Abstract
Dead wood is an important microsite for seedling regeneration in forest ecosystems. Although recent studies have found important associations between fungal wood decay type (white rot and brown rot) and both density and species composition of regenerating seedlings, its abiotic and biotic mechanisms are unknown. In the present study, pot experiments were conducted with the seedlings of two ectomycorrhizal tree species (Abies veitchii and Betula ermanii) and two arbuscular mycorrhizal tree species (Chamaecyparis obtusa and Cryptomeria japonica) to evaluate their growth using three substrates: brown rot wood, white rot wood, and soil. Results showed that the shoot growth of B. ermanii grown in white rot wood was greater than in other substrates, but this effect disappeared in sterilized substrates, suggesting some biotic positive effects occur in white rot wood. The seedling weights of Cr. japonica and Ch. obtusa were found to be greater in soil than in wood, and this may be partly attributable to the high mycorrhizal rate of their roots in soil. Colonization of arbuscular and ectomycorrhizal fungi was restricted to the seedlings in unsterilized soil. These results demonstrate the importance of the biological mechanisms affecting seedlings’ preferences for a variety of regeneration microsites and illustrate the need for future experiments to include larger sets of seedling species.
1. Introduction
Dead wood is an important microsite for seedling regeneration in forest ecosystems [1]. Several abiotic and biotic environmental conditions found in dead wood benefit seedlings of especially small-seeded tree species in boreal, subalpine, temperate, and tropical ecosystems [2]. For example, in boreal to temperate forests, the seedlings of the genera Picea, Abies, Tsuga, Cryptomeria, and Chamaecyparis among gymnosperms, and Betula, Clethra, and Sorbus among angiosperms, are often reported to regenerate on dead wood [1,3,4,5,6,7,8]. The growth is stimulated by several factors, including more sunlight, stable moisture levels, fewer pathogens, less litter cover, and less root competition, which allow the seedlings to grow and survive better on dead wood than on the ground soil [1,7]. Increasing the understanding of the factors that drive seedling establishment on dead wood and their mechanisms is essential for enabling the improved prediction of forest dynamics.
Recently, Fukasawa [2] reported that properties of dead wood associated with fungal species-specific wood decay activities, (i.e., decay type) were critical to determining seedling density and community composition on dead wood. Fungi, particularly in the class basidiomycetes, are strong decomposers of wood structural components, and their decay type is traditionally categorized into brown rot and white rot. This reflects different fungal species’ preferences for wood lignin and carbohydrates [9]. In brown rot, fungi decay only carbohydrates with little modification of lignin. In white rot, fungi decay both lignin and carbohydrates. It is quite typical that brown rot wood becomes blocky in texture, brown in color due to the accumulation of lignin, and acidic because of organic acid production by brown rot fungi [10]. In contrast, white rot wood becomes fiber-like in texture, and pale in color due to delignification. In addition to texture, pH, and nutrient content [11,12], fungal [13,14] and bacterial [15,16,17] communities differ between brown and white rot wood. However, it is unclear how and which of these properties affect seedling regeneration on dead wood.
A seedling’s mycorrhizal type is a critical factor in determining its preference and performance in regeneration sites, and affects their population dynamics [18,19,20]. In forest trees, the two important mycorrhizal types, arbuscular mycorrhiza and ectomycorrhiza, exhibit regeneration niche preferences among their seedlings in relation to mycorrhizal capacity for providing nutrients and protection from soil-borne diseases [19]. Fukasawa [2] reviewed the results of previous field studies on seedling regeneration on dead wood and found a pattern that seedling densities of the species associated with arbuscular mycorrhizal fungi were greater on brown rot logs than on white rot logs, whereas the densities of seedlings associated with ectomycorrhizal fungi were lower on brown rot logs than on white rot logs. Since ectomycorrhizal fungi have the ability to decompose organic matter [21], it is reasonable to consider that they colonize and utilize the white rot logs which have abundant delignified carbohydrates. In contrast, arbuscular mycorrhizal fungi are poor decomposers of organic matter and therefore cannot be dominant in environments with abundant available carbohydrates where strong decomposers dominate [22], i.e., white rot wood in this study. Thus, they may prefer brown rot wood for colonization. Since communities of mycorrhizal fungi develop well in logs in late stages of decay [23], we predict that these mycorrhizal communities affect seedling establishment and are associated with wood decay type. However, this idea has not yet been tested in controlled experiments.
In the present study, pot experiments were conducted for the seedlings of two arbuscular mycorrhizal tree species, Cryptomeria japonica and Chamaecyparis obtusa, and two ectomycorrhizal tree species, Betula ermanii and Abies veitchii, to evaluate their growth on three substrates (brown rot wood and white rot wood of Pinus densiflora, and soil from a pine-dominated forest) in laboratory conditions. Seedlings of all these four trees are known to regenerate on dead wood [8,13,24,25]. Nutrient ion concentrations and fungal communities were also analyzed in the three substrates and their potential effects on seedling growth were discussed. We hypothesized that growth of arbuscular mycorrhizal trees (Cr. japonica and Ch. obtusa) would be better on brown rot wood than on white rot wood, whereas the growth of ectomycorrhizal trees (B. ermanii and A. veitchii) would be better on white rot wood than on brown rot wood. Forest soil was prepared as a standard substrate for purpose of comparison.
2. Materials and Methods
2.1. Tree Species
Seeds of two ectomycorrhizal tree species (A. veitchii, B. ermanii), and seeds of two arbuscular mycorrhizal tree species (Ch. obtusa, Cr. japonica) were used for pot experiments. All the seeds were obtained from the Forest Tree Breeding Center of FFPRI (Hitachi, Ibaraki, Japan). The seeds were surface sterilized with hydrogen peroxide: submerged in 70% ethanol (v/v) for 1 min to wet the surface, sterilized for 50 s in a solution of 30% hydrogen peroxide (v/v), and then rinsed with sterile deionized water. The surface sterilized seeds were then kept in darkness at 2 °C in moistened autoclaved cotton for 1 month to break dormancy. At the point seeds were surface sterilized again as described above, and placed onto moistened autoclaved cotton to induce germination (20 °C, 13 h light, 20% luminescence) in a Biotron system (NK system, Osaka, Japan). Aseptically germinated seedlings were then individually transported to pots prepared as described below.
2.2. Pot Experiment
In July 2019, well-decayed (decay class IV in the five-decay class system [7]), pine (P. densiflora) dead wood was collected from two sites of mixed secondary forest dominated by P. densiflora and Quercus serrata in Miyagi, Japan (38°37.2 N, 140°48.6 E). The dead wood was then categorized into white rot and brown rot by visual criteria after Araya [26]. These dead wood samples were then sieved through 6 mm of mesh twice. Samples from three dead woods were combined and mixed well to make one sample for each of white rot wood and brown rot wood. Samples were also collected from the top 10 cm of soil at three locations in the same collection sites of as the dead wood. These were sieved through 5 mm mesh, and then mixed to make one soil sample. Invertebrates in the sample were carefully hand-sorted and removed. Half volumes of each sample of white rot wood, brown rot wood, and soil were then autoclaved for 20 min at 121 °C. Autoclaving was repeated three times with 1 day intervals. These six types of substrates (sterilized and non-sterilized soil, white rot wood, and brown rot wood) were then mixed with equal volumes of vermiculite and used as substrates for the pot experiment.
Individual germinants were planted into pots (4 cm diameter, 4.2 cm height; polypot, Tokorozawa Ueki Bachi Center, Saitama, Japan) filled with substrates prepared as described above. The pots were incubated at 20 °C, 13 h light, 100% luminescence (average photon density = 345 µmol m−2·s−1) in a Biotron for 193 days, except for A. veitchii which was incubated for 188 days (Figure S1). For B. ermanii, Cr. japonica, and Ch. obtusa, 15 replicates were prepared, although only eight replicates were prepared for A. veitchii due to a low germination rate. Pots were watered every 4 days during the incubation period. Seedling shoot length (length from the substrate surface to the top growth point) was measured at three time periods during incubation: day 20, day 141, and day 193 (end of incubation) for Cr. japonica, Ch. obtusa, and B. ermanii, and day 15, day 136, and day 188 for A. veitchii. Seedling shoot growth was measured as the difference from the first measurement. After the incubation period, seedlings were harvested, weighed in fresh state, and dried at 70 °C to measure dry weight. Before drying, the mycorrhizal colonization rate was measured as follows. For arbuscular mycorrhizal trees (Cr. japonica and Ch. obtusa), root samples were separated into two subsamples: one for measuring mycorrhizal colonization rate, and the other for measuring water content of the roots to calculate dry weight of the whole root system using their fresh weight.
2.3. Mycorrhizal Colonization Rate
The mycorrhizal fungi colonization rate (%) of root systems of individual germinants was measured after incubation. For arbuscular mycorrhizal trees (Cr. japonica and Ch. obtusa), roots (>10 cm length) were washed with 0.005% aerosol OT (Wako, Osaka, Japan) in vortex for 1 min, then washed with hypersonic for 10 min, rinsed with deionized water twice, and cleared by heating in 10% KOH at 100 °C for more than 1 h. Cleared roots were rinsed with deionized water, bleached in 0.5% H2O2 for 20 min, rinsed with deionized water and fixed in 2% HCl for more than 10 min. The fixed roots were stained with trypan blue and stored in lactoglycerol (lactic acid 525 mL, glycerin 37.8 mL, deionized water 37.2 mL). Colonization was assessed (following McGonigle et al. [27]) under 200× magnification to obtain percentages of root length colonized by arbuscular mycorrhizal fungal structures, specifically arbuscules, coils, and vesicles (Figure S2A–C).
Among the ectomycorrhizal trees (A. veitchii and B. ermanii), the colonization rate (%) of ectomycorrhizal fungi was measured as the percentage of ectomycorrhizal root tips (Figure S2D,E) relative to the total number of root tips (120 on average) by direct observation of root systems under binocular of <45× magnification (SZ2–ILST, Olympus, Tokyo, Japan).
2.4. Chemical Properties of Substrates
For each of the six substrates, pH and concentrations of cations (Na+, NH4+, Ca+, K+, Mg+) and anions (NO3−, SO42−, Cl−) were measured. Fresh subsamples (ca. 80 mL, five replicates for each substrate) were extracted with 200 mL deionized water in 250 mL polyethylene bottles and shaken for 1 h (100 rpm; Shaker MK201, Yamato Scientific, Tokyo, Japan). The pH of the extract was measured using a potable pH meter (LAQUAtwin-pH-11B, HORIBA, Kyoto, Japan), and was filtered using filter paper (5C; ADVANTEC, Tokyo, Japan) and a syringe filter (DISMIC25CS; ADVANTEC, Tokyo, Japan). The filtrate was then analyzed using an Ion Chromatography system (Shim-pack IC, Shimadzu, Kyoto, Japan) with 0.6 mM Na2CO3/12 mM NaHCO3 as the anion eluent and 2.5 mM oxalic acid as the cation eluent, 40 °C at the separation column. Concentrations of ions were calculated as 100 g-substrate bases.
2.5. Fungal Metabarcoding of Substrates
Whole DNA was extracted from 0.2 g white rot wood and brown rot wood subsamples, and 0.3 g soil subsamples (five replicates for each substrate, not including sterilized ones) using ISOIL for Beads Beating (Nippon Gene, Tokyo, Japan) following the manufacturer’s protocol. Prior to extraction, freeze-dried wood samples were milled using Multi-beads Shocker (Yasui Kikai, Osaka, Japan) for 30 s three times each. For sequencing of the fungal internal transcribed spacer 1 (ITS1) region using the MiSeq sequencing platform with 250 × 2 paired-end reads (Illumina, San Diego, CA, USA), a two-step PCR protocol with ITS1F_KYO1/ITS2_KYO2 primers [28] in the primary amplification containing tails for adding indices and Illumina flow cell adapters in the secondary amplification was conducted. Both positive and negative controls were used for PCR, and positive controls were used for MiSeq sequencing. Note that the ITS region has been proposed as the formal fungal barcode [29]. Details of the sample preparation for MiSeq sequencing (using MiSeq Reagent Nano Kit v2 for 2 × 250 bp PE) are provided in the Supplementary file.
A total of 675,854 reads were obtained after MiSeq sequencing and chimera check (deposited in the Sequence Read Archive of the DNA Data Bank of Japan, accession number DRA014309). The sequence reads were trimmed with a minimum quality value of 30, and the 5′- and 3′-primer sequences were then removed from the trimmed reads. Next, the trimmed reads were denoised with Claident [30] using Assams-assembler 0.2.2015.05.10 [31]. A chimera check was conducted with Claident software using the UNITE database (https://unite.ut.ee, accessed on 5 May 2022). Then, the quality-filtered sequences were classified into molecular operational taxonomic units (OTU), and taxonomically identified using the Claident software original fungal ITS database (fungi_its_genus) which is structured after the International Nucleotide Sequence Database (http://www.insdc.org, accessed on 5 May 2022), at a threshold similarity of 97%, which is widely used for the fungal ITS region [32]. One soil sample with less than 1000 reads was removed. For each sample, OTUs with less than 0.1% of the total number of reads per sample were removed. After the filtering process, a total of 567,516 reads were obtained. The OTU numbers were saturated in all samples after the filtering process (Figure S3). Singleton OTUs were removed from the downstream analyses. Consequently, each of the 133 filtered OTUs was searched using the FUNGuild database and assigned to one of eight functional groups: arbuscular mycorrhizal, brown-rot, ectomycorrhizal, plant pathogen, soft-rot, undefined saprotroph, white-rot, and wood decay with unknown decay type (https://github.com/UMNFuN/FUNGuild, accessed on 5 May 2022 [33]) (Table S1).
2.6. Statistical Analysis
All the analyses were performed using R 4.0.5 [34]. Nutrient ion concentrations were compared among the six substrates using the Nemenyi post-hoc test. Because seedling growth data (shoot growth for 121 days and whole dried weight after the harvest) include some ties, comparisons among the six substrates were applied using the Brunner–Munzel test with the Holm adjustment of probability values. Colonization rates of arbuscular mycorrhizal fungi (arbuscule, coil, and vesicle) and ectomycorrhizal fungi were compared among the six substrates using the chi-square test and Ryan’s post-hoc test (http://aoki2.si.gunma-u.ac.jp/R/p_multi_comp.html, accessed on 5 May 2022). The pH was not compared among the substrates because five replicates had the same values.
The occurrence (presence/absence) of fungal OTUs was recorded for each substrate sample regardless of the number of sequence reads due to the quantitative issues in read numbers resulting from amplicon sequencing [35], and was used as binary data for all statistical analyses. The dissimilarities in the fungal communities between the substrate samples were calculated using Raup–Crick metrics (vegdist command) in the vegan package [36]. Significant differences in the community composition among the samples were determined by permutational multivariate analysis of variance (PERMANOVA [37]) with 10,000 permutations (adonis command). Additionally, the community variance between samples (calculated using the betadisper command with Raup-Crick metrics) was compared between substrates using analysis of variance (anova command).
The occurrence frequency of fungal functional groups was compared across the three unsterilized substrates using Fisher’s exact test. Fungal OTU numbers for the samples were compared among the three substrates using the Brunner–Munzel test with Holm adjustment of probability values. Furthermore, indicator OTUs for each substrate were searched using the multipatt command in the indicspecies package [38].
Pearson’s correlation coefficients were calculated between seedling growth (shoot growth for 121 days, and whole dried weight after the harvest) and colonization of mycorrhizal fungi.
3. Results
3.1. Chemical Properties of Substrates
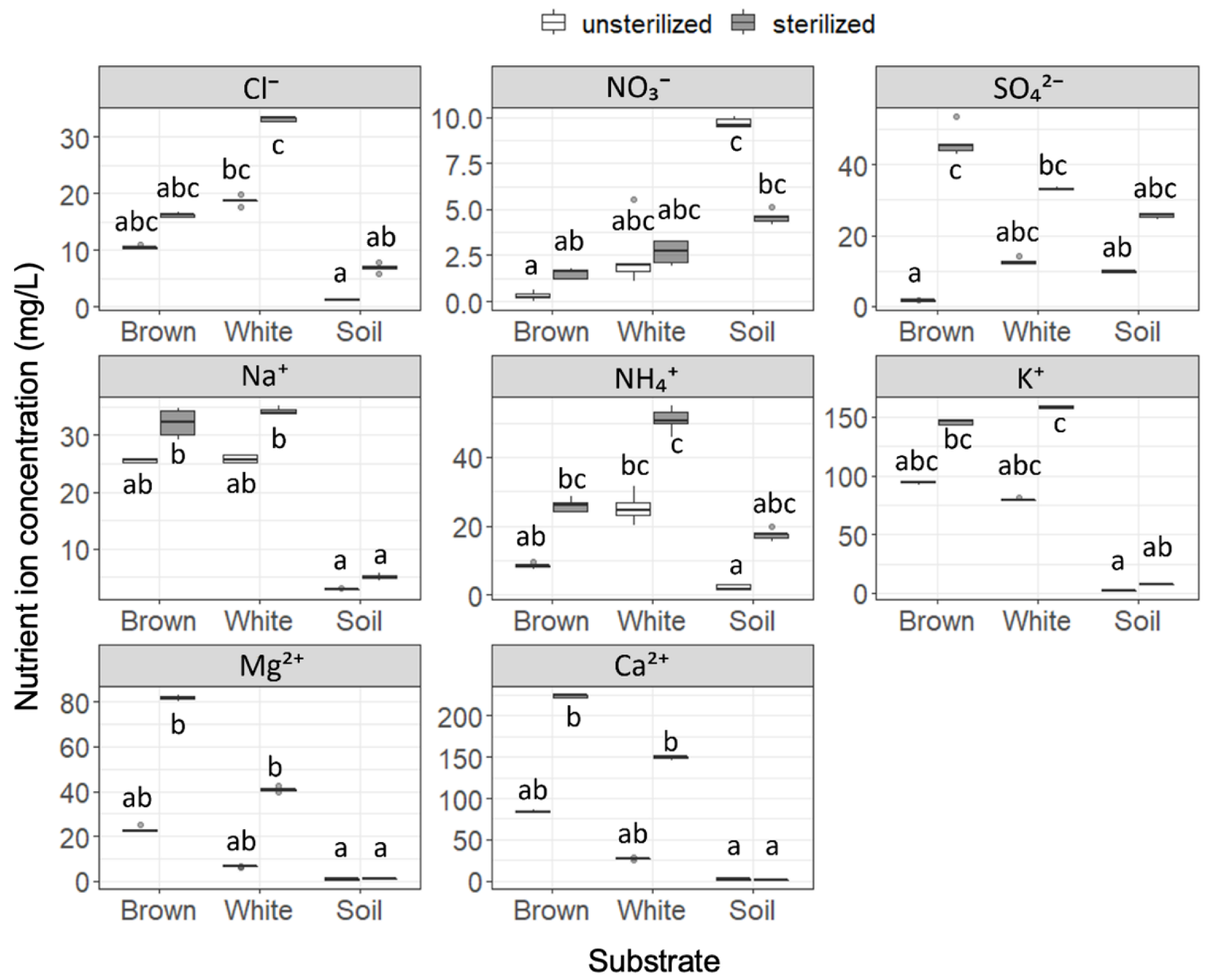
Nutrient ion concentrations were found to be significantly different among the six substrates (Figure 1), and sterilization was seen to increase the nutrient ion concentrations in all except for NO3− in soil. Cl− concentration was significantly higher in white rot compared to soil, regardless of sterilization. The NO3− concentration was significantly higher in soil than in brown rot in unsterilized substrates, but this difference disappeared after sterilization. In contrast, the SO42− concentration was significantly increased by sterilization in brown rot, but was not in white rot or soil. Na+ concentrations in wood samples (brown rot and white rot) were higher than seen in soil, and the difference was significant after the samples were sterilized. NH4+ concentration was higher in white rot compared to soil, but this difference disappeared after sterilization. Similar to Na+, concentrations of K+, Mg2+, and Ca2+ in wood samples were not significantly different from those in soil, but were significantly higher than in soil after sterilization. Concentrations of NO2−, Br−, and PO43− were too low to be detected as meaningful data. The five replicates showed almost equal pH values within substrates: 3.6–3.7, 4.2, and 5.0 for unsterilized brown rot, white rot, and soil, respectively, and 3.4–3.5, 4.0–4.1, and 5.5–5.6 for sterilized brown rot, white rot, and soil, respectively. While we did not perform statistical analysis, the pH increased in this order, and was altered by sterilization (slightly decreased in wood samples and increased in soil).
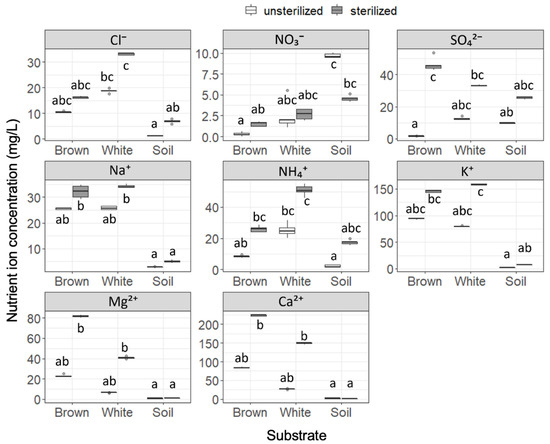
Figure 1.
Nutrient ion concentrations in unsterilized and sterilized substrates (brown rot wood, white rot wood, and soil). Box plots display median, 95% confidence interval, and minimum and maximum values and points outside the box represent outliers. Different letters indicate significant difference among the six substrates (p < 0.05, Brunner–Munzel test with Holm adjustment of probability values). n = 5.
3.2. Fungal Community in Substrates
In total, 132 fungal OTUs were detected (Table S1). The mean OTU richness per log ranged from 29.2 in brown rot logs to 54.3 in soil (Table 1), and was larger in the soil than in the logs (BM test, brown rot vs. soil p = 0.048; white rot vs. soil p = 0.048). No significant difference was detected in total OTU richness between brown rot and white rot logs. Among the 132 OTUs, 52 OTUs were assigned to one of the eight functional groups (Table S1 and Table 1). Ectomycorrhizal fungi included the largest 19 OTUs, mainly detected among soil samples (Table 1), followed by undefined saprotrophs (15 OTUs), and soft rot fungi (seven OTUs). White rot fungi and wood decay fungi with an unknown decay type include four OTUs for each, and arbuscular mycorrhizal, brown rot fungi, and plant pathogens include one OTU for each. Eighty OTUs were functionally unknown. Results of PERMANOVA analysis suggested that fungal communities were significantly different among the three substrates, while dispersions were not significantly different among the substrates (Table 2). Indicator species analyses suggested that seven OTUs were indicative of brown rot wood, 11 OTUs were indicative of white rot wood, and 39 OTUs were indicative of soil (Table 3). Seven OTUs indicative of brown rot wood consisted of one brown rot fungi (Pseudomerulius curtisii), two soft rot fungi in the genus Scytalidium, and four OTUs functionally unidentified. Eleven OTUs indicative of white rot wood consisted of three white rot fungi (Gymnopilus picreus, Xeromphalina sp., and Sistotremastrum sp.), a wood decay fungi with an unknown decay type (Botryobasidium sp.), a soft rot fungi (Scytalidium album), and six OTUs which were functionally unidentified. Among the 39 OTUs associated with soil, 16 OTUs were ectomycorrhizal fungi, six OTUs were undefined saprotrophs, one of each were arbuscular mycorrhizal (Archaeospora sp.) and soft rot fungi (Trichoderma sp.), and 15 OTUs were functionally unidentified. Ectomycorrhizal OTUs associated with soil were dominated by the genera Russula (three OTUs), Inocybe (two OTUs), Sebacina (two OTUs), and Tomentella (two OTUs). Cenococcum geophilum, Clavulina sp., Cortinarius sp., Delastria sp., Elaphomyces sp., Laccaria bicolor, and Thelephora sp. were also detected.

Table 1.
OTU richness of fungal functional groups in brown rot wood, white rot wood, and soil.

Table 2.
Results of PERMANOVA comparing dissimilarities in fungal OTU composition, and ANOVA comparing dispersion.

Table 3.
Results of indicator species analysis for brown rot wood, white rot wood, and soil. OTUs with p < 0.01 were shown. See footnote in Table 1 for abbreviations of function.
3.3. Seedling Growth
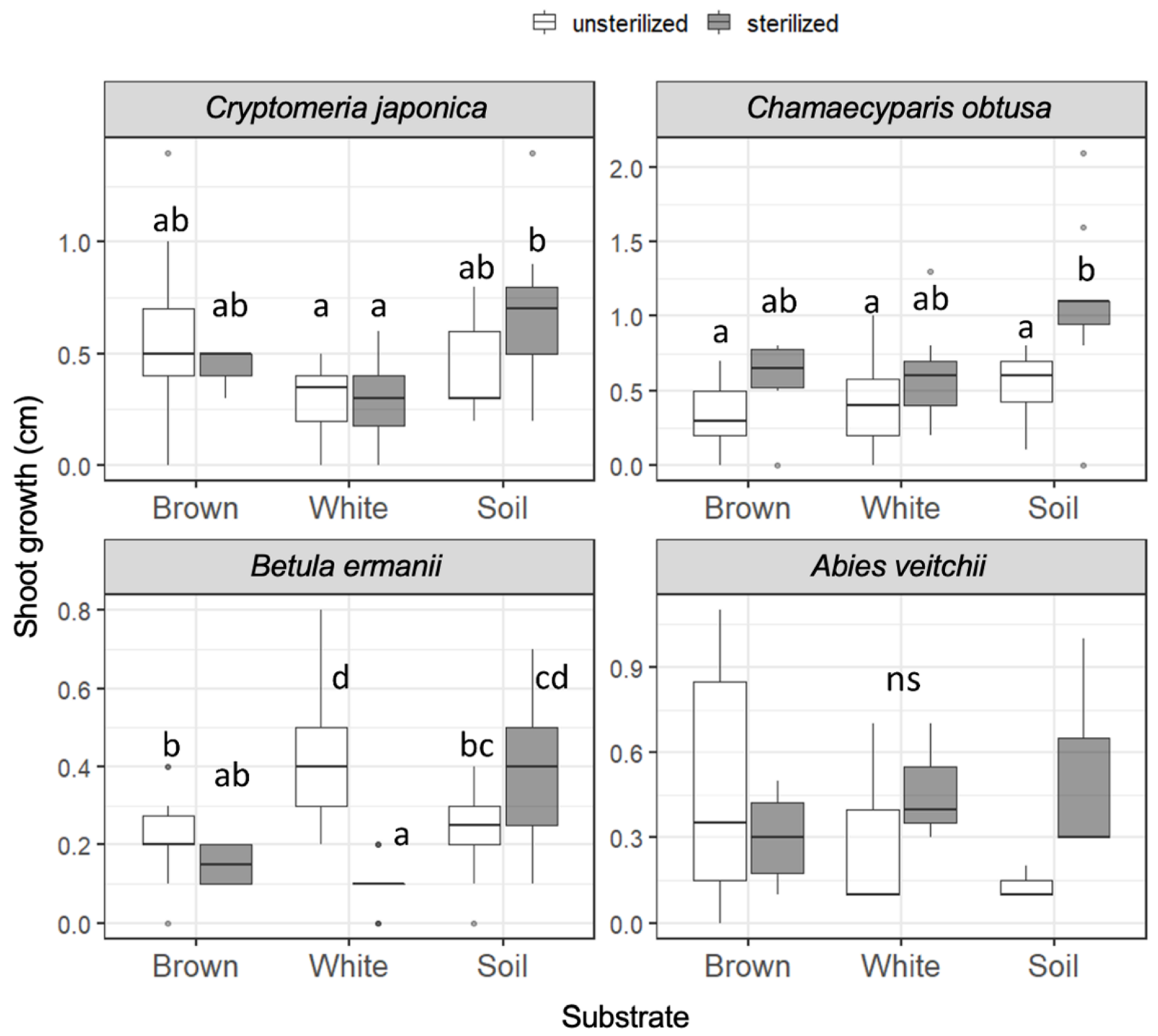
The growth of the seedling shoots during 121 days from day 20 to day 141 (day 15 to day 136 for A. veitchii) is shown in Figure 2. In Cr. japonica, the greatest growth rate was observed in unsterilized brown rot and in sterilized soil (1.4 cm). However, the shoot growth of Cr. japonica was not significantly different among unsterilized substrates, and growth in soil was significantly greater than that in white rot only when the substrates were sterilized. Ch. obtusa growth was not significantly different among the substrates regardless of sterilization, although the growth was stimulated by sterilization in soil, and the greatest growth (2.1 cm) was recorded in sterilized soil. B. ermanii growth was significantly greater in white rot (max. 0.8 cm) than in brown rot and soil in unsterilized substrates. However, growth in white rot was significantly reduced when the substrates were sterilized, and growth in soil became significantly greater than that in wood (brown rot and white rot). A. veitchii growth (max. 1.1 cm) was not different among the substrates. Seedling shoot growth during 173 days from day 20 to day 193 (day 15 to day 188 for A. veitchii), covering the entire period, is shown in Figure S4. The significant differences in shoot growth described above were disappeared except for the negative effect of sterilization on B. ermanii seedlings in white rot wood.
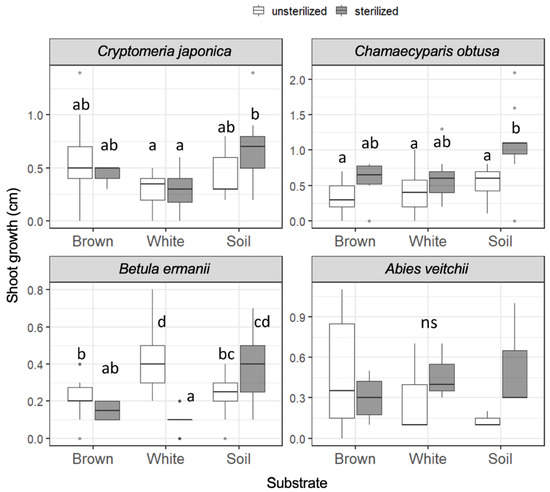
Figure 2.
Seedling shoot growth of the four tree species during 121 days on unsterilized and sterilized substrates (brown rot wood, white rot wood, and soil). Box plots display median, 95% confidence interval, and the minimum and maximum values and points outside the box represent outliers. Different letters indicate significant differences among the six substrates (p < 0.05, Permuted Brunner–Munzel test with Holm adjustment of probability values). ns, not significant. n = 15 except for A. veitchii (n = 8).
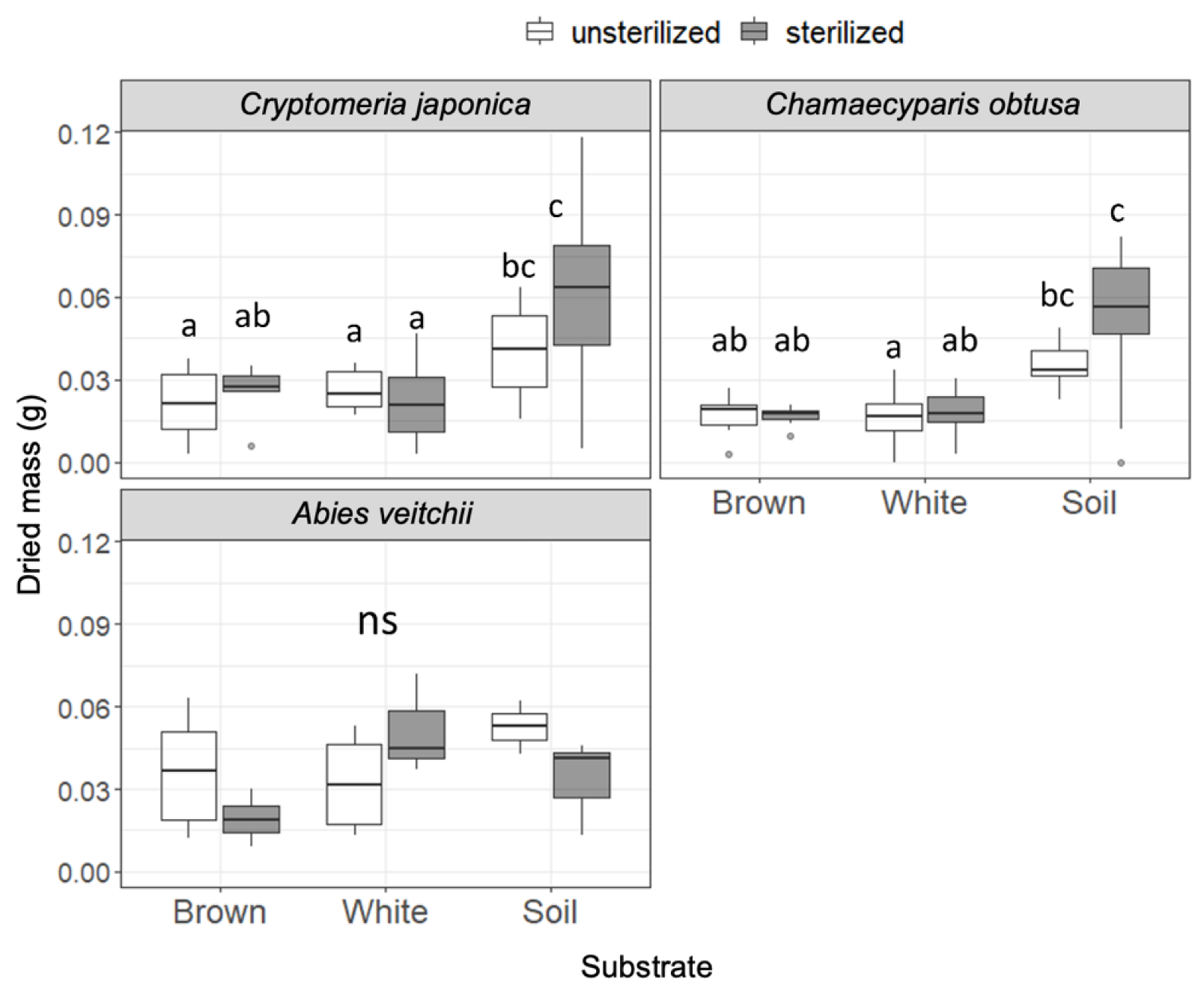
Dried masses of the seedlings are shown in Figure 3. In Cr. japonica, seedling mass was significantly greater in soil than in wood regardless of the sterilization. Similarly, seedling mass of Ch. obtusa was greater in soil than in wood, particularly in sterilized substrates. Seedling mass of A. veitchii did not differ among substrates. No effect of sterilization was observed in all substrates for all species. The seedling mass of B. ermanii was too small to measure.
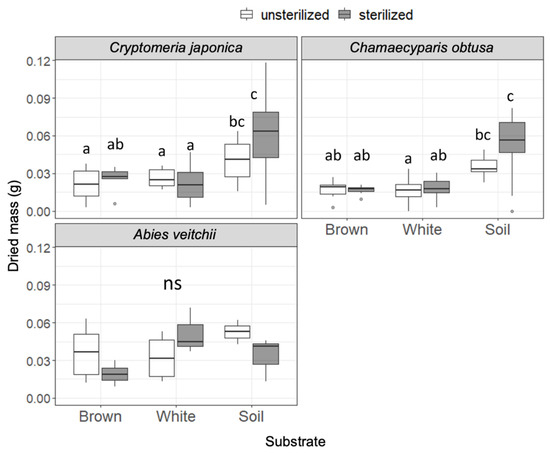
Figure 3.
Seedling dry mass of the three tree species after incubation on unsterilized and sterilized substrates (brown rot wood, white rot wood, and soil). Box plots display median, 95% confidence interval, and the minimum and maximum values and points outside the box represent outliers. Different letters indicate significant differences among the six substrates (p < 0.05, Permuted Brunner–Munzel test with Holm adjustment of probability values). ns, not significant. n = 15 except for A. veitchii (n = 8).
3.4. Mycorrhizal Colonization in Seedling Roots
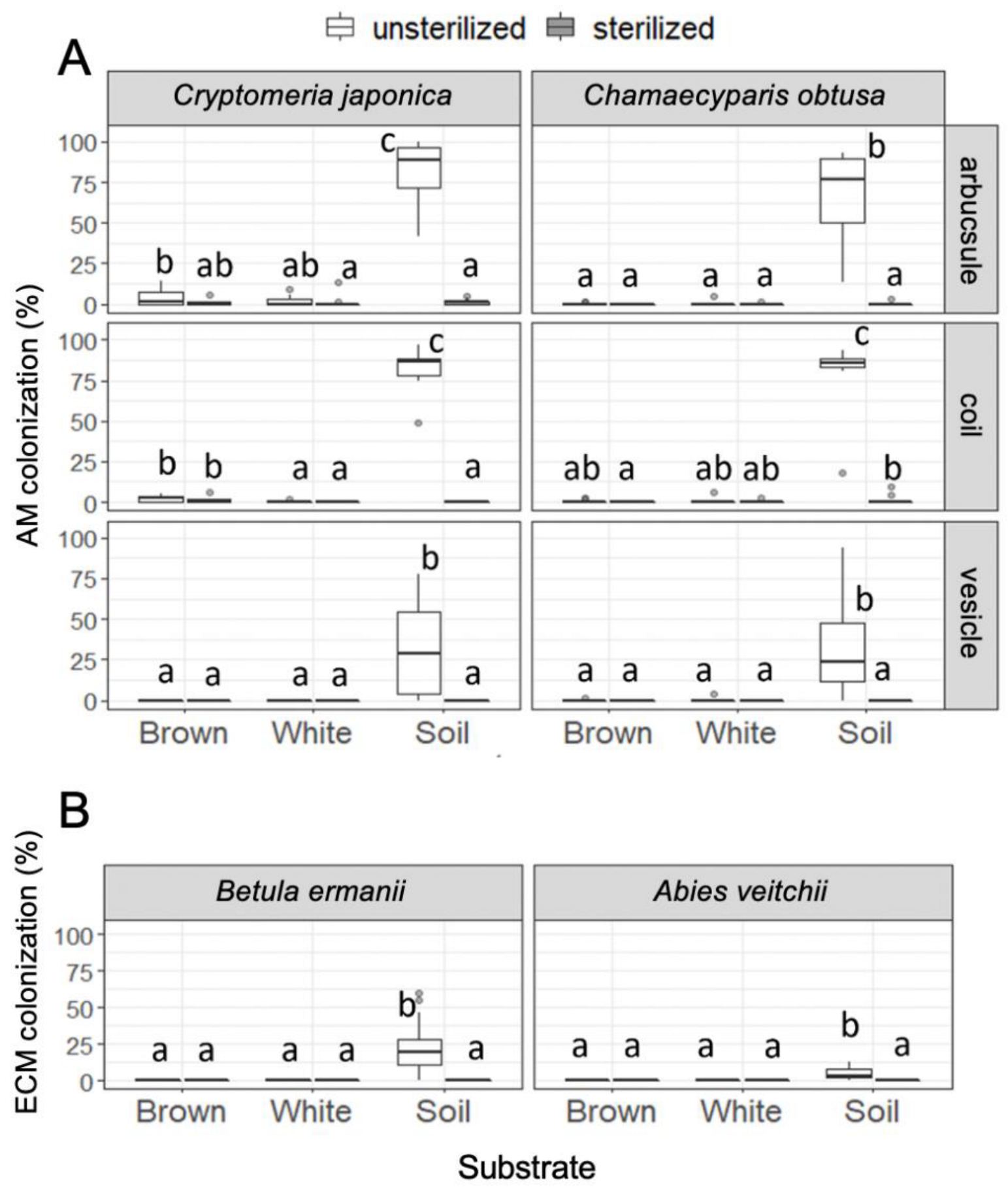
Colonization of arbuscular mycorrhizal fungi was almost entirely restricted to the seedlings in unsterilized soil, although there was some colonization of arbuscule and coil in Cr. japonica roots in brown rot, and of coil in Ch. obtusa roots in sterilized soil (Figure 4). Colonization of ectomycorrhizal fungi was also entirely restricted to the seedlings in unsterilized soil. All observed ectomycorrhizal root tips were Cenococcum-type (Figure S2D,E). There was no significant relationship observed between seedling growth (shoot growth and dry mass) and mycorrhizal colonization rate (data not shown).
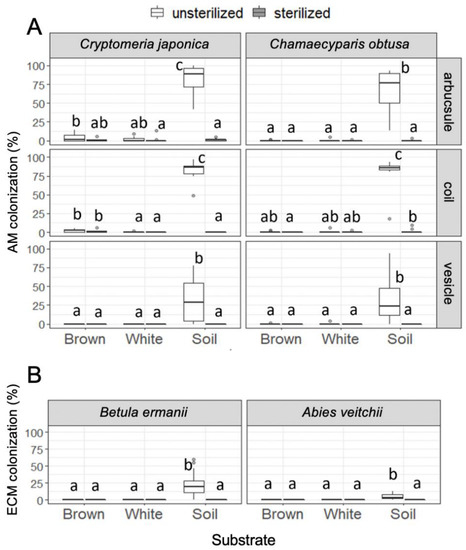
Figure 4.
(A) Arbuscular mycorrhizal colonization in Cr. japonica and Ch. obtusa roots and (B) ectomycorrhizal colonization in B. ermanii and A. veitchii roots in unsterilized and sterilized substrates (brown rot wood, white rot wood, and soil). Different letters indicate significant difference among the six substrates (p < 0.05, Permuted Brunner–Munzel test with Holm adjustment of probability values). n = 15 except for A. veitchii (n = 8).
4. Discussion
The present study showed significant effects of substrate and sterilization on seedling growth. Because the nutrient concentrations differ greatly between sterilized and unsterilized substrates (Figure 1), it would be difficult to discuss the biotic factors without these large differences. Rather, it would be better to sort out the differences in growth responses within the sterilized and unsterilized conditions, respectively. The greater shoot growth seen in B. ermanii in unsterilized white rot wood, compared with brown rot wood and soil, might be attributable to some positive effects of microorganisms in white rot wood. This is likely because this advantage of white rot wood disappeared when it was sterilized (Figure 2). Since sterilization generally increased nutrient concentrations in the substrates (Figure 1), this positive effect of white rot wood cannot be explained by the nutrient status of the substrates. Although fungal communities were significantly different across the substrates (Table 2), any dominance of ectomycorrhizal fungi (Table 1 and Table 3) and their colonization (Figure 4) which may facilitate the growth of B. ermanii [39], was not detected in white rot wood. Instead of the species richness detected in ectomycorrhizal fungi in soil, the observed ectomycorrhizal root tips were dominated by Cenococcum-type fungi, indicating that conditions in the Biotron growth chamber somewhat stressed the seedlings, impacted their growth, and were unsuitable for colonization by a variety of ectomycorrhizal fungi. It is reported that Cenococcum species are stress-tolerant and dominate root systems of ectomycorrhizal trees under stressful conditions such as drought [40,41]. Even if more ECM fungal OTUs were detected in the substrate, their activity may have been limited because they were not reflected in the field conditions. For example, the watering was done once every 4 days, but the water holding capacity of soil and wood must be different. These results do not identify the reason why shoot growth of B. ermanii was greater in unsterilized white rot wood than in other substrates. A possibility is that fungi with unknown functions facilitated B. ermanii growth. For example, species in Helotiales include many root endophytes [42], and some endophytic species, such as Phialocephala fortinii, have positive effects on plant growth [43]. In the present study, Helotiales OTU_307 was detected only in white rot wood (Table S1), although its occurrence was not significantly indicative in white rot wood (Table 3). However, Mayerhofer et al. [44] reported that colonization of Helotiales could reduce plant growth depending on environmental conditions. Future studies should measure colonization rates of not only mycorrhizal fungi but also root endophytes to allow comparison among different substrates.
The effects of substrates on the growth of arbuscular mycorrhizal trees Ch. obtusa and Cr. japonica was clear in seedling weight, with both being greater in soil than in wood (Figure 3). The higher colonization rate of arbuscular mycorrhizal fungi (Figure 4) in soil might be a cause of this effect. Arbuscular mycorrhizal fungi are important for plants, enabling absorption of phosphorus [45], which is generally absorbed by Al and Fe in soil [46], and is insoluble to water and was not detected in the present study. However, the effect is not so simple, because the growth, particularly shoot growth of Ch. obtusa, was significantly increased by substrate sterilization, particularly in soil (Figure 2) even though the colonization rates of arbuscular mycorrhizal fungi were dramatically reduced (Figure 4). The observed increase in nutrients after sterilization (Figure 1) might compensate for the reduction of mycorrhizal colonization in this case. Note that Ch. obtusa and Cr. japonica are late-successional tree species. Gehring [47] mentions that late-successional species have a slower response to AM-induced growth. For this reason, it is supposed that the six-month experimental period was not enough to make a difference between the substrates. Another possibility for the positive effect of sterilization on seedling growth is the reduction of the negative effects of pathogenic fungi. However, no OTUs of dominant plant pathogens and parasitic fungi were detected in the soil.
Brown rot wood substrates used in the present study appear to be decayed primarily by a single brown rot fungus, Pseudomerulius curtisii, because this fungus was detected in 100% of the brown rot samples and there were no other brown rot fungi detected. Whereas in white rot wood, decay appears to be the result of a combination of several white rot fungi, such as Gymnopilus picreus, Sistotremastrum sp., and Xeromphalina sp., which were detected as indicator species in white rot wood. Previous studies reported other brown rot fungi from the genera Calocera, Dacrymyces, Fibroporia, Fomitopsis, Gloeophyllum, Leptoporus, Neolentinus, and Serpula; and white rot fungi from the genera Ceriporia, Coniochaeta, Hyphoderma, Hyphodontia, Hypochnicium, Mycoasia, Nigroporus, Phanerochaete, Phlebia, Phlebiopsis, Pholiota, Pluteus, Rigidoporus, Skeletocutis, Trichaptum, and Tyromyces in the dead wood of Pinus densiflora [48]. Additionally, the physicochemical properties and microbial communities might differ among the wood decayed by the different fungi, even within the same decay type [11,49]. Our previous study using substrates from another study site, where different species of brown rot fungi Neolentinus lepideus dominated on P. densiflora logs, showed that white rot wood contains higher concentrations of potassium, magnesium, and calcium than brown rot wood, and soil contained higher concentrations of SO42− than wood. These findings are inconsistent with the findings of the present study. These results suggest that studies using substrates decayed by different fungal species may have different effects on the seedlings, and thus wood decayed by known fungal species should be used as the substrates in future research.
To summarize, seedling growth of the two arbuscular mycorrhizal trees Cr. japonica and Ch. obtusa, and an ectomycorrhizal tree B. ermanii, were significantly different among the three substrates: white rot wood, brown rot wood, and soil. Particularly, the shoot growth of B. ermanii was significantly greater in white rot wood than in the other two substrates, supporting our hypothesis. However, the growth of Cr. japonica and Ch. obtusa was not significantly different between brown rot and white rot wood. These results suggest that good growth of B. ermanii on white rot wood could be a reason for their regeneration success on white rot logs observed in the field [13], although its mechanism is still unknown. In contrast, the dominance of Cr. japonica and Ch. obtusa seedlings on brown rot logs [8,24] might be attributable to mechanisms in other stages of seedling development, such as germination, or survival, and should be tested in future field experiments. Furthermore, we did not detect any effects associated with the substrates and sterilization in A. veitchii seedlings, suggesting that responses to the substrates and sterilization differ among seedling species within the same mycorrhizal type. Therefore, further research is needed involving many tree species, including ectomycorrhizal and arbuscular mycorrhizal trees, to test their performance on different substrates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13071036/s1, Supplementary methods; Figure S1: Situation of pot experiments in Biotron; Figure S2: Photographic images of arbuscular mycorrhizal and ectomycorrhizal structures; Figure S3: Sample-based rarefaction curves of fungal OTUs detected in the samples; Figure S4: Seedling growth of the four tree species during 173 days on unsterilized and sterilized substrates; Table S1: List of fungal OTUs and their frequencies detected from ITS1 metabarcoding of DNA extracted from substrates.
Author Contributions
Conceptualization, Y.F.; methodology, Y.F.; validation, Y.F.; formal analysis, H.K.; investigation, Y.F.; data curation, Y.F. and H.K.; writing—original draft preparation, Y.F.; writing—review and editing, Y.F.; visualization, H.K.; project administration, Y.F.; funding acquisition, Y.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Inamori Foundation.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors thank Yoshimi Yokoyama for running ion chromatography system.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Harmon, M.E.; Franklin, J.F.; Swanson, F.J.; Sollins, P.; Gregory, S.V.; Lattin, J.D.; Anderson, N.H.; Cline, S.P.; Aumen, N.G.; Sedell, J.R.; et al. Ecology of Coarse Woody Debris in Temperate Ecosystems. Adv. Ecol. Res. 1986, 15, 133–302. [Google Scholar]
- Fukasawa, Y. Ecological Impacts of Fungal Wood Decay Types: A Review of Current Knowledge and Future Research Directions. Ecol. Res. 2021, 36, 910–931. [Google Scholar] [CrossRef]
- Kubota, Y.; Konno, Y.; Hiura, T. Stand Structure and Growth Patterns of Understorey Trees in a Coniferous Forest, Taisetsuzan National Park, Northern Japan. Ecol. Res. 1994, 9, 333–341. [Google Scholar] [CrossRef]
- Noguchi, M.; Yoshida, T. Tree Regeneration in Partially Cut Conifer-Hardwood Mixed Forests in Northern Japan: Roles of Establishment Substrate and Dwarf Bamboo. For. Ecol. Manag. 2004, 190, 335–344. [Google Scholar] [CrossRef]
- Mori, A.; Mizumachi, E.; Osono, T.; Doi, Y. Substrate-Associated Seedling Recruitment and Establishment of Major Conifer Species in an Old-Growth Subalpine Forest in Central Japan. For. Ecol. Manag. 2004, 196, 287–297. [Google Scholar] [CrossRef]
- Katsumata, N.; Okitsu, S.; Minami, Y. Influence of Regional Climates on the Availability of Decaying-Log Microsite for Conifer Establishment on Vancouver Island, Canada. Hort Res. 2009, 63, 55–60. [Google Scholar]
- Fukasawa, Y. Effects of Wood Decomposer Fungi on Tree Seedling Establishment on Coarse Woody Debris. For. Ecol. Manag. 2012, 266, 232–238. [Google Scholar] [CrossRef]
- Fukasawa, Y. Pine Stumps Act as Hotspots for Seedling Regeneration after Pine Dieback in a Mixed Natural Forest Dominated by Chamaecyparis obtusa. Ecol. Res. 2018, 33, 1169–1179. [Google Scholar] [CrossRef]
- Eaton, R.A.; Hale, M.D.C. Wood: Decay, Pests and Protection; Chapman & Hall: Tokyo, Japan, 1993. [Google Scholar]
- Espejo, E.; Agosin, E. Production and Degradation of Oxalic Acid by Brown Rot Fungi. Appl. Environ. Microbiol. 1991, 57, 1980–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrofsky, A.; Jellison, J.; Smith, K.T.; Shortle, W.C. Changes in Cation Concentration in Red Spruce Wood Decayed by Brown Rot and White Rot Fungi. Can. J. For. Res. 1997, 27, 567–571. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Komagata, Y.; Kawakami, S.-I. Nutrient Mobilization by Plasmodium of Myxomycete Physarum Rigidum in Deadwood. Fungal Ecol. 2017, 29, 42–44. [Google Scholar] [CrossRef]
- Tedersoo, L.; Suvi, T.; Jairus, T.; Kõljalg, U. Forest Microsite Effects on Community Composition of Ectomycorrhizal Fungi on Seedlings of Picea abies and Betula pendula. Environ. Microbiol. 2008, 10, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Lindner, D.L.; Vasaitis, R.; Kubartová, A.; Allmer, J.; Johannesson, H.; Banik, M.T.; Stenlid, J. Initial Fungal Colonizer Affects Mass Loss and Fungal Communty Development in Picea abies Logs 6 Yr after Inoculation. Fungal Ecol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Jurgensen, M.F.; Larsen, M.J.; Wolosiewicz, M.; Harvey, A.E. A Comparison of Dinitrogen Fixation Rates in Wood Litter Decayed by White-Rot and Brown-Rot Fungi. Plant Soil 1989, 115, 117–122. [Google Scholar] [CrossRef]
- Hoppe, B.; Kahl, T.; Karasch, P.; Wubet, T.; Bauhus, J.; Buscot, F.; Krüger, D. Network Analysis Reveals Ecological Links between N-Fixing Bacteria and Wood-Decaying Fungi. PLoS ONE 2014, 9, e88141. [Google Scholar]
- Haq, I.U.; Hillmann, B.; Moran, M.; Willard, S.; Knights, D.; Fixen, K.R.; Schilling, J.S. Bacterial Communities Associated with Wood Rot Fungi that Use Distinct Decomposition Mechanisms. ISME Commun. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Seiwa, K.; Negishi, Y.; Eto, Y.; Hishita, M.; Masaka, K.; Fukasawa, Y.; Matsukura, K.; Suzuki, M. Successful Seedling Establishment of Arbuscular Mycorrhizal-Compared to Ectomycorrhizal-Associated Hardwoods in Arbuscular Cedar Plantations. For. Ecol. Manag. 2020, 468, 118155. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Zobel, M. How Mycorrhizal Associations Drive Plant Population and Community Biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Masaka, K.; Fukasawa, Y.; Matsukura, K.; Seiwa, K. Gap Creation Alters the Mode of Conspecific Distance-Dependent Seedling Establishment via Changes in the Relative Influence of Pathogens and Mycorrhizae. Oecologia 2020, 192, 449–462. [Google Scholar]
- Fernandez, C.W.; Kennedy, P.G. Revisiting the ‘Gadgil Effect’: Do Interguild Fungal Interactions Control Carbon Cycling in Forest Soils? New Phytol. 2015, 209, 1382–1394. [Google Scholar] [CrossRef]
- Steidinger, B.S.; Crowther, T.W.; Liang, J.; Van Nuland, M.E.; Werner, G.D.A.; Reich, P.B.; Nabuurs, G.; de-Miguel, S.; Zhou, M.; Picard, N.; et al. Climatic Controls of Decomposition Drive the Global Biogeography of Forest-Tree Symbioses. Nature 2019, 569, 404–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajala, T.; Tuomivirta, T.; Pennanen, T.; Makipaa, R. Habitat Models of Wood-Inhabiting Fungi along a Decay Gradient of Norway Spruce Logs. Fungal Ecol. 2015, 18, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, Y.; Komagata, Y.; Ushijima, S. Fungal Wood Decomposer Activity Induces Niche Separation between Two Dominant Tree Species Seedlings Regenerating on Coarse Woody Debris. Can. J. For. Res. 2017, 47, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, Y.; Ando, Y.; Oishi, Y.; Suzuki, S.N.; Matsukura, K.; Okano, K.; Song, Z. Does Typhoon Disturbance in Subalpine Forest Have Long-Lasting Impacts on Saproxylic Fungi, Bryophytes, and Seedling Regeneration on Coarse Woody Debris? For. Ecol. Manag. 2018, 432, 309–318. [Google Scholar] [CrossRef]
- Araya, K. Relationship between the Decay Types of Dead Wood and Occurrence of Lucanid Beetles (Coleoptera: Lucanidae). Appl. Entomol. Zool. 1993, 28, 27–33. [Google Scholar] [CrossRef] [Green Version]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A New Method Which Gives an Objective Measure of Colonization of Roots by Vescular-Arbuscular Mycorrhizal Fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-Coverage ITS Primers for the DNA-Based Identification of Ascomycetes and Basidiomycetes in Environmental Samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Fungal Barcoding Consortium Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, A.S.; Toju, H. Two New Computational Methods for Universal DNA Barcoding: A Benchmark Using Barcode Sequences of Bacteria, Archaea, Animals, Fungi and Land Plants. PLoS ONE 2013, 8, e76910. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, A.S. Assams-Assembler v0.2.2015.08.08, Software Distributed by the Author. Available online: http://www.fifthdimension.jp/ (accessed on 29 April 2022).
- Osono, T. Metagenomic approach yields insights into fungal diversity and functioning. In Species Diversity and Community Structure: Novel Patterns and Processes in Plants, Insects, and Fungi; Sota, T., Kagata, H., Ando, Y., Utsumi, S., Osono, T., Eds.; Springer: Berlin, Germany, 2014; pp. 1–23. [Google Scholar]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Skelton, J.; Jusino, M.A.; Carlson, P.S.; Smith, K.; Banik, M.T.; Lindner, D.L.; Palmer, J.M.; Hulcr, J. Relationships among Wood-Boring Beetles, Fungi, and the Decomposition of Forest Biomass. Mol. Ecol. 2019, 28, 4971–4986. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. Vegan: Community Ecology Package. R Package Version 2.3.4. 2016. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 29 April 2022).
- Anderson, M.J. A New Method for Non Parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- De Cáceres, M.; Jansen, F.; Dell, N. Package ‘Indicspecies’ (ver 1.7.12). Available online: https://cran.r-project.org/web/packages/indicspecies/indicspecies.pdf (accessed on 29 April 2022).
- Nara, K.; Hogetsu, T. Ectomycorrhizal Fungi on Established Shrubs Facilitate Subsequent Seedling Establishment of Successional Plant Species. Ecology 2004, 85, 1700–1707. [Google Scholar] [CrossRef]
- Fernandez, C.W.; Koide, R.T. The Function of Melanin in the Ectomycorrhizal Fungus Cenococcum geophilum under Water Stress. Fungal Ecol. 2013, 6, 479–486. [Google Scholar] [CrossRef]
- Herzog, C.; Peter, M.; Pritsch, K.; Günthardt-Goerg, M.S.; Egli, S. Drought and Air Warming Affects Abundance and Exoenzyme Profiles of Cenococcum geophilum Associated with Quercus robur, Q. petraea and Q. pubescens. Plant Biol. 2013, 15, 230–237. [Google Scholar] [CrossRef]
- Toju, H.; Yamamoto, S.; Sato, H.; Tanabe, A.S.; Gilbert, G.S.; Kadowaki, K. Community Composition of Root-Associated Fungi in a Quercus-Dominated Temperate Forest. “Codominance” of Mycorrhizal and Root-Endophytic Fungi. Ecol. Evol. 2013, 3, 1281–1293. [Google Scholar] [CrossRef]
- Newsham, K.K. A Meta-Analysis of Plant Responses to Dark Septate Root Endophytes. New Phytol. 2011, 190, 783–793. [Google Scholar] [CrossRef]
- Mayerhofer, M.S.; Kernaghan, G.; Harper, K.A. The Effects of Fungal Root Endophytes on Plant Growth: A Meta-Analysis. Mycorrhiza 2013, 23, 119–128. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008; p. 787. [Google Scholar]
- Nanzyo, M. Infrared Spectra of Phosphate Sorbed on Iron Hydroxide Gel and the Sorption Products. Soil Sci. Plant Nutr. 1986, 32, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Gehring, C.A. Growth Responses to Arbuscular Mycorrhizae by Rain Forest Seedlings Vary with Light Intensity and Tree Species. Plant Ecol. 2003, 167, 127–139. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Matsuoka, S. Communities of Wood-Inhabiting Fungi in Dead Pine Logs along a Geographical Gradient in Japan. Fungal Ecol. 2015, 18, 75–82. [Google Scholar] [CrossRef]
- Johnston, S.R.; Hiscox, J.; Savoury, M.; Boddy, L.; Weightman, A.J. Highly Competitive Fungi Manipulate Bacterial Communities in Decomposing Beech Wood (Fagus sylvatica). FEMS Microb. Ecol. 2019, 95, fiy225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).