Genetic Parameters of Diameter Growth Dynamics in Norway Spruce Clones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Material
2.2. Modelling Approach
2.3. Genetic Parameters
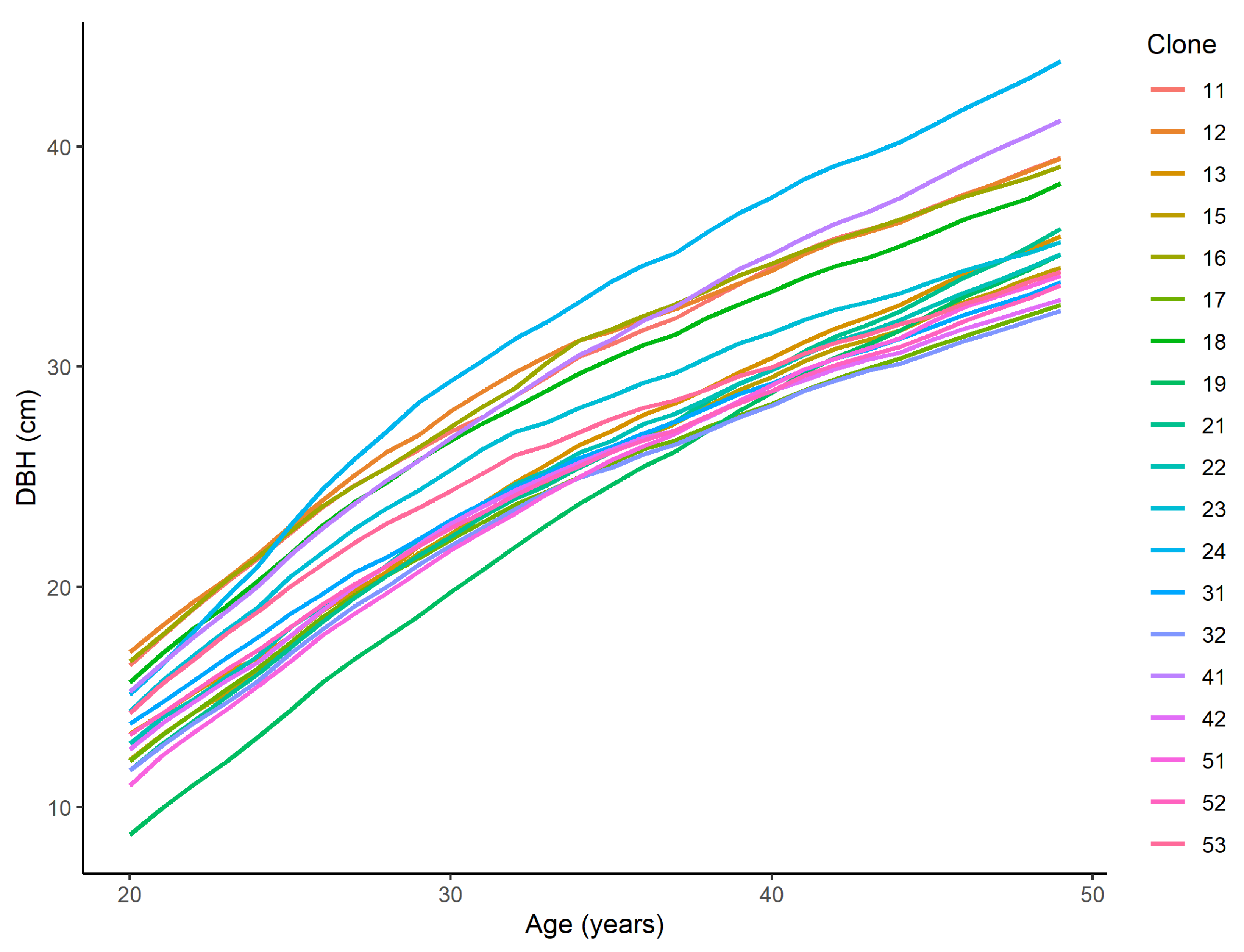
3. Results
3.1. The Growth Model
3.2. Dynamics of Genetic Parameters
4. Discussion
4.1. Dynamics of Clone-Specific Diameter Growth and Its Genetic Parameters over Time
4.2. Age–Age Genotypic Correlations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ruotsalainen, S. Increased forest production through forest tree breeding. Scand. J. For. Res. 2014, 29, 333–344. [Google Scholar] [CrossRef]
- Jansons, Ā.; Donis, J.; Danusevičius, D.; Baumanis, I. Differential analysis for next breeding cycle for Norway spruce in Latvia. Balt. For. 2015, 21, 285–297. [Google Scholar]
- Jansson, G.; Hansen, J.K.; Haapanen, M.; Kvaalen, H.; Steffenrem, A. The genetic and economic gains from forest tree breeding programmes in Scandinavia and Finland. Scand. J. For. Res. 2017, 32, 273–286. [Google Scholar] [CrossRef]
- Ahtikoski, A.; Ahtikoski, R.; Haapanen, M.; Hynynen, J.; Kärkkäinen, K. Economic performance of genetically improved reforestation material in joint production of timber and carbon sequestration: A case study from Finland. Forests 2020, 11, 847. [Google Scholar] [CrossRef]
- Haapanen, M. Performance of genetically improved Norway spruce in one-third rotation-aged progeny trials in southern Finland. Scand. J. For. Res. 2020, 35, 221–226. [Google Scholar] [CrossRef]
- Katrevičs, J.; Džeriņa, B.; Neimane, U.; Desaine, I.; Bigača, Z. Jansons Production and profitability of low density Norway spruce (Picea abies (L.) Karst.) plantation at 50 years of age: Case study from eastern Latvia. Agron. Res. 2018, 16, 113–121. [Google Scholar] [CrossRef]
- Pfister, O.; Wallentin, C.; Nilsson, U.; Ekö, P.M. Effects of wide spacing and thinning strategies on wood quality in Norway spruce (Picea abies) stands in southern Sweden. Scand. J. For. Res. 2007, 22, 333–343. [Google Scholar] [CrossRef]
- Routa, J.; Kilpeläinen, A.; Ikonen, V.-P.; Asikainen, A.; Venäläinen, A.; Peltola, H. Effects of intensified silviculture on timber production and its economic profitability in boreal Norway spruce and Scots pine stands under changing climatic conditions. For. Int. J. For. Res. 2019, 92, 648–658. [Google Scholar] [CrossRef] [Green Version]
- Arhipova, N.; Gaitnieks, T.; Donis, J.; Stenlid, J.; Vasaitis, R. Butt rot incidence, causal fungi, and related yield loss in Picea abies stands of Latvia. Can. J. For. Res. 2011, 41, 2337–2345. [Google Scholar] [CrossRef]
- Allikmäe, E.; Laarmann, D.; Korjus, H. Vitality assessment of visually healthy trees in Estonia. Forests 2017, 8, 223. [Google Scholar] [CrossRef] [Green Version]
- Donis, J.; Kitenberga, M.; Šņepsts, G.; Dubrovskis, E.; Jansons, Ā. Factors affecting windstorm damage at the stand level in hemiboreal forests in Latvia: Case study of 2005 winter storm. Silva Fenn. 2018, 52, 10009. [Google Scholar] [CrossRef] [Green Version]
- Zeltiņš, P.; Katrevičs, J.; Gailis, A.; Maaten, T.; Bāders, E.; Jansons, Ā.; Zeltiņš, P.; Katrevičs, J.; Gailis, A.; Maaten, T.; et al. Effect of Stem Diameter, Genetics, and Wood Properties on Stem Cracking in Norway Spruce. Forests 2018, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- de Groot, M.; Diaci, J.; Ogris, N. Forest management history is an important factor in bark beetle outbreaks: Lessons for the future. For. Ecol. Manag. 2019, 433, 467–474. [Google Scholar] [CrossRef]
- Rosvall, O.; Jansson, G.; Andersson, B. Predicted genetic gain from existing and future seed orchards and clone mixes in Sweden. In Proceedings of the Integrating Tree Breeding and Forestry—Proceedings of the Nordic Group for Management of Genetic Resources of Trees, Mekrijärvi, Finland, 23–27 March 2001; Haapanen, M., Mikola, J., Eds.; Finnish Forest Research Institute: Vantaa, Finland, 2002; Volume 1, pp. 23–27. [Google Scholar]
- Mullin, T.J.; Morgenstern, E.K.; Park, Y.S.; Fowler, D.P. Genetic parameters from a clonally replicated test of black spruce (Picea mariana). Can. J. For. Res. 1992, 22, 24–36. [Google Scholar] [CrossRef]
- Wu, H.X. Benefits and risks of using clones in forestry—A review. Scand. J. For. Res. 2019, 34, 352–359. [Google Scholar] [CrossRef]
- Chen, Z.Q.; Hai, H.N.T.; Helmersson, A.; Liziniewicz, M.; Hallingbäck, H.R.; Fries, A.; Berlin, M.; Wu, H.X. Advantage of clonal deployment in Norway spruce (Picea abies (L.) H. Karst.). Ann. For. Sci. 2020, 77, 14. [Google Scholar] [CrossRef] [Green Version]
- Rosvall, O. Using Norway spruce clones in Swedish forestry: General overview and concepts. Scand. J. For. Res. 2019, 34, 336–341. [Google Scholar] [CrossRef]
- Liziniewicz, M.; Berlin, M. Differences in growth and areal production between Norway spruce (Picea abies L. Karst.) regeneration material representing different levels of genetic improvement. For. Ecol. Manag. 2019, 435, 158–169. [Google Scholar] [CrossRef]
- Rosvall, O. Using Norway spruce clones in Swedish forestry: Swedish forest conditions, tree breeding program and experiences with clones in field trials. Scand. J. For. Res. 2019, 34, 342–351. [Google Scholar] [CrossRef]
- Sabatia, C.O. Stand Dynamics, Growth, and Yield of Genetically Enhanced Loblolly Pine (Pinus taeda L.). Ph.D. Thesis, Virginia Tech, Blacksburg, VA, USA, 2011. Volume 1. [Google Scholar]
- Egbäck, S.; Nilsson, U.; Nyström, K.; Högberg, K.-A.; Fahlvik, N. Modeling early height growth in trials of genetically improved Norway spruce and Scots pine in southern Sweden. Silva Fenn. 2017, 51, 5662. [Google Scholar] [CrossRef] [Green Version]
- Fahlvik, N.; Nyström, K.; Nystrom, K. Models for predicting individual tree height increment and tree diameter in young stands in southern Sweden. Scand. J. For. Res. 2006, 21, 16–28. [Google Scholar] [CrossRef]
- Ahtikoski, A.; Ojansuu, R.; Haapanen, M.; Hynynen, J.; Kärkkäinen, K. Financial performance of using genetically improved regeneration material of Scots pine (Pinus sylvestris L.) in Finland. New For. 2012, 43, 335–348. [Google Scholar] [CrossRef]
- Bravo, F.; Fabrika, M.; Ammer, C.; Barreiro, S.; Bielak, K.; Coll, L.; Fonseca, T.; Kangur, A.; Löf, M.; Merganičová, K.; et al. Modelling approaches for mixed forests dynamics prognosis. Research gaps and opportunities. For. Syst. 2019, 28, eR002. [Google Scholar] [CrossRef] [Green Version]
- Gould, P.J.; Marshall, D.D. Estimation and Application of Genetic- Gain Multipliers for Douglas-Fir Height and Diameter Growth. For. Sci. 2008, 54, 588–596. [Google Scholar]
- Rehfeldt, G.E.; Wykoff, W.R.; Hoff, R.J.; Steinhoff, R.J. Genetic gains in growth and simulated yield of Pinus monticola. For. Sci. 1991, 37, 326–342. [Google Scholar]
- Carson, S.D.; Garcia, O.; Hayes, J.D. Realized gain and prediction of yield with genetically improved Pinus radiata in New Zealand. For. Sci. 1999, 45, 186–200. [Google Scholar]
- Buford, M.A.; Burkhart, H.E. Genetic improvement effects on growth and yield of loblolly pine plantations. For. Sci. 1987, 33, 707–724. [Google Scholar] [CrossRef]
- Newton, P.F. Systematic review of yield responses of four North American conifers to forest tree improvement practices. For. Ecol. Manag. 2003, 172, 29–51. [Google Scholar] [CrossRef]
- Vergara, R.; White, T.L.; Huber, D.A.; Shiver, B.D.; Rockwood, D.L. Estimated realized gains for first-generation slash pine (Pinus elliottii var. elliottii) tree improvement in the southeastern United States. Can. J. For. Res. 2004, 34, 2587–2600. [Google Scholar] [CrossRef]
- Adams, J.P.; Matney, T.G.; Land, S.B.; Belli, K.L.; Duzan, H.W. Incorporating genetic parameters into a loblolly pine growth-and-yield model. Can. J. For. Res. 2006, 36, 1959–1967. [Google Scholar] [CrossRef]
- Adame, P.; Hynynen, J.; Cañellas, I.; del Río, M. Individual-tree diameter growth model for rebollo oak (Quercus pyrenaica Willd.) coppices. For. Ecol. Manag. 2008, 255, 1011–1022. [Google Scholar] [CrossRef]
- Subedi, N.; Sharma, M. Individual-tree diameter growth models for black spruce and jack pine plantations in northern Ontario. For. Ecol. Manag. 2011, 261, 2140–2148. [Google Scholar] [CrossRef]
- Timilsina, N.; Staudhammer, C.L. Individual tree-based diameter growth model of slash pine in Florida using nonlinear mixed modeling. For. Sci. 2013, 59, 27–37. [Google Scholar] [CrossRef]
- Xu, H.; Sun, Y.; Wang, X.; Fu, Y.; Dong, Y.; Li, Y. Nonlinear mixed-effects (NLME) diameter growth models for individual China-fir (Cunninghamia lanceolata) trees in southeast China. PLoS ONE 2014, 9, e104012. [Google Scholar] [CrossRef]
- Anderson, B.D.; Russell, M.B.; Windmuller-Campione, M.A.; Palik, B.J.; Kastendick, D.N. Development and evaluation of black spruce (Picea mariana (Miller) B.S.P.) Diameter increment models across silvicultural treatments in northern Minnesota, USA. Forests 2018, 9, 747. [Google Scholar] [CrossRef] [Green Version]
- Hallingbäck, H.R.; Högberg, K.A.; Säll, H.; Lindeberg, J.; Johansson, M.; Jansson, G. Optimal timing of early genetic selection for sawn timber traits in Picea abies. Eur. J. For. Res. 2018, 137, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Liziniewicz, M.; Berlin, M.; Karlsson, B. Early assessments are reliable indicators for future volume production in Norway spruce (Picea abies L. Karst.) genetic field trials. For. Ecol. Manag. 2018, 411, 75–81. [Google Scholar] [CrossRef]
- Rosvall, O.; Bradshaw, R.H.W.; Egertsdotter, U.; Ingvarsson, P.K.; Mullin, T.J.; Wu, H. Using Norway spruce clones in Swedish forestry: Implications of clones for management. Scand. J. For. Res. 2019, 34, 390–404. [Google Scholar] [CrossRef]
- White, T.L.; Adams, W.T.; Neale, D.B. Forest Genetics; CABI Publishing: Wallingford, UK, 2007; ISBN 9780851990835. [Google Scholar]
- Buss, K. Forest ecosystem classification in Latvia. Proc. Latv. Acad. Sci. Sect. B 1997, 51, 204–218. [Google Scholar]
- Harris, I.; Osborn, T.J.; Jones, P.; Lister, D. Version 4 of the CRU TS monthly high-resolution gridded multivariate climate dataset. Sci. Data 2020, 7, 109. [Google Scholar] [CrossRef] [Green Version]
- Spiecker, H.; Hansen, N.; Schinker, M.G. High-Frequency Densitometry-A New Method for the Rapid Evaluation of Wood Density Variations. IAWA J. 2003, 24, 231–239. [Google Scholar] [CrossRef]
- Holmes, R. Computer-Assisted Quality Control in Tree-Ring Dating and Measurement. Tree-Ring Bull. 1983, 43, 51–67. [Google Scholar]
- Rohner, B.; Bugmann, H.; Bigler, C. Estimating the age-diameter relationship of oak species in Switzerland using nonlinear mixed-effects models. Eur. J. For. Res. 2013, 132, 751–764. [Google Scholar] [CrossRef] [Green Version]
- Richards, F.J. A flexible growth function for empirical use. J. Exp. Bot. 1959, 10, 290–301. [Google Scholar] [CrossRef]
- Bolker, B.M.; Gardner, B.; Maunder, M.; Berg, C.W.; Brooks, M.; Comita, L.; Crone, E.; Cubaynes, S.; Davies, T.; de Valpine, P.; et al. Strategies for fitting nonlinear ecological models in R, AD Model Builder, and BUGS. Methods Ecol. Evol. 2013, 4, 501–512. [Google Scholar] [CrossRef] [Green Version]
- Zeide, B. Analysis of Growth Equations. For. Sci. 1993, 39, 594–616. [Google Scholar] [CrossRef]
- Sharma, R.P.; Vacek, Z.; Vacek, S.; Jansa, V.; Kučera, M. Modelling individual tree diameter growth for Norway spruce in the Czech Republic using a generalized algebraic difference approach. J. For. Sci. 2017, 63, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, J.; Bates, D. Mixed-Effects Models in S and S-PLUS; Springer Science & Business Media: Cham, Switzerland, 2000. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. The R Core Team Nlme: Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-143; 2019. Available online: https://cran.r-project.org/web/packages/nlme/nlme.pdf (accessed on 21 March 2022).
- Montgomery, D.; Peck, E.; Vining, G. Introduction to Linear Regression Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Falconer, D.S.; Mackay, T.F. Introduction to Quantitative Genetics, 4th ed.; Longman Group Ltd.: London, UK, 1996. [Google Scholar]
- Savageau, M.A. Growth of complex systems can be related to the properties of their underlying determinants. Proc. Natl. Acad. Sci. USA 1979, 76, 5413–5417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivancich, H.; Martínez Pastur, G.J.; Lencinas, M.V.; Cellini, J.M.; Peri, P.L. Proposals for Nothofagus antarctica diameter growth estimation: Simple vs. global models. J. For. Sci. 2014, 60, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Meredith, M.P.; Stehman, S.V. Repeated measures experiments in forestry: Focus on analysis of response curves. Can. J. For. Res. 1991, 21, 957–965. [Google Scholar] [CrossRef]
- Fekedulegn, D.; Mac Siurtain, M.P.; Colbert, J.J. Parameter estimation of nonlinear growth models in forestry. Silva Fenn. 1999, 33, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Nothdurft, A.; Kublin, E.; Lappi, J. A non-linear hierarchical mixed model to describe tree height growth. Eur. J. For. Res. 2006, 125, 281–289. [Google Scholar] [CrossRef]
- Harrison, X.A.; Donaldson, L.; Correa-Cano, M.E.; Evans, J.; Fisher, D.N.; Goodwin, C.E.D.; Robinson, B.S.; Hodgson, D.J.; Inger, R. A brief introduction to mixed effects modelling and multi-model inference in ecology. PeerJ 2018, 2018, e4794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatia, C.O.; Burkhart, H.E. Modeling height development of loblolly pine genetic varieties. For. Sci. 2013, 59, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Knowe, S.A.; Foster, G.S. Application of Growth Models For Simulating Genetic Gain of Loblolly Pine. For. Sci. 1989, 35, 211–228. [Google Scholar]
- Nance, W.L.; Wells, O.O. Site index models for height growth of planted loblolly pine (Pinus taeda L.) seed sources. In Proceedings of the 16th Southern Forest Tree Improvement Conference, Blacksburg, VA, USA, 27–28 May 1981; 1981; pp. 86–96. [Google Scholar]
- Sprinz, P.T.; Talbert, C.B.; Strub, M.R. Height-age trends from an Arkansas seed source study. For. Sci. 1989, 35, 677–691. [Google Scholar] [CrossRef]
- Tang, S.; Meng, F.R.; Bourque, C.P.A. Analyzing parameters of growth and yield models for Chinese fir provenances with a linear mixed model approach. Silvae Genet. 2001, 50, 140–145. [Google Scholar]
- Der Chung, J.; Chien, C.T.; Nigh, G.; Ying, C.C. Genetic variation in growth curve parameters of Konishii fir (Cunninghamia lanceolata (Lamb.) Hook. var. konishii). Silvae Genet. 2009, 58, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Schielzeth, H.; Nakagawa, S. Nested by design: Model fitting and interpretation in a mixed model era. Methods Ecol. Evol. 2013, 4, 14–24. [Google Scholar] [CrossRef]
- Oddi, F.J.; Miguez, F.E.; Ghermandi, L.; Bianchi, L.O.; Garibaldi, L.A. A nonlinear mixed-effects modeling approach for ecological data: Using temporal dynamics of vegetation moisture as an example. Ecol. Evol. 2019, 9, 10225–10240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatia, C.O.; Burkhart, H.E. Height and Diameter Relationships and Distributions in Loblolly Pine Stands of Enhanced Genetic Material. For. Sci. 2013, 59, 278–289. [Google Scholar] [CrossRef]
- Hannrup, B.; Cahalan, C.; Chantre, G.; Grabner, M.; Karlsson, B.; Le Bayon, I.; Jones, G.L.; Müller, U.; Pereira, H.; Rodrigues, J.C.; et al. Genetic Parameters of Growth and Wood Quality Traits in Picea abies. Scand. J. For. Res. 2004, 19, 14–29. [Google Scholar] [CrossRef]
- Chen, Z.Q.; Gil, M.R.G.; Karlsson, B.; Lundqvist, S.O.; Olsson, L.; Wu, H.X. Inheritance of growth and solid wood quality traits in a large Norway spruce population tested at two locations in southern Sweden. Tree Genet. Genomes 2014, 10, 1291–1303. [Google Scholar] [CrossRef]
- Hong, Z.; Fries, A.; Wu, H.X. Age trend of heritability, genetic correlation, and efficiency of early selection for wood quality traits in Scots pine. Can. J. For. Res. 2015, 825, 817–825. [Google Scholar] [CrossRef]
- Wahid, N.; Rainville, A.; Lamhamedi, M.S.; Margolis, H.A.; Beaulieu, J.; Deblois, J. Genetic parameters and performance stability of white spruce somatic seedlings in clonal tests. For. Ecol. Manag. 2012, 270, 45–53. [Google Scholar] [CrossRef]
- Isik, K.; Kleinschmit, J.; Steiner, W. Age-age correlations and early selection for height in a clonal genetic test of Norway spruce. For. Sci. 2010, 56, 212–221. [Google Scholar] [CrossRef]
- Chen, Z.-Q. Quantitative Genetics of Norway Spruce in Sweden; Sveriges Lantbruksuniversitet: Uppsala, Sweden, 2016. [Google Scholar]
- Mihai, G.; Mirancea, I. Age trends in genetic parameters for growth and quality traits in Abies alba. IForest 2016, 9, 954–959. [Google Scholar] [CrossRef] [Green Version]
- Hiraoka, Y.; Miura, M.; Fukatsu, E.; Iki, T.; Yamanobe, T.; Kurita, M.; Isoda, K.; Kubota, M.; Takahashi, M. Time trends of genetic parameters and genetic gains and optimum selection age for growth traits in sugi (Cryptomeria japonica) based on progeny tests conducted throughout Japan. J. For. Res. 2019, 24, 303–312. [Google Scholar] [CrossRef]
- Jansson, G.; Kerr, R.; Dutkowski, G.; Kroon, J. Competition effects in breeding value prediction of forest trees. Can. J. For. Res. 2021, 51, 1002–1014. [Google Scholar] [CrossRef]
- Radtke, P.J.; Westfall, J.A.; Burkhart, H.E. Conditioning a distance-dependent competition index to indicate the onset of inter-tree competition. For. Ecol. Manag. 2003, 175, 17–30. [Google Scholar] [CrossRef]
- Amateis, R.L.; Burkhart, H.E. Rotation-age results from a loblolly pine spacing trial. South. J. Appl. For. 2012, 36, 11–18. [Google Scholar] [CrossRef]
- Franklin, E.C. Model relating levels of genetic variance to stand development of four north American conifers. Silvae Genet. 1979, 28, 207–212. [Google Scholar]
- Sato, Y.; Fukatsu, E.; Hiraoka, Y.; Watanabe, A.; Takahashi, M. The effect of genotype and planting density on the growth patterns and selection of local varieties of Sugi (Cryptomeria japonica). Nihon Ringakkai Shi/J. Jpn. For. Soc. 2016, 98, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Štícha, V.; Sharma, R.P.; Vacek, Z.; Vacek, S.; Nuhlíček, O. Timber and branch volume prediction: Effects of stand and site characteristics on dendromass and timber-to-branch volume ratio of Norway spruce in managed forests. Forests 2019, 10, 144. [Google Scholar] [CrossRef] [Green Version]
- Zubizarreta Gerendiain, A.; Peltola, H.; Pulkkinen, P.; Jaatinen, R.; Pappinen, A.; Kellomäki, S. Differences in growth and wood property traits in cloned Norway spruce (Picea abies). Can. J. For. Res. 2007, 37, 2600–2611. [Google Scholar] [CrossRef]
- Jayawickrama, K.J.; McKeand, S.E.; Jett, J.B. Rootstock effects on scion growth and reproduction in 8-year-old grafted loblolly pine. Can. J. For. Res. 1997, 27, 1781–1787. [Google Scholar] [CrossRef]
- Olesen, P.O. On cyclophysis and topophysis. Silvae Genet. 1978, 27, 173–178. [Google Scholar]
- Greenwood, M.S.; Hutchison, K.W. Maturation as a Developmental Process. In Clonal Forestry I; Springer: Berlin/Heidelberg, Germany, 1993; pp. 14–33. ISBN 978-3-642-84177-4. [Google Scholar]
- Viherä-Aarnio, A.; Ryynänen, L. Seed production of micropropagated plants, grafts and seedlings of birch in a seed orchard. Silva Fenn. 1994, 28, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Wendling, I.; Trueman, S.J.; Xavier, A. Maturation and related aspects in clonal forestry-Part I: Concepts, regulation and consequences of phase change. New For. 2014, 45, 449–471. [Google Scholar] [CrossRef]
- Cannell, M.; Sheppard, L.; Cahalan, C. C effects and second generation clone performance in Picea sitchensis and Pinus contorta. Silvae Genet. 1988, 37, 15–19. [Google Scholar]
- Huston, M.; DeAngelis, D.; Post, W. New Computer Models Unify Ecological Theory. Bioscience 1988, 38, 682–691. [Google Scholar] [CrossRef]
- Johnson, G.R.; Sniezko, R.A.; Mandel, N.L. Age trends in Douglas-fir genetic parameters and implications for optimum selection age. Silvae Genet. 1998, 46, 349–358. [Google Scholar]
- Kroon, J.; Ericsson, T.; Jansson, G.; Andersson, B. Patterns of genetic parameters for height in field genetic tests of Picea abies and Pinus sylvestris in Sweden. Tree Genet. Genomes 2011, 7, 1099–1111. [Google Scholar] [CrossRef]
- Zubizarreta-Gerendiain, A.; Gort-Oromi, J.; Mehtätalo, L.; Peltola, H.; Venäläinen, A.; Pulkkinen, P. Effects of cambial age, clone and climatic factors on ring width and ring density in Norway spruce (Picea abies) in southeastern Finland. For. Ecol. Manag. 2012, 263, 9–16. [Google Scholar] [CrossRef]
- Cieszewski, C.J.; Bailey, R.L. Generalized Algebraic Difference Approach: A New Methodology for Derivation of Biologically Based Dynamic Site Equations. For. Sci. 2000, 46, 116–126. [Google Scholar]
- Diéguez-Aranda, U.; Burkhart, H.E.; Amateis, R.L. Dynamic site model for loblolly pine (Pinus taeda L.) plantations in the United States. For. Sci. 2006, 52, 262–272. [Google Scholar] [CrossRef]




| Random Parameter in Model | Parameter Estimates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β10 | β20 | β30 | AIC | BIC | logLik | |||||
| β1 | 46.403 | 0.0466 | 1.610 | 16.417 | n/a | n/a | 24.253 | −73.140 | 96.806 | 61.57 |
| β1, β2 | 46.630 | 0.0472 | 1.640 | 25.116 | 3.53 × 10−5 | n/a | 23.509 | −184.040 | −0.498 | 119.02 |
| β1, β3 | 46.262 | 0.0471 | 1.650 | 15.141 | n/a | 0.0436 | 23.0467 | −153.329 | 30.213 | 103.66 |
| β1, β2, β3 | 46.991 | 0.0467 | 1.624 | 26.686 | 6.37 × 10−5 | 0.0371 | 21.678 | −189.152 | 14.783 | 124.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeltiņš, P.; Kangur, A.; Katrevičs, J.; Jansons, Ā. Genetic Parameters of Diameter Growth Dynamics in Norway Spruce Clones. Forests 2022, 13, 679. https://doi.org/10.3390/f13050679
Zeltiņš P, Kangur A, Katrevičs J, Jansons Ā. Genetic Parameters of Diameter Growth Dynamics in Norway Spruce Clones. Forests. 2022; 13(5):679. https://doi.org/10.3390/f13050679
Chicago/Turabian StyleZeltiņš, Pauls, Ahto Kangur, Juris Katrevičs, and Āris Jansons. 2022. "Genetic Parameters of Diameter Growth Dynamics in Norway Spruce Clones" Forests 13, no. 5: 679. https://doi.org/10.3390/f13050679







