. [Google Scholar]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zobel, B.J.; Van Buijtenen, J.P. Wood Variation: Its Causes and Control; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Hong, J.K.; Yang, J.C.; Lee, Y.M.; Kim, J.H. Molecular phylogenetic study of Pinus in Korea based on chloroplast DNA psbA-trnH and atpF-H sequences data. Korean J. Plant Taxon. 2014, 44, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Omer, G.; Mutanga, O.; Abdel-Rahman, E.M.; Adam, E. Performance of Support Vector Machines and Artificial Neural Network for Mapping Endangered Tree Species Using WorldView-2 Data in Dukuduku Forest, South Africa. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 4825–4840. [Google Scholar] [CrossRef]
- Cho, M.A.; Malahlela, O.; Ramoelo, A. Assessing the utility WorldView-2 imagery for tree species mapping in South African subtropical humid forest and the conservation implications: Dukuduku forest patch as case study. Int. J. Appl. Earth Obs. Geoinformation ITC J. 2015, 38, 349–357. [Google Scholar] [CrossRef]
- Karlson, M.; Ostwald, M.; Reese, H.; Bazié, H.R.; Tankoano, B. Assessing the potential of multi-seasonal WorldView-2 imagery for mapping West African agroforestry tree species. Int. J. Appl. Earth Obs. Geoinformation ITC J. 2016, 50, 80–88. [Google Scholar] [CrossRef]
- Okada, T.; Afendi, F.M.; Amin, A.U.; Takahashi, H.; Nakamura, K.; Kanaya, S. Metabolomics of medicinal plants: The importance of multivariate analysis of analytical chemistry data. Curr. Comput. Aided-Drug Des. 2010, 6, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.C.; Acunha, T.D.; Perin, E.C.; Rombaldi, C.V.; Galli, V.; Chaves, F.C. Untargeted metabolomics of strawberry (Fragaria x ananassa ‘Camarosa’) fruit from plants grown under osmotic stress conditions. J. Sci. Food Agric. 2019, 99, 6973–6980. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oh, D.G.; Singh, D.; Lee, H.J.; Kim, G.R.; Lee, S.; Lee, J.S.; Lee, C.H. Untargeted Metabolomics toward systematic characterization of antioxidant compounds in Betulaceae family plant extracts. Metabolites 2019, 9, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Cova, M.; Tauler, R.; Jaumot, J. Adverse Effects of Arsenic Uptake in Rice Metabolome and Lipidome Revealed by Untargeted Liquid Chromatography Coupled to Mass Spectrometry (LC-MS) and Regions of Interest Multivariate Curve Resolution. Separations 2022, 9, 79. [Google Scholar] [CrossRef]


| Model | Train Set (70%) | Test Set (30%) | New Test Set | |||||
|---|---|---|---|---|---|---|---|---|
| Total | Co | Lk | Pd | Pk | Pt | |||
| Support Vector Machine (SVM) | 98.48% | 92.89% | 52/52 | 39/39 | 33/36 | 46/49 | 39/49 | 90.22% |
| 100% | 100% | 91.67% | 93.88% | 79.59% | ||||
| Random Forest (RF) | 99.81% | 94.22% | 52/52 | 39/39 | 32/36 | 49/49 | 40/49 | 93.33% |
| 100% | 100% | 88.89% | 100% | 81.63% | ||||
| Artificial Neural Network (ANN) | 100% | 98.22% | 52/52 | 39/39 | 33/36 | 49/49 | 48/49 | 98.67% |
| 100% | 100% | 91.67% | 100% | 97.96% | ||||
| Model | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|
| Support Vector Machine (SVM) | 0.929 | 0.927 | 0.930 | 0.929 | 0.904 |
| Random Forest (RF) | 0.942 | 0.938 | 0.941 | 0.939 | 0.964 |
| Artificial Neural Network (ANN) | 0.982 | 0.982 | 0.979 | 0.981 | 0.984 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, G.; Lee, Y.-G.; Yoon, Y.-S.; Ahn, J.-Y.; Lee, J.-W.; Jang, Y.-P. Machine Learning-Based Species Classification Methods Using DART-TOF-MS Data for Five Coniferous Wood Species. Forests 2022, 13, 1688. https://doi.org/10.3390/f13101688
Park G, Lee Y-G, Yoon Y-S, Ahn J-Y, Lee J-W, Jang Y-P. Machine Learning-Based Species Classification Methods Using DART-TOF-MS Data for Five Coniferous Wood Species. Forests. 2022; 13(10):1688. https://doi.org/10.3390/f13101688
Chicago/Turabian StylePark, Geonha, Yun-Gyo Lee, Ye-Seul Yoon, Ji-Young Ahn, Jei-Wan Lee, and Young-Pyo Jang. 2022. "Machine Learning-Based Species Classification Methods Using DART-TOF-MS Data for Five Coniferous Wood Species" Forests 13, no. 10: 1688. https://doi.org/10.3390/f13101688
APA StylePark, G., Lee, Y.-G., Yoon, Y.-S., Ahn, J.-Y., Lee, J.-W., & Jang, Y.-P. (2022). Machine Learning-Based Species Classification Methods Using DART-TOF-MS Data for Five Coniferous Wood Species. Forests, 13(10), 1688. https://doi.org/10.3390/f13101688







