Leaf and Root Litter Species Identity Influences Bacterial Community Composition in Short-Term Litter Decomposition
Abstract
:1. Introduction
2. Materials and Methods
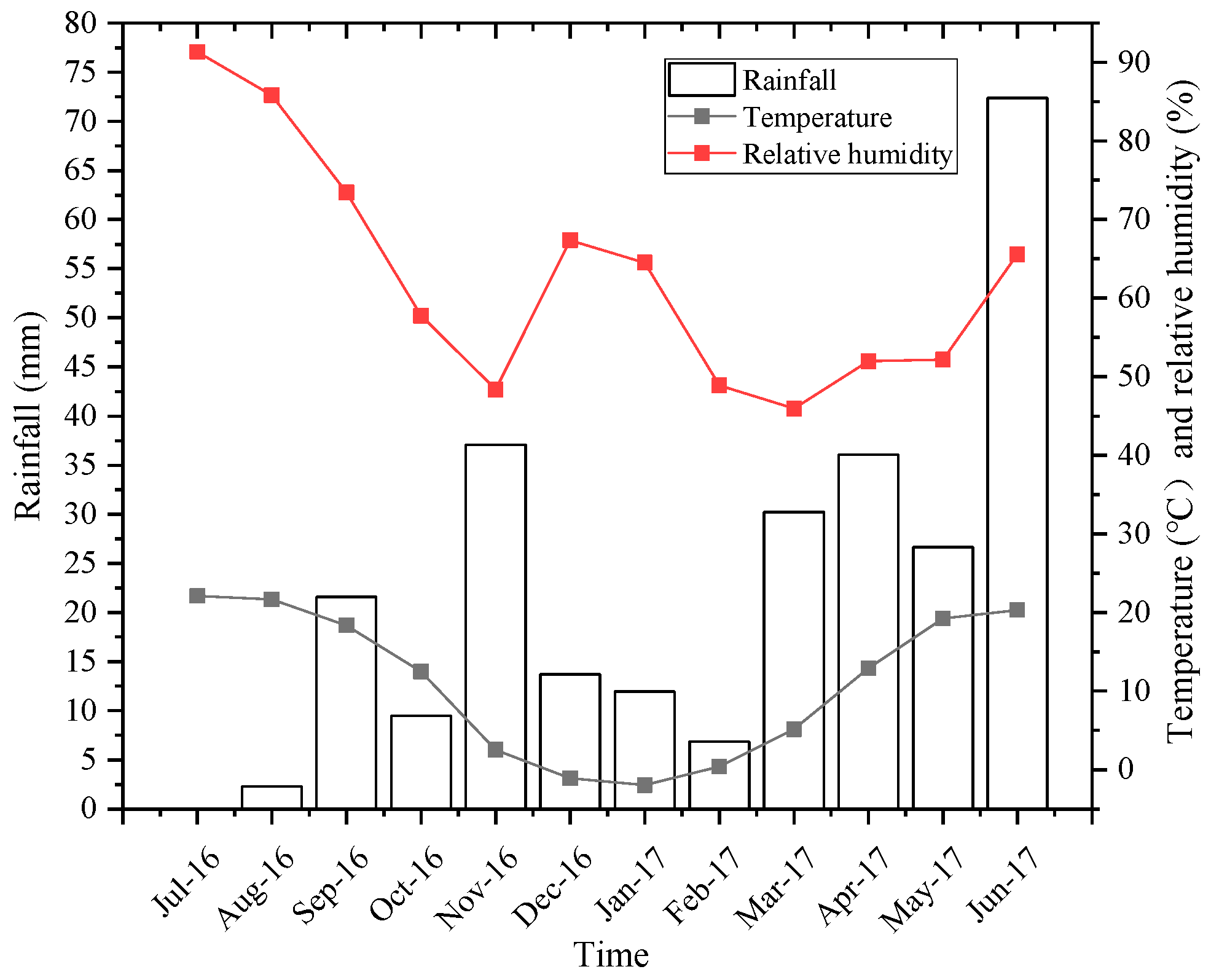
2.1. Study Site
2.2. Experimental Design and Litter Bag Collection
2.3. Litter Chemical Analysis
2.4. DNA Extraction and Sequencing
2.5. Data Analysis
3. Results
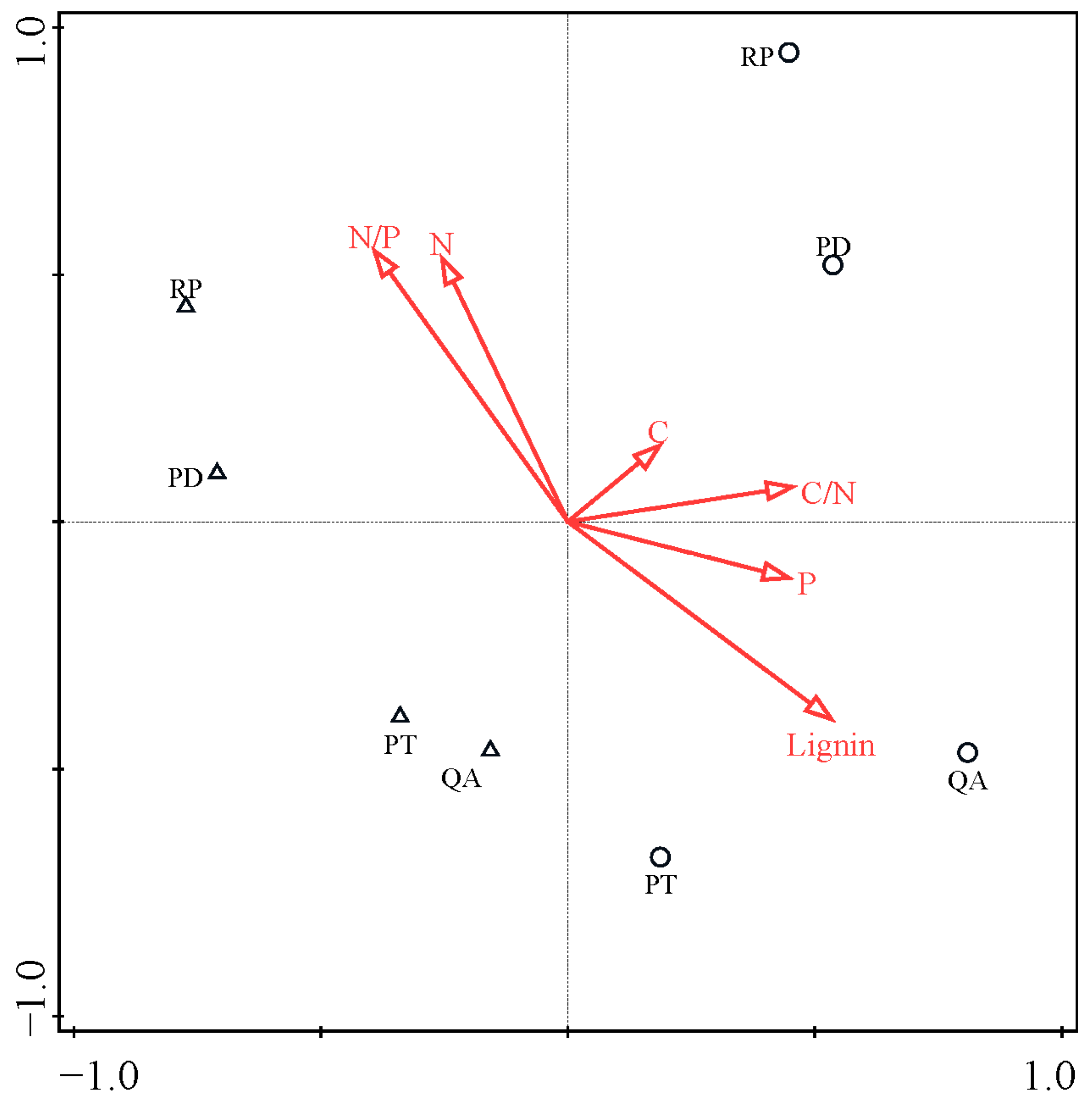
3.1. Initial Litter Chemistry and Decomposition Rate
3.2. Bacterial Alpha Diversity
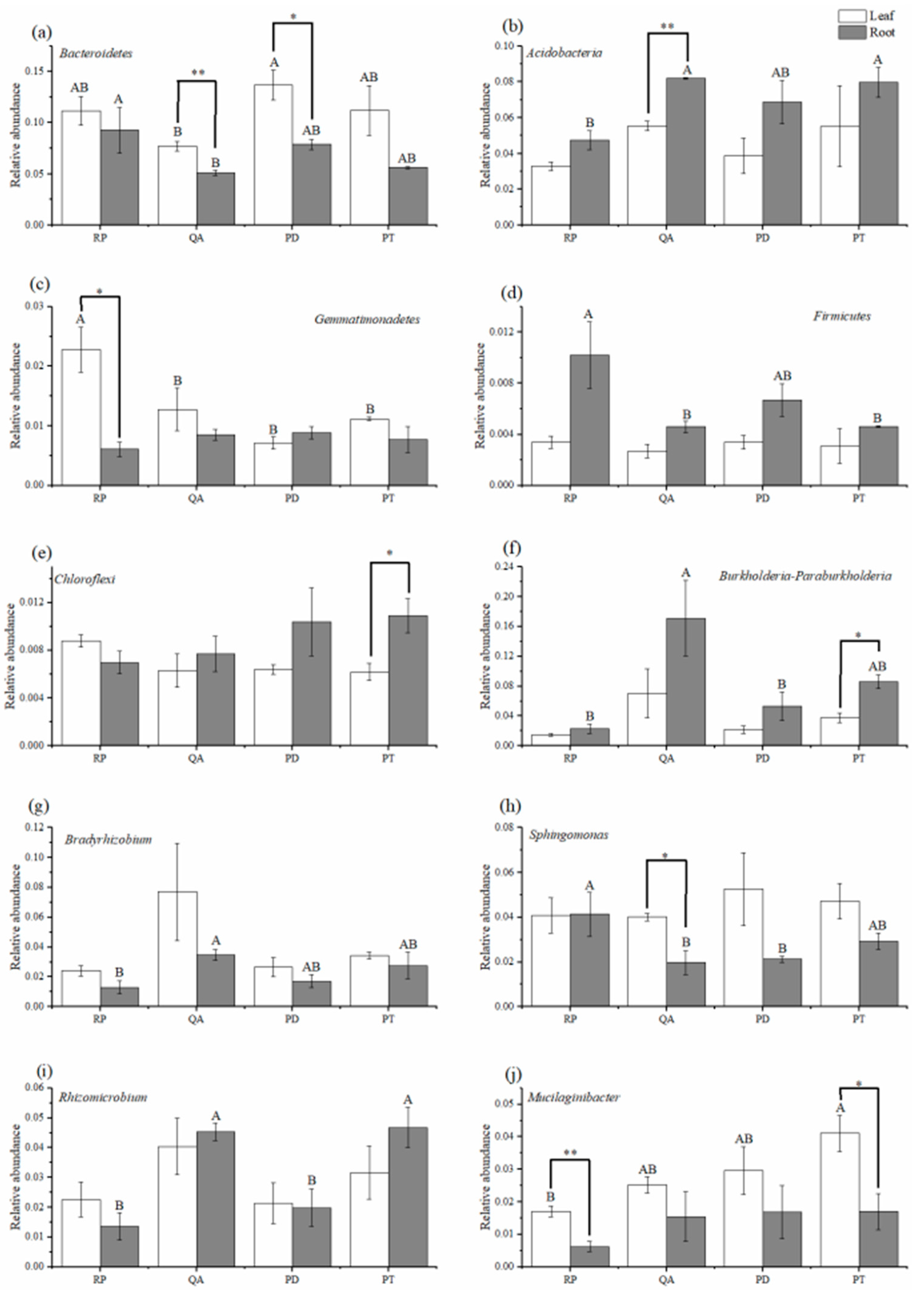
3.3. Relative Abundance of Dominant Bacterial Phyla and Genera
3.4. Bacterial Community Composition
4. Discussion
4.1. Effect of Litter Tissues and Species on Litter Decomposition
4.2. Effect of Litter Tissues and Species on the Relative Abundances of Dominant Bacterial Phyla
4.3. Effect of Leaf and Root Litter Species on the Relative Abundance of Dominant Bacterial Genera
4.4. Effect of Litter Tissues and Species on the Bacterial Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pei, G.T.; Liu, J.; Peng, B.; Gao, D.C.; Wang, C.; Dai, W.W.; Jiang, P.; Bai, E. Nitrogen, lignin, C/N as important regulators of gross nitrogen release and immobilization during litter decomposition in a temperate forest ecosystem. For. Ecol. Manag. 2019, 440, 61–69. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, L.G.; Fei, S.X.; Zhang, J.W.; Jiang, X.M.; Wang, Y.; Yu, X. Responses of Soil Organic Carbon Mineralization and Microbial Communities to Leaf Litter Addition under Different Soil Layers. Forests 2021, 12, 170. [Google Scholar] [CrossRef]
- Chapman, S.K.; Koch, G.W. What type of diversity yields synergy during mixed litter decomposition in a natural forest ecosystem? Plant Soil 2007, 299, 153–162. [Google Scholar] [CrossRef]
- Mooshammer, M.; Wanek, W.; Schnecker, J.; Wild, B.; Leitner, S.; Hofhansl, F.; Blochl, A.; Hammerle, I.; Frank, A.H.; Fuchslueger, L.; et al. Stoichiometric controls of nitrogen and phosphorus cycling in decomposing beech leaf litter. Ecology 2012, 93, 770–782. [Google Scholar] [CrossRef]
- Liu, S.J.; Behm, J.E.; Wan, S.Q.; Yan, J.H.; Ye, Q.; Zhang, W.; Yang, X.D.; Fu, S.L. Effects of canopy nitrogen addition on soil fauna and litter decomposition rate in a temperate forest and a subtropical forest. Geoderma 2021, 382, 114703. [Google Scholar] [CrossRef]
- Lin, D.M.; Pang, M.; Fanin, N.; Wang, H.J.; Qian, S.H.; Zhao, L.; Yang, Y.C.; Mi, X.C.; Ma, K.P. Fungi participate in driving home-field advantage of litter decomposition in a subtropical forest. Plant Soil 2019, 434, 467–480. [Google Scholar] [CrossRef]
- Baietto, A.; Hirigoyen, A.; Hernandez, J.; del Pino, A. Comparative Dynamics of Nutrient Release through Litter Decomposition in Eucalyptus grandis Hill ex Maiden and Pinus taeda L. Stands. Forests 2021, 12, 1227. [Google Scholar] [CrossRef]
- Shen, Y.F.; Wang, N.; Cheng, R.M.; Xiao, W.F.; Yang, S.; Guo, Y.; Lei, L.; Zeng, L.X.; Wang, X.R. Characteristics of Fine Roots of Pinus massoniana in the Three Gorges Reservoir Area, China. Forests 2017, 8, 183. [Google Scholar] [CrossRef]
- Luo, D.; Cheng, R.M.; Shi, Z.M.; Wang, W.X. Decomposition of Leaves and Fine Roots in Three Subtropical Plantations in China Affected by Litter Substrate Quality and Soil Microbial Community. Forests 2017, 8, 412. [Google Scholar] [CrossRef]
- Huangfu, C.H.; Hui, D.F.; Qi, X.X.; Li, K.L. Plant interactions modulate root litter decomposition and negative plant-soil feedback with an invasive plant. Plant Soil 2019, 437, 179–194. [Google Scholar] [CrossRef]
- Zwetsloot, M.J.; Ucros, J.M.; Wickings, K.; Wilhelm, R.C.; Sparks, J.; Buckley, D.H.; Bauerle, T.L. Prevalent root-derived phenolics drive shifts in microbial community composition and prime decomposition in forest soil. Soil Biol. Biochem. 2020, 145, 107797. [Google Scholar] [CrossRef]
- Wang, Q.K.; Yu, Y.Z.; He, T.X.; Wang, Y.P. Aboveground and belowground litter have equal contributions to soil CO2 emission: An evidence from a 4-year measurement in a subtropical forest. Plant Soil 2017, 421, 7–17. [Google Scholar] [CrossRef]
- Cao, J.B.; He, X.X.; Chen, Y.Q.; Chen, Y.P.; Zhang, Y.J.; Yu, S.Q.; Zhou, L.X.; Liu, Z.F.; Zhang, C.L.; Fu, S.L. Leaf litter contributes more to soil organic carbon than fine roots in two 10-year-old subtropical plantations. Sci. Total Environ. 2020, 704, 135341. [Google Scholar] [CrossRef]
- Sun, T.; Hobbie, S.E.; Berg, B.; Zhang, H.G.; Wang, Q.K.; Wang, Z.W.; Hattenschwiler, S. Contrasting dynamics and trait controls in first-order root compared with leaf litter decomposition. Proc. Natl. Acad. Sci. USA 2018, 115, 10392–10397. [Google Scholar] [CrossRef]
- Wang, Y.X.; Wang, H.M.; Dai, X.Q.; Kou, L.; Meng, S.W.; Fu, X.L. Decoupled responses of leaf and root decomposition to nutrient deposition in a subtropical plantation. Soil Biol. Biochem. 2022, 168, 108643. [Google Scholar] [CrossRef]
- Chen, Y.C.; Ma, S.Q.; Jiang, H.M.; Hu, Y.; Lu, X.Y. Influences of litter diversity and soil moisture on soil microbial communities in decomposing mixed litter of alpine steppe species. Geoderma 2020, 377, 114577. [Google Scholar] [CrossRef]
- Liu, J.L.; Dang, P.; Gao, Y.; Zhu, H.L.; Zhu, H.N.; Zhao, F.; Zhao, Z. Effects of tree species and soil properties on the composition and diversity of the soil bacterial community following afforestation. For. Ecol. Manag. 2018, 427, 342–349. [Google Scholar] [CrossRef]
- Leloup, J.; Baude, M.; Nunan, N.; Meriguet, J.; Dajoz, I.; Le Roux, X.; Raynaud, X. Unravelling the effects of plant species diversity and aboveground litter input on soil bacterial communities. Geoderma 2018, 317, 1–7. [Google Scholar] [CrossRef]
- Otsing, E.; Barantal, S.; Anslan, S.; Koricheva, J.; Tedersoo, L. Litter species richness and composition effects on fungal richness and community structure in decomposing foliar and root litter. Soil Biol. Biochem. 2018, 125, 328–339. [Google Scholar] [CrossRef]
- Xiao, W.Y.; Chen, H.Y.H.; Kumar, P.; Chen, C.; Guan, Q.W. Multiple interactions between tree composition and diversity and microbial diversity underly litter decomposition. Geoderma 2019, 341, 161–171. [Google Scholar] [CrossRef]
- Li, Y.B.; Bezemer, T.M.; Yang, J.J.; Lu, X.T.; Li, X.Y.; Liang, W.J.; Han, X.G.; Li, Q. Changes in litter quality induced by N deposition alter soil microbial communities. Soil Biol. Biochem. 2019, 130, 33–42. [Google Scholar] [CrossRef]
- Kennedy, A.C. Bacterial diversity in agroecosystems. Agric. Ecosyst. Environ. 1999, 74, 65–76. [Google Scholar] [CrossRef]
- Morgan, J.A.W.; Bending, G.D.; White, P.J. Biological costs and benefits to plant-microbe interactions in the rhizosphere. J. Exp. Bot. 2005, 56, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setala, H.; van der Putten, W.H.; Wall, D.H. Ecological linkages between aboveground and belowground biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Prescott, C.E.; Grayston, S.J. Tree species influence on microbial communities in litter and soil: Current knowledge and research needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Jia, T.; Liang, X.X.; Guo, T.Y.; Wu, T.H.; Chai, B.F. Bacterial community succession and influencing factors for Imperata cylindrica litter decomposition in a copper tailings area of China. Sci. Total Environ. 2022, 815, 152908. [Google Scholar] [CrossRef]
- Hong, C.; Si, Y.X.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar] [CrossRef]
- Sauvadet, M.; Fanin, N.; Chauvat, M.; Bertrand, I. Can the comparison of above-and below-ground litter decomposition improve our understanding of bacterial and fungal successions? Soil Biol. Biochem. 2019, 132, 24–27. [Google Scholar] [CrossRef]
- Iiyama, K.; Wallis, A.F.A. An improved acetyl bromide procedure for determining lignin in woods and wood pulps. Wood Sci. Technol. 1988, 22, 271–280. [Google Scholar] [CrossRef]
- Li, R.S.; Zhang, Y.Z.; Yu, D.; Wang, Y.; Zhao, X.X.; Zhang, R.H.; Zhang, W.D.; Wang, Q.K.; Xu, M.; Chen, L.C.; et al. The decomposition of green leaf litter is less temperature sensitive than that of senescent leaf litter: An incubation study. Geoderma 2021, 381, 114691. [Google Scholar] [CrossRef]
- Gupta, S.R.; Singh, J.S. The effect of plant-species, weather variables and chemical-composition of plant-material on decomposition in a tropical grassland. Plant Soil 1981, 59, 99–117. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; Idbella, M.; Zotti, M.; Santorufo, L.; Motti, R.; Maisto, G.; De Marco, A. Decomposition and temperature sensitivity of fine root and leaf litter of 43 mediterranean species. Plant Soil 2021, 464, 453–465. [Google Scholar] [CrossRef]
- Guo, L.L.; Deng, M.F.; Yang, S.; Liu, W.X.; Wang, X.; Wang, J.; Liu, L.L. The coordination between leaf and fine root litter decomposition and the difference in their controlling factors. Glob. Ecol. Biogeogr. 2021, 30, 2286–2296. [Google Scholar] [CrossRef]
- Van Huysen, T.L.; Harmon, M.E.; Perakis, S.S.; Chen, H. Decomposition and nitrogen dynamics of N-15-labeled leaf, root, and twig litter in temperate coniferous forests. Oecologia 2013, 173, 1563–1573. [Google Scholar] [CrossRef]
- Reich, P.B. The world-wide ‘fast-slow’ plant economics spectrum: A traits manifesto. J. Ecol. 2014, 102, 275–301. [Google Scholar] [CrossRef]
- McLaren, J.R.; Turkington, R. Plant functional group identity differentially affects leaf and root decomposition. Glob. Chang. Biol. 2010, 16, 3075–3084. [Google Scholar] [CrossRef]
- Zhao, B.Y.; Xing, P.; Wu, Q.L.L. Microbes participated in macrophyte leaf litters decomposition in freshwater habitat. Fems Microbiol. Ecol. 2017, 93, fix108. [Google Scholar] [CrossRef]
- Pichon, N.A.; Cappelli, S.L.; Soliveres, S.; Holzel, N.; Klaus, V.H.; Kleinebecker, T.; Allan, E. Decomposition disentangled: A test of the multiple mechanisms by which nitrogen enrichment alters litter decomposition. Funct. Ecol. 2020, 34, 1485–1496. [Google Scholar] [CrossRef] [Green Version]
- Gui, H.; Purahong, W.; Hyde, K.D.; Xu, J.C.; Mortimer, P.E. The Arbuscular Mycorrhizal Fungus Funneliformis mosseae Alters Bacterial Communities in Subtropical Forest Soils during Litter Decomposition. Front. Microbiol. 2017, 8, 1120. [Google Scholar] [CrossRef]
- Xu, M.P.; Lu, X.Q.; Xu, Y.D.; Zhong, Z.K.; Zhang, W.; Ren, C.J.; Han, X.; Yang, G.H.; Feng, Y.Z. Dynamics of bacterial community in litter and soil along a chronosequence of Robinia pseudoacacia plantations. Sci. Total Environ. 2020, 703, 135613. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Andersson, S.G.E.; Battin, T.J.; Prosser, J.I.; Schimel, J.P.; Whitman, W.B.; Hallin, S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010, 8, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.J.; Liu, W.B.; Zhu, C.; Luo, G.W.; Kong, Y.L.; Ling, N.; Wang, M.; Dai, J.Y.; Shen, Q.R.; Guo, S.W. Bacterial rather than fungal community composition is associated with microbial activities and nutrient-use efficiencies in a paddy soil with short-term organic amendments. Plant Soil 2018, 424, 335–349. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.W.; Yan, Q.Y.; Yang, S.; Van Nostrand, J.D.; Wang, Z.R.; He, Z.L.; Zhou, J.Z.; Jiang, Y.; Deng, Y. Soil microbial beta-diversity is linked with compositional variation in aboveground plant biomass in a semi-arid grassland. Plant Soil 2018, 423, 465–480. [Google Scholar] [CrossRef]
- Fanin, N.; Fromin, N.; Bertrand, I. Functional breadth and home-field advantage generate functional differences among soil microbial decomposers. Ecology 2016, 97, 1023–1037. [Google Scholar] [CrossRef]
- Osburn, E.D.; Hoch, P.J.; Lucas, J.M.; McBride, S.G.; Strickland, M.S. Evaluating the roles of microbial functional breadth and home-field advantage in leaf litter decomposition. Funct. Ecol. 2022, 36, 1258–1267. [Google Scholar] [CrossRef]
- Sauvadet, M.; Chauvat, M.; Cluzeau, D.; Maron, P.A.; Villenave, C.; Bertrand, I. The dynamics of soil micro-food web structure and functions vary according to litter quality. Soil Biol. Biochem. 2016, 95, 262–274. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of Plant Polymers on the Distribution and Cultivation of Bacteria in the Phylum Acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [Green Version]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Z.; Ren, C.J.; Zhang, L.; Han, X.H.; Yang, G.H.; Wang, J. Changes in soil microbial community are linked to soil carbon fractions after afforestation. Eur. J. Soil Sci. 2018, 69, 370–379. [Google Scholar] [CrossRef]
- Aislabie, J.; Deslippe, J.R. Soil Microbes and Their Contribution to Soil Services; Manaaki Whenua Press: Lincoln, New Zealand, 2013; pp. 143–161. [Google Scholar]
- Yamada, T.; Sekiguchi, Y. Cultivation of Uncultured Chloroflexi Subphyla: Significance and Ecophysiology of Formerly Uncultured Chloroflexi ‘Subphylum I’ with Natural and Biotechnological Relevance. Microbes Environ. 2009, 24, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.B.; Chen, Y.C.; He, T.B.; Liao, R.J.; Liu, R.L.; Yi, M.; Huang, L.; Yang, Z.M.; Fu, T.L.; Li, X.Y. Soil nitrogen leaching decreases as biogas slurry DOC/N ratio increases. Appl. Soil Ecol. 2017, 111, 105–113. [Google Scholar] [CrossRef]
- Nie, Y.X.; Wang, M.C.; Zhang, W.; Ni, Z.; Hashidoko, Y.; Shen, W.J. Ammonium nitrogen content is a dominant predictor of bacterial community composition in an acidic forest soil with exogenous nitrogen enrichment. Sci. Total Environ. 2018, 624, 407–415. [Google Scholar] [CrossRef]
- Ko, Y.; Hwang, W.M.; Kim, M.; Kang, K.; Ahn, T.Y. Sphingomonas silvisoli sp nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2704–2710. [Google Scholar] [CrossRef] [PubMed]
- Janssens, I.A.; Dieleman, W.; Luyssaert, S.; Subke, J.A.; Reichstein, M.; Ceulemans, R.; Ciais, P.; Dolman, A.J.; Grace, J.; Matteucci, G.; et al. Reduction of forest soil respiration in response to nitrogen deposition. Nat. Geosci. 2010, 3, 315–322. [Google Scholar] [CrossRef]
- Joly, F.X.; Fromin, N.; Kiikkila, O.; Hattenschwiler, S. Diversity of leaf litter leachates from temperate forest trees and its consequences for soil microbial activity. Biogeochemistry 2016, 129, 373–388. [Google Scholar] [CrossRef]
- Zhang, W.W.; Yang, K.; Lyu, Z.T.; Zhu, J.J. Microbial groups and their functions control the decomposition of coniferous litter: A comparison with broadleaved tree litters. Soil Biol. Biochem. 2019, 133, 196–207. [Google Scholar] [CrossRef]
- Gunina, A.; Smith, A.R.; Godbold, D.L.; Jones, D.L.; Kuzyakov, Y. Response of soil microbial community to afforestation with pure and mixed species. Plant Soil 2017, 412, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Baah-Acheamfour, M.; Carlyle, C.N.; Bissett, A.; Richardson, A.E.; Siddique, T.; Bork, E.W.; Chang, S.X. Determinants of bacterial communities in Canadian agroforestry systems. Environ. Microbiol. 2016, 18, 1805–1816. [Google Scholar] [CrossRef] [PubMed]



| Elevation (m) | Slope Degree (°) | Slope Aspect | Soil Layer Depth (cm) | pH | C (%) | N (%) | Soil Organic Carbon (g/kg) | |
|---|---|---|---|---|---|---|---|---|
| Forest-free area | 730 | 23 | south | 26.08 | 5.00 | 2.13 | 0.15 | 9.32 |
| Organ | Species | C% | N% | P% | C:N | N:P | Lignin% |
|---|---|---|---|---|---|---|---|
| Leaf | RP | 45.57 ± 0.13 c** | 1.86 ± 0.05 a** | 0.41 ± 0.01 c** | 24.58 ± 0.57 b** | 4.55 ± 0.27 a** | 28.84 ± 0.36 c |
| QA | 48.23 ± 0.84 b | 1.21 ± 0.03 b** | 0.43 ± 0.01 c** | 39.91 ± 1.03 a* | 2.82 ± 0.08 c** | 33.63 ± 0.57 b | |
| PD | 50.64 ± 0.15 a** | 2.02 ± 0.01 a** | 0.59 ± 0.01 a** | 25.10 ± 0.16 b** | 3.42 ± 0.08 b** | 22.83 ± 0.54 d** | |
| PT | 50.34 ± 0.51 a | 1.90 ± 0.08 a** | 0.54 ± 0.01 b | 26.64 ± 0.90 b** | 3.50 ± 0.16 b** | 37.21 ± 0.61 a | |
| Root | RP | 48.77 ± 0.33 c | 3.36 ± 0.002 a | 0.56 ± 0.01 b | 14.51 ± 0.09 d | 6.05 ± 0.16 a | 29.59 ± 0.47 c |
| QA | 46.39 ± 0.17 d | 1.08 ± 0.01 b | 0.63 ± 0.01 a | 43.02 ± 0.17 c | 1.73 ± 0.04 b | 33.78 ± 0.60 b | |
| PD | 54.65 ± 0.17 a | 0.38 ± 0.01 d | 0.50 ± 0.01 c | 142.48 ± 3.72 a | 0.76 ± 0.02 c | 38.34 ± 0.30 a | |
| PT | 49.96 ± 0.13 b | 0.85 ± 0.003 c | 0.53 ± 0.004 bc | 59.04 ± 0.19 b | 1.61 ± 0.01 b | 37.78 ± 0.15 a |
| Organ | Species | Average Value | ||||
|---|---|---|---|---|---|---|
| RP | QA | PD | PT | |||
| Observed species | Leaf | 2000 ± 9.8 a* | 1946 ± 47.6 a | 1832 ± 10.4 b** | 1776 ± 21.0 b** | 1888 ± 29.1 ** |
| Root | 2149 ± 40.7 b | 2155 ± 197.2 b | 2759 ± 14.4 a | 2568 ± 22.2 a | 2407 ± 90.8 | |
| Chao1 | Leaf | 2729.5 ± 53.4 a* | 2672.9 ± 11 a* | 2221.5 ± 86.4 b** | 2275.8 ± 88.7 b** | 2474.9 ± 258.2 ** |
| Root | 3227.5 ± 161.3 b | 2824.2 ± 51.1 c | 3544.7 ± 29.1 a | 3395.0 ± 1.3 ab | 3247.8 ± 308.4 | |
| Phylogenetic diversity (PD) | Leaf | 145.3 ± 0.4 a** | 142.5 ± 3.4 a | 134.3 ± 2.6 b** | 132.8 ± 0.4 b** | 138.7 ± 6.4 ** |
| Root | 159.2 ± 2.4 b | 147.8 ± 4.4 c | 198.6 ± 3.0 a | 193.1 ± 1.9 a | 174.7 ± 23.1 | |
| Shannon | Leaf | 9.13 ± 0.06 a | 8.38 ± 0.04 d | 8.97 ± 0.03 b | 8.78 ± 0.03 c | 8.82 ± 0.30 |
| Root | 8.43 ± 0.35 a | 8.40 ± 0.28 a | 8.80 ± 0.20 a | 8.76 ± 0.16 a | 8.60 ± 0.42 | |
| C% | N% | P% | C:N | N:P | Lignin% | |
|---|---|---|---|---|---|---|
| Observed species | 0.480 * | −0.535 ** | −0.022 | 0.784 ** | −0.558 ** | 0.517 ** |
| Chao1 | 0.305 | −0.294 | −0.077 | 0.618 ** | −0.303 | 0.499 * |
| Phylogenetic diversity (PD) | 0.526 ** | −0.500 * | −0.056 | 0.769 ** | −0.521 ** | 0.561 ** |
| Shannon | 0.114 | −0.038 | −0.227 | 0.028 | 0.053 | −0.229 |
| Dominant phylum | C% | N% | P% | Lignin% | C:N | N:P | Remaining Mass% |
|---|---|---|---|---|---|---|---|
| Proteobacteria | −0.191 | 0.201 | 0.170 | −0.038 | −0.148 | 0.131 | −0.396 |
| Actinobacteria | 0.095 | −0.229 | −0.195 | 0.115 | 0.159 | −0.147 | 0.278 |
| Bacteroidetes | 0.056 | 0.420 * | −0.043 | −0.542 ** | −0.313 | 0.425 * | 0.113 |
| Acidobacteria | 0.175 | −0.469 * | 0.275 | 0.558 ** | 0.382 | −0.543 ** | 0.082 |
| Planctomycetes | 0.278 | −0.477 * | −0.201 | 0.356 | 0.408 * | −0.440 * | 0.482 * |
| Gemmatimonadetes | −0.460 * | −0.039 | −0.692 ** | −0.110 | −0.154 | 0.209 | −0.024 |
| Cyanobacteria | 0.251 | −0.028 | 0.051 | 0.314 | 0.051 | −0.049 | 0.061 |
| Verrucomicrobia | −0.083 | −0.291 | 0.034 | 0.046 | 0.003 | −0.255 | 0.205 |
| Firmicutes | 0.172 | 0.350 | 0.215 | 0.025 | 0.133 | 0.252 | −0.296 |
| Chloroflexi | 0.174 | −0.377 | −0.179 | 0.275 | 0.427 * | −0.331 | 0.085 |
| Mass remaining | 0.644 ** | −0.636 ** | −0.254 | 0.272 | 0.619 ** | −0.610 ** | 1 |
| Dominant Genus | C% | N% | P% | Lignin% | C:N | N:P | Remaining Mass% |
|---|---|---|---|---|---|---|---|
| Burkholderia-Paraburkholderia | −0.159 | −0.431 * | 0.357 | 0.359 | 0.155 | −0.498 * | −0.067 |
| Bradyrhizobium | −0.264 | −0.191 | −0.301 | 0.094 | −0.123 | −0.110 | 0.159 |
| Massilia | 0.074 | 0.122 | 0.139 | −0.433 * | −0.171 | 0.089 | 0.128 |
| Sphingomonas | −0.088 | 0.458 * | −0.116 | −0.440 * | −0.474 * | 0.487 * | 0.002 |
| Caulobacter | 0.249 | −0.081 | −0.283 | −0.005 | 0.325 | −0.021 | 0.115 |
| Rhizomicrobium | −0.237 | −0.434 * | 0.076 | 0.360 | −0.009 | −0.434 * | 0.016 |
| Rhizobium | 0.104 | 0.322 | 0.140 | −0.310 | −0.239 | 0.268 | −0.091 |
| Pseudoxanthomonas | −0.023 | 0.385 | 0.090 | −0.083 | −0.108 | 0.339 | −0.265 |
| Chitinophaga | 0.118 | 0.470 * | 0.201 | −0.459 * | −0.156 | 0.381 | −0.065 |
| Mucilaginibacter | 0.171 | −0.100 | 0.051 | 0.035 | −0.114 | −0.109 | 0.353 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Zhang, L.; Li, K.; Ni, R.; Han, R.; Li, C.; Zhang, C.; Shen, W.; Zhang, Z. Leaf and Root Litter Species Identity Influences Bacterial Community Composition in Short-Term Litter Decomposition. Forests 2022, 13, 1402. https://doi.org/10.3390/f13091402
Lu Y, Zhang L, Li K, Ni R, Han R, Li C, Zhang C, Shen W, Zhang Z. Leaf and Root Litter Species Identity Influences Bacterial Community Composition in Short-Term Litter Decomposition. Forests. 2022; 13(9):1402. https://doi.org/10.3390/f13091402
Chicago/Turabian StyleLu, Ying, Liudong Zhang, Kun Li, Ruiqiang Ni, Rongchu Han, Chuanrong Li, Caihong Zhang, Weixing Shen, and Zhongjun Zhang. 2022. "Leaf and Root Litter Species Identity Influences Bacterial Community Composition in Short-Term Litter Decomposition" Forests 13, no. 9: 1402. https://doi.org/10.3390/f13091402
APA StyleLu, Y., Zhang, L., Li, K., Ni, R., Han, R., Li, C., Zhang, C., Shen, W., & Zhang, Z. (2022). Leaf and Root Litter Species Identity Influences Bacterial Community Composition in Short-Term Litter Decomposition. Forests, 13(9), 1402. https://doi.org/10.3390/f13091402










